Abstract
1-(β-d-Ribofuranosyl)-2,5,6-trichlorobenzimidazole (TCRB) and its 2-bromo analog, BDCRB, are potent and selective inhibitors of human cytomegalovirus (HCMV) DNA processing and packaging. Since they are readily metabolized in vivo, analogs were synthesized to improve biostability. One of these, 1-(β-l-ribofuranosyl)-2-isopropylamino-5,6-dichlorobenzimidazole (1263W94; maribavir), inhibits viral DNA synthesis and nuclear egress. Resistance to maribavir was mapped to UL97, and this viral kinase was shown to be a direct target of maribavir. In the present study, an HCMV strain resistant to TCRB and BDCRB was passaged in increasing concentrations of maribavir, and resistant virus was isolated. This strain (G2) grew at the same rate as the wild-type virus and was resistant to both BDCRB and maribavir. Resistance to BDCRB was expected, because the parent strain from which G2 was isolated was resistant due to known mutations in UL56 and UL89. However, no mutations were found in UL97 or other relevant open reading frames that could explain resistance to maribavir. Because sequencing of selected HCMV genes did not identify the resistance mutation, a cosmid library was made from G2, and a series of recombinant G2 wild-type viruses were constructed. Testing the recombinants for sensitivity to maribavir narrowed the locus of resistance to genes UL26 to UL32. Sequencing identified a single coding mutation in ORF UL27 (Leu335Pro) as the one responsible for resistance to maribavir. These results establish that UL27 is either directly or indirectly involved in the mechanism of action of maribavir. They also suggest that UL27 could play a role in HCMV DNA synthesis or egress of HCMV particles from the nucleus.
HCMV is one of eight human herpesviruses that causes widespread infection. In the United States an estimated 40 to 100% of the population is infected with HCMV (32). In healthy individuals HCMV infections are typically asymptomatic, while in immunocompromised populations, such as AIDS patients (27), bone marrow recipients (39), and solid organ transplant recipients (17), they are often life threatening. HCMV is also a leading cause of birth defects and infections in children (7).
Currently there are five drugs approved for use in the United States for treatment of HCMV infection. They include ganciclovir (10), its prodrug valganciclovir (11), cidofovir (18), foscarnet (9), and the phosphorothioate oligonucleotide complementary to HCMV IE2 mRNA, fomivirsen (29). There are disadvantages associated with the use of each of these drugs, including poor oral bioavailability and toxicity (2, 10, 12, 18). Numerous clinical strains resistant to ganciclovir, foscarnet, and cidofovir have been isolated (14, 15). Since these drugs share the same molecular target, HCMV DNA polymerase, the emergence of cross-resistant strains also has been reported (14, 15). Therefore, there is a need for new compounds with better oral bioavailability, safer pharmacological profiles, and novel mechanisms of action.
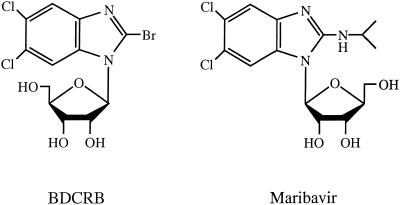
In 1995, we reported the synthesis and antiviral activity of TCRB and its 2-bromo analog, BDCRB (34) (Fig. 1). These drugs inhibit HCMV replication via a novel mechanism by blocking the processing of concatemeric DNA into genome-length pieces (24, 37). Resistance of HCMV to these drugs was mapped to two genes involved in this process: UL89 and UL56 (24, 37). Although TCRB and BDCRB show excellent activity in vitro, their glycosidic bonds are hydrolyzed in vivo to less active metabolites (16). This prompted the design and synthesis of benzimidazole analogs with more-stable glycosidic bonds (35). One of these analogs, an l-ribofuranosyl benzimidazole (maribavir [Fig. 1]), is active against both laboratory strains and clinical isolates of HCMV. HCMV strains resistant to ganciclovir, foscarnet, and BDCRB are sensitive to maribavir (5). Like TCRB and BDCRB, maribavir has no significant activity against herpes simplex virus types 1 and 2, varicella- zoster virus, MCMV, human immunodeficiency virus, hepatitis B virus, or human papilloma virus (5). Unlike TCRB, it is active against Epstein-Barr virus (42). Preclinical safety and pharmacokinetic studies with maribavir found it was well tolerated and established that the drug has good oral bioavailability and low toxicity compared to currently available HCMV drugs (21, 25). Human studies yielded a good safety profile and established that maribavir reduced titers of HCMV in semen and urine from patients in a phase I/II clinical trial (25). Therefore, this drug is an outstanding candidate for further clinical development.
FIG. 1.
Structures of benzimidazole ribonucleosides BDCRB and maribavir.
The structural similarity between maribavir and TCRB or BDCRB led to an initial hypothesis that its antiviral mechanism of action would be through inhibition of cleavage of concatemeric viral DNA. However, in contrast to TCRB and BDCRB, maribavir does not inhibit this process; rather, it reduces HCMV DNA synthesis (5). Unlike GCV, valganciclovir, foscarnet, and cidofovir it does not block DNA synthesis by inhibiting viral DNA polymerase nor is it phosphorylated in virus-infected or uninfected cells (5). A strain of HCMV resistant to maribavir isolated by in vitro selection under drug pressure had a mutation in UL97 kinase, suggesting that this was the target for the drug (5). It also was found that maribavir inhibits the activity of purified recombinant glutathione S-transferase-UL97 fusion protein (4), confirming that this viral kinase is a direct target of maribavir. A separate recent study showed that maribavir not only inhibits HCMV DNA synthesis but also prevents nuclear egress of HCMV particles (23).
In order to better understand the mechanism of action of benzimidazole ribonucleosides, we isolated an HCMV strain resistant to both BDCRB and maribavir. Resistance gene mapping results showed that a single mutation in HCMV UL27 is necessary and sufficient for resistance of HCMV to maribavir. This is the first time that ORF UL27 has been shown to be involved in the mechanism of action of an antiviral drug.
(Portions of this work were reported at the 16th International Conference on Antiviral Research, Savannah, Ga., April 2003).
MATERIALS AND METHODS
Abbreviations.
The following abbreviations have been used: BDCRB, 1-(β-d-ribofuranosyl)-2-bromo-5,6-dichlorobenzimidazole; CCMV, chimpanzee cytomegalovirus; GCV, ganciclovir; HCMV, human cytomegalovirus; HFF, human foreskin fibroblasts; HHV-6, human herpesvirus 6; HHV-7, human herpesvirus 7; IC50 or IC90, 50% or 90% inhibitory concentration; maribavir, 1-(β-l-ribofuranosyl)-2-isopropylamino-5,6-dichloro-benzimidazole or 1263W94; MCMV, murine cytomegalovirus, MEM(E), minimal essential media with Earle's salts; MOI, multiplicity of infection; ORF, open reading frame; TCRB, 1-(β-d-ribofuranosyl)-2,5,6-trichlorobenzimidazole; THV, Tupaia herpesvirus.
Chemicals.
BDCRB was synthesized in the laboratory of L. B. Townsend as previously described (34). Maribavir was synthesized at GlaxoSmithKline (22) and was provided through the courtesy of K. K. Biron. GCV was provided by Hoffman La Roche (Palo Alto, Calif.).
Cell culture procedures.
HFF, derived in our laboratory, were grown in minimal essential medium with Earle's salts with 10% fetal bovine serum. They were grown at 37°C in a humidified atmosphere of 3% CO2-97% air and were regularly passaged at 1:2 dilutions using conventional procedures with 0.05% trypsin plus 0.02% EDTA in HEPES-buffered saline (31, 36).
Viral strains and virological procedures.
The HCMV Towne strain was kindly provided by M. F. Stinski, University of Iowa. HCMV strains C4 and D10 strains were derived in our laboratory by passaging HCMV Towne in the presence of TCRB (24). HCMV AD169-RV was obtained by harvesting viral progeny from HFF cells transfected with AD169-BAC and plasmids expressing Cre recombinase (pBRep-Cre) and HCMV pp71 (pCGN71). Access to AD169-BAC and Escherichia coli strain DY380 were kindly provided by U. H. Koszinowski, Ludwig-Maximilians-Universität München, Munich, Germany (6, 19), and Donald Court, National Cancer Institute, Frederick, Md., respectively. Plasmid pBRep-Cre was constructed by W. Brune (19), University of Wuerzburg, Wuerzburg, Germany. Plasmid pCGN71 was constructed in the laboratory of T. Shenk, Princeton University, Princeton, N.J. Stocks of HCMV were prepared by infecting HFF cells at a MOI of 0.01 PFU per cell, and stock viral titers were determined using monolayer cultures of HFF cells as described previously (30, 36).
HCMV antiviral assays.
For plaque reduction assay, HFF cells were planted at 85,000 cells per well in 24-well cluster dishes. Three days later they were infected with HCMV at 100 PFU per well in minimal essential medium with Earle's salts. One to two hours postinfection, media containing selected drug dilutions and final fetal bovine serum concentration of 3% and 0.5% methylcellulose were added. All drug dilutions were tested at least in duplicate using five to six drug concentrations. After incubation at 34°C for 9 to 11 days, cell monolayers were stained with crystal violet, and plaques were enumerated by light microscopy. The number of plaques observed in the presence of each drug concentration was compared to the number observed in the absence of drug in order to determine drug effects.
For yield reduction assays, HFF cells were planted at 10,000 cells per well in 96-well cluster dishes, incubated overnight, and infected with HCMV at a MOI of 0.5. After virus adsorption, medium was replaced with fresh medium containing test compounds in eight 1:3 dilutions from a starting drug concentration of 100 μM. Plates were incubated for 7 days and subjected to one cycle of freezing and thawing, and titers were determined by transferring 100-μl aliquots from each of the wells to a fresh 96-well monolayer culture of HFF cells followed by serial dilution across the plate. Cultures were incubated for 7 days, cells were stained, and the number of plaques was determined (30, 36).
Dose-response relationships were used to quantify drug effects. For plaque reduction assay, the percent inhibition of plaque number was plotted against log10 drug concentrations. For yield reduction assay, the log10 of the percent inhibition of titer of virus was plotted against the log10 drug concentrations. Fifty percent inhibitory concentrations (IC50) and ninety percent inhibitory concentrations (IC90) were interpolated from the linear portions of the regression lines.
Selection of maribavir-resistant virus.
HFF cells were infected at a MOI of 0.01 with HCMV strain C4 (23), and the virus was grown in 25-cm2 tissue culture flask in the presence of 4 μM maribavir for 2 weeks. BDCRB-resistant C4 rather than Towne was used as the HCMV strain to passage in the presence of maribavir so that newly selected virus would have a related drug resistance marker as an internal control. Supernatant progeny virus then was passaged in the presence of first 8-, then 32-, and finally 64-μM concentrations of maribavir. The duration of each passage was 2 weeks. The resulting virus was further passaged in 64 μM maribavir for 3 months. It was then frozen in liquid nitrogen, and the titer was determined. The resulting viral stock was purified by the Klein limiting dilution method (20) and termed G2 HCMV.
Growth characteristics.
HFF cells were planted at 100,000 cells per well in 24-well cell culture plates. The next day they were infected with either wild-type Towne or G2 HCMV at a MOI of 0.01. At time points spaced ∼12 h apart over a period of 10 days, the cultures were removed from the 37°C incubator and frozen at −80°C. Experiments were carried out in duplicate for all time points. After all cultures were collected, they were thawed, and titers were determined in duplicate across 96-well plates (30). Seven days later they were stained with crystal violet, and titers were calculated.
DNA sequencing.
Primers for PCR amplification and sequencing were designed using the PRIME program in the GCG package (Wisconsin Package, version 10.3; Accelrys Inc., San Diego, Calif.) and were based on the published sequence of the AD169 strain of HCMV. A list of primers is available on our laboratory website (http://sitemaker.umich.edu/drachlab). After PCR amplification, products were run on a 0.8% agarose gel, extracted, and purified using a QIAquick gel extraction kit (Qiagen, Valencia, Calif.). Sequencing PCR was done with a BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.) in a manner similar to that previously reported (24) except that sequencing reactions were separated and detected with an ABI Prism 310 genetic analyzer. Sequences were aligned and edited using Sequencher software (GeneCodes, Ann Arbor, Mich.).
Cosmid library construction.
A G2 HCMV cosmid library was constructed using the SuperCos 1 cosmid vector kit (Stratagene, La Jolla, Calif.). The SuperCos 1 vector was modified by inserting a PmeI-BamHI-PmeI adapter (GTTTAAACGGATCCGTTTAAAC) in place of the original BamHI-cut site in the cloning region. Since PmeI does not cut anywhere in the genome of HCMV, the entire cosmid insert can be cut out later with this enzyme. Modified SuperCos 1 vector was digested with XbaI restriction endonuclease, dephosphorylated with shrimp alkaline phosphatase, and then digested with BamHI. HCMV G2 genomic DNA was isolated from cell-free virus by standard extraction methods (33) with some modifications. Briefly, supernatant virus from HCMV-infected cells was pelleted at ∼10,000 × g for 2.5 h. Virus pellets were suspended in proteinase K buffer (0.01 M Tris [pH 7.8], 0.005 M EDTA, 0.5% sodium dodecyl sulfate) and digested overnight with proteinase K (100 μg/ml) at 37°C. The resulting DNA-containing solution was gently extracted eight times with 1 volume of phenol-chloroform-isoamylalcohol (24:23:1) and once with 1 volume of chloroform. The DNA was precipitated overnight at −20°C with 2.5 volumes of ice-cold 95% ethanol and 0.1 volume of 3 M NaOAc (pH 5.2). Precipitates were collected by centrifugation at ∼12,000 × g for 30 min, and the DNA pellet was washed once with 70% ethanol and finally dissolved in 10 mM Tris-HCl [pH 7.5] and 1 mM EDTA by standing undisturbed at room temperature for 3 days. It was then partially digested with 0.01 U of Sau 3AI restriction endonuclease at 37°C for 1 h. The digested genomic DNA was dephosphorylated with shrimp alkaline phosphatase and ligated into the XbaI- and BamHI-digested SuperCos 1 vector with T4 ligase at 15°C overnight. Next, the ligation mixture was packaged into phage using Gigapack III Gold Packaging Extract (Stratagene) as described in the manufacturer's instructions. The cosmid-containing phages were then used to infect E. coli by mixing cosmid packaging reaction with prepared XL1-Blue MR E. coli cells as described in Stratagene's instructions. The cosmid library was screened by sequencing the ends of the cosmids and doing a BLAST search in order to map their coordinates in the HCMV genome. To verify that the cosmids had the expected DNA content, they were also digested with EcoRI restriction enzyme, and the resulting maps were compared with the expected pattern based on the published EcoRI map for HCMV strain Towne. Forty-eight cosmids were mapped in this way. Suggestions for cosmid library construction were kindly provided by George Kemble (Aviron), Mark Underwood (GlaxoSmithKline), and Gilbert Mulamba (Harvard University).
Recombinant HCMV construction.
The genome of the HCMV AD169 strain cloned as a bacterial artificial chromosome (AD169-BAC) (6, 19) in Koszinowski's laboratory (Ludwig-Maximilians-Universität München, Munich, Germany) was used in recombinant HCMV construction. It was electroporated into DY380 E. coli cells (26) (D. L. Court, Frederick Cancer Research and Development center, Frederick, Md.) by W. Brune (University of Wuerzburg, Wuerzburg, Germany) and kindly provided to us. The zeocin resistance gene driven by the bacterial EM7 promoter was amplified by PCR from pZeo (13) using primers which at each end had 50 bp of sequence homology to the target in AD169-BAC. These PCR products were electroporated into DY380 E. coli cells containing AD169-BAC, and mutants were selected by growing bacteria on Luria-Bertani agar plates supplemented with the antibiotic Zeocin (25 μg/ml). Preparation of DY380 electroporation-competent cells and electroporation conditions were described previously by Yu et al. (41). Six AD169-BAC mutants, termed UL13-UL35Δ, UL13-UL46Δ, UL46-UL68Δ, UL20-UL46Δ, UL26-UL46Δ, and UL32-UL46Δ, were constructed in this way. Genome coordinates of the sequences that were deleted and replaced with the Zeocin resistance gene are as follows: 19919 to 47921, 19919 to 60055, 60055 to 97770, 25655 to 60055, 33395 to 60055, and 41512 to 60055 respectively. To confirm that these viruses had the expected deletions in their genome, genomic DNA was digested with either EcoRI or HindIII restriction endonucleases and compared to the wild-type digest.
In order to rescue deletion viruses, the five AD169-BAC mutants (2 to 4 μg) were electroporated into HFF cells together with their corresponding G2 cosmids (2 to 3 μg) plus plasmids pBRep-Cre and pCGN71 (1 μg each). Genome coordinates of the cosmids used are the following: 3153 to 49921, 18469 to 61455, 58673 to 99170, 23606 to 65417, 31346 to 68484, and 39463 to 80461. Electroporation was performed by using a Bio-Rad gene pulser II set at 260 V and capacitance extender at 975 μF. Medium was changed the next day, and virus was harvested 2 to 3 days after 100% cytopathogenic effect was observed, usually 3 to 4 weeks postelectroporation.
Nucleotide and amino acid sequences were obtained from GenBank. Accession numbers are as follows: HCMV AD169, X17430; HCMV AD169 UL27, NP_039961.1; CCMV UL27, NP_612671.1; HHV6 U7, NP_050187.1; HHV6 SSL1, JQ1650; HHV7 U7, AAC40723.1; HHV7 U5, NP_043761.1; THV T27, NP_116370.1; rat cytomegalovirus pR27, NP_064132.1. HCMV strain Towne open reading frames UL51, UL52, UL56, UL77, UL83, UL89 exon 1, UL89 exon 2, UL97, UL104, and UL105 are as follows: AF039234, AF047521, AF047523, AF047522, M67443, AF047525, AF047526, UO7355, AF047524, and U51564.
Web resources.
BLAST searches were done through the National Center for Biotechnology Information web browser (http://www.ncbi.nlm.nih.gov/BLAST/), and CLUSTAL W (1.82) multiple sequence alignment was done through the EMBL European Bioinformatics Institute web browser (http://www.ebi.ac.uk/clustalw/).
A list of the primers used to sequence the ORFs mentioned in this study and a list of sequence differences in these genes between Towne and AD169 are provided on our laboratory website (http://sitemaker.umich.edu/drachlab).
Nucleotide sequence accession numbers.
The sequences from HCMV strain Towne genes as reported herein for the first time are as follows: UL26, AY223526; UL27, AY223527; UL28, AY223528; UL29; AY223529; UL30, AY223530; UL31, AY223531; UL32, AF309642; UL33, AY335906; UL34, AY335907; UL35, AY335908; UL36, AY335909; UL37 exon 1, AY223525; UL44, AY223523; UL57, AY223524; UL93, AY223533; UL98, AF318210. The accession number for UL27 from the HCMV strain G2 is AY223532.
RESULTS
Isolation and initial characterization of resistant virus.
In order to investigate the mode of action of d- and l-ribosyl benzimidazoles, we isolated an HCMV strain which is resistant to both BDCRB and maribavir (Fig. 1). This was done by propagating BDCRB-resistant HCMV strain C4 (24) in the presence of increasing concentrations of maribavir, up to 64 μM. Strain C4 had been previously isolated from another BDCRB-resistant strain, D10, which in turn was isolated from wild-type Towne HCMV (24). The resulting virus strain, termed G2, was purified by the Klein purification method (20) and was resistant to maribavir in yield reduction assays (Table 1). The variability of the activity of maribavir observed with the Towne strain in these yield-reduction assays also has been observed in plaque-reduction assays and is discussed separately by Williams et al. (38). As expected, the G2 strain was also resistant to BDCRB due to the known mutations in the UL89 and UL56 genes, which were confirmed by DNA sequencing. The mutations in these genes have been previously described and are responsible for resistance of C4 HCMV to BDCRB (24). (No additional mutations in these genes were found in strain G2 [Table 2]). The C4 strain was resistant to BDCRB as expected, but it also showed some variable resistance to maribavir. As indicated by standard deviations of the IC90s for the activity of maribavir against all three viruses, the amount of resistance to this compound was somewhat variable, but it was observed consistently in four separate experiments. We have no explanation for the apparent resistance of strain C4 to this drug. In contrast, all three HCMV strains were sensitive to GCV (Table 1).
TABLE 1.
Resistance of HCMV isolates to benzimidazole ribonucleosides and ganciclovir
| HCMV strain | IC90 (μM)a
|
||
|---|---|---|---|
| BDCRB | Maribavir | GCV | |
| Towne | 0.75 ± 0.2 | 2.6 ± 3.1 | 2.5 ± 1.2 |
| C4 | 19 ± 5.7 | 17.7 ± 12.9 | 2.8 ± 1.3 |
| G2 | 22.5 ± 0.7 | 60 ± 32 | 3 ± 1.3 |
Results from yield reduction assays performed in duplicate using the indicated strains of HCMV as described in the text. IC90s ± standard deviations were calculated from two to four experiments using at least five drug concentrations each. Isolate C4 was shown previously to have the mutations in UL56 and UL89 that are responsible for resistance to BDCRB (24).
TABLE 2.
Sequencing data for G2 HCMVa
| ORF | Description or function | Coding mutation in G2 HCMV |
|---|---|---|
| UL97 | Kinase | NMb |
| UL44 | DNA processivity factor | NM |
| UL57 | DNA binding protein | NM |
| UL105 | Helicase | NM |
| UL37 exon 1 | Transactivator, apoptosis | NM |
| UL77 | DNA cleavage and packaging | NM |
| UL51 | DNA cleavage and packaging | NM |
| UL52 | DNA cleavage and packaging | NM |
| UL93 | DNA cleavage and packaging | NM |
| UL56 | DNA cleavage and packaging | Q204Rc |
| UL89 | DNA cleavage and packaging | D344Ec |
| UL104 | DNA cleavage and packaging | L21Fc |
| UL32 | Tegument protein | NM |
| UL83 | Tegument protein | NM |
| UL98 | Alkaline nuclease | NM |
The ORFs listed were amplified from G2 HCMV and sequenced. They were compared to wild-type HCMV Towne sequence determined at the same time.
NM, no mutations were found.
Mutation also in strain C4, from which G2 was derived.
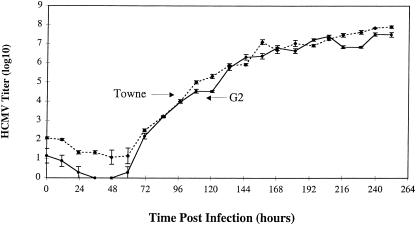
To determine if the newly acquired resistance to maribavir would change growth characteristics of G2 HCMV, HFF cells were infected at a MOI of 0.01 and its growth was monitored over a period of 10 days (Fig. 2). This study revealed that G2 HCMV was not growth deficient compared to wild-type Towne HCMV.
FIG. 2.
Growth study comparing wild-type HCMV Towne and maribavir-resistant HCMV isolate G2. HFF cells were infected at a MOI of 0.01 PFU/cell, incubated at 37°C, and harvested at the times indicated over the course of 10 days. After all samples were collected, titers were determined as described in Materials and Methods.
Sequence analysis of G2 HCMV DNA.
After determining that the G2 strain was resistant to maribavir, two approaches were used to search for the mutation(s) responsible for its resistance: sequencing of selected genes and resistance gene mapping. Genes for sequencing were selected based upon the known action of maribavir (5). A maribavir-resistant HCMV strain previously isolated was known to be resistant due to a Leu397Arg mutation in UL97 kinase (5). Based on this finding, the UL97 gene from G2 HCMV was sequenced, but it had no mutations compared to wild-type virus from which G2 was derived (Table 2). Because maribavir inhibits phosphorylation of HCMV DNA processivity factor (UL44) by UL97 (K. K. Biron, personal communication) and inhibits HCMV DNA synthesis (5), UL44, and other genes involved in DNA synthesis (single-stranded DNA binding protein [UL57], HCMV helicase [UL105], and UL37 exon 1) were sequenced. No mutations were found in any of these genes. Another recent report indicated that UL97 plays a role late in the viral replication cycle (40). This prompted sequencing of genes involved in DNA cleavage and packaging (UL77, UL51, UL52, UL93, UL56, UL89, and UL104). Again, no mutations that could explain resistance of G2 to maribavir were found in these genes. A novel mutation, Leu21Phe, was found in UL104, but the same mutation also was present in parent strains C4 and D10 and therefore was not responsible for HCMV resistance to maribavir. The role of this mutation in BDCRB resistance has been investigated and will be presented elsewhere. Genes encoding tegument proteins UL32 and UL83 as well as the gene encoding alkaline nuclease (UL98) also were sequenced. No coding mutations were found in any of these ORFs (Table 2).
Resistance gene mapping.
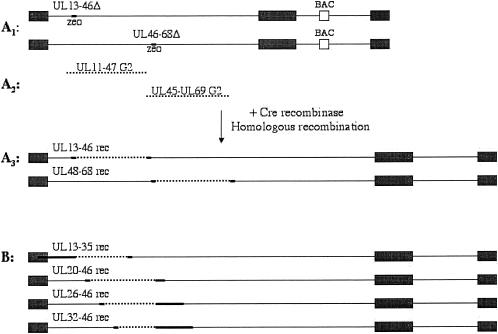
Since the HCMV genome of ∼235 kb is one of the largest viral genomes with more than 200 open reading frames, it would be difficult to sequence the entire genome in order to find a mutation responsible for resistance of G2 virus to maribavir. Therefore, recombinant viruses were constructed in which portions of wild-type DNA were replaced with G2 DNA fragments. By testing these viruses for sensitivity to maribavir, the region of genome with resistance mutation(s) was determined. In order to do this, G2 genomic DNA was cloned as a cosmid library. Then, genomic sequences that were going to be replaced with G2 cosmids were deleted from the wild-type genome. In order to do this, an HCMV genome recently cloned as a bacterial artificial chromosome (AD169-BAC) (19) was utilized. First, two AD169-BAC deletion mutants were constructed in which genes UL13 to UL46 or UL46 to UL68 (genome coordinates 19919 to 60055 or 60055 to 97770) were replaced by the Zeocin resistance gene driven by the bacterial EM7 promoter (Fig. 3A1). Because both of these viruses were missing a number of essential genes, they were not expected to replicate. They were rescued by homologous recombination with the corresponding G2 cosmids (Fig. 3A2). The genome coordinates of the first cosmid were 18469 to 61455, and those of the second cosmid were 58673 to 99170. Thus, each had the missing DNA fragment from the deletion mutant plus ∼1.4 kb of DNA at each end for homologous recombination. The two recombinant viruses made in this fashion (UL13-46rec and UL46-68rec) (Fig. 3A3) were tested for sensitivity to benzimidazole ribonucleosides. UL13-46rec HCMV was resistant to maribavir, whereas UL46-68rec HCMV was not (Table 3). These results narrowed the region of maribavir resistance to genes UL13 to UL46. Resistance of UL46-68rec to BDCRB was expected, because its UL56 gene had come from G2 HCMV and therefore carried the BDCRB resistance mutation Gln204Arg.
FIG. 3.
Construction of recombinant viruses. Solid lines represent wild-type HCMV genomic DNA, dotted lines represent G2 HCMV DNA, and bold line segments represent either wild-type or G2 HCMV DNA. (A1) AD169-BAC with either genes UL13-46 or UL46-68 replaced with the Zeocin resistance cassette. (A2) G2 HCMV cosmids. (A3) Recombinant viruses in which the Zeocin resistance cassette was replaced with either UL13-46 or UL46-68 genes from G2 HCMV. (B) Recombinant viruses in which genes UL13 to UL35, UL20 to UL46, UL26 to UL46, or UL32 to UL46 were from G2 HCMV. Details on construction of these viruses are provided in Materials and Methods.
TABLE 3.
Resistance of HCMV strains and recombinants to benzimidazole ribonucleosides
| HCMV strain or recombinant | IC50 ± SD (μM)a
|
|
|---|---|---|
| 1263W94 | BDCRB | |
| AD169-RVb | 1.6 ± 2 | 0.28 ± 0.04 |
| UL13-46rec | 25.4 ± 13.4 | 0.29 ± 0.07 |
| UL46-68rec | 1.15 ± 1.2 | 2.8 ± 0.28 |
| UL13-35rec | 18 | 0.34 |
| UL20-46rec | 27.8 ± 14.4 | 0.28 |
| UL26-46rec | 33 ± 17 | 0.22 |
| UL32-46rec | 4.3 ± 3.9 | 0.31 ± 0.04 |
Results are from plaque reduction assays performed in duplicate or triplicate using the indicated strains of HCMV as described in the text. IC50s ± standard deviation were calculated from two to nine experiments using at least five drug concentrations each. Data without standard deviations are from a single experiment with assays performed in duplicate or triplicate for UL13-35 rec.
AD169-RV is wild-type AD169-BAC from which the bacterial artificial chromosome cassette has been excised by Cre recombinase. Other recombinant viruses are designated by which genes from isolate G2 were recombined into AD169-RV. See Fig. 3 and the text for details.
Once the UL13-UL46 region of the HCMV G2 genome was identified as containing gene(s) responsible for resistance to maribavir, the region responsible was narrowed further, since this was deemed too large of a fragment to sequence (∼40 kb). Four more cosmids from the G2 cosmid library which contained smaller portions of this region were selected to make recombinant viruses. These recombinants would have either genes UL13 to UL35, UL20 to UL46, UL26 to UL46, or UL32 to UL46 replaced with G2 genomic DNA. The corresponding genes were deleted from AD169-BAC (genome coordinates 19919 to 47921, 25655 to 60055, 33395 to 60055, or 41512 to 60055), and the deletion viruses were rescued with the corresponding G2 cosmids (genome coordinates 3153 to 49921, 23606 to 65417, 31346 to 68484, or 39463 to 80461) as outlined above. The recombinant viruses so constructed (UL13-35rec, UL20-46rec, UL26-46rec, and UL32-46rec) (Fig. 3B) were tested for sensitivity to BDCRB and maribavir. All four viruses were sensitive to BDCRB as expected (Table 3). UL13-35rec, UL20-46rec, and UL26-46rec were resistant to maribavir, whereas UL32-46rec was sensitive. This established that the maribavir resistance mutation(s) must be in DNA fragment 31346 to 39463, which corresponds to a portion of UL25, genes UL26 to UL31, and a portion of UL32. This fragment was sequenced, and a single coding mutation T1004C (Leu335Pro) was identified in the UL27 ORF. This mutation was not present in the C4 or Towne HCMV strains from which G2 was derived and therefore is responsible for resistance of this strain to maribavir. To exclude the possibility that a second mutation in ORFs UL33 to UL36 was required but not sufficient for resistance, a DNA fragment from the G2 virus containing these ORFs also was sequenced, and no mutations were found. This confirmed that the T1004C mutation in UL27 was sufficient for resistance.
Comparison of UL27 homologues in other herpesviruses.
To our knowledge there is nothing reported in the literature on the function of UL27. As an initial approach to understanding its function, a BLAST search was preformed to find homologues. Three other cytomegaloviruses—chimpanzee, rat, and murine—were identified that had genes with high homology to HCMV UL27 (Table 4). Other herpesviruses, HHV-6, HHV-7, and THV, also had homologues of HCMV UL27. Using the CLUSTAL W program, a multiple sequence alignment was done in order to compare these genes. This alignment revealed that the amino acid that mutated to give resistance to maribavir, leucine 335, is highly conserved (Table 4). The change of this conserved amino acid, however, did not appear to change viability of the G2 HCMV strain, as demonstrated in the growth study (Fig. 2). Except for the report that MCMV M27 plays a role in growth and virulence of MCMV in mice (1), little has been reported in the literature on the function of any of the HCMV UL27 homologues that would give insight on the role of this open reading frame in HCMV replication. Consequently, this shall be pursued in additional studies.
TABLE 4.
Comparison of amino acid sequences implied for UL27 homologues in other herpesviruses
| Protein, virus | Positiona | Sequenceb |
|---|---|---|
| UL27, HCMV G2 | 329 | H L G A F H P P A I R H L |
| UL27, HCMV C4 | 329 | H L G A F H L P A I R H L |
| UL27, HCMV Towne | 329 | H L G A F H L P A I R H L |
| UL27, HCMV AD169 | 329 | H L G A F H L P A I R H L |
| UL27, CCMV | 330 | H L G A F H L P A I R H L |
| pR27, RCMV | 261 | E L G R L R L P R I R H L |
| M27, MCMV | 322 | N I G V L R L P K I R H L |
| U7, HHV-6 | 610 | M I G S I R L P A F L H L |
| SSL1, HHV-6 | 293 | M I G S I R L P A F L H L |
| U7, HHV-7 | 579 | A L G R I D L P I F P H L |
| U5, HHV-7 | 573 | A L G R I D L P I F P H L |
| T27, THV | 304 | E L G V L R L P C L R H L |
Position number is given for the first amino acid listed.
Amino acid sequences for G2, C4, and Towne were obtained by sequencing in our laboratory. Others were obtained from GenBank. Amino acid change is shown in bold type.
DISCUSSION
Prior studies have established that maribavir inhibits HCMV DNA synthesis and egress of viral nucleocapsids from the nucleus (5, 23). HCMV UL97 kinase, which plays a role in both of these processes, is known to be a direct target of maribavir (3, 4, 23). Furthermore, HCMV resistant to maribavir had a mutation (Leu397Arg) in this ORF (5). In the present study we were surprised to find that there were no mutations in UL97 of a viral isolate selected for resistance to maribavir. After sequencing selected other HCMV genes involved in DNA synthesis and packaging and not finding any coding mutations in their ORFs, we constructed a series of recombinant G2 wild- type HCMV viruses in order to map resistance. This was done by cloning the genome of G2 HCMV as a cosmid library, deleting sequences corresponding to selected cosmids from the wild-type HCMV strain cloned as a bacterial artificial chromosome (AD169-BAC) (6, 19), and then recombining selected cosmids with their corresponding AD169-BAC deletion viruses. Since each AD169-BAC deletion virus had a number of HCMV essential genes deleted from its genome, these isolates could grow only if the missing genes were provided by a G2 cosmid. This approach had the distinct advantage that there was no need to plaque purify resulting viruses using drug to suppress growth of wild-type virus. It also demonstrated that herpesvirus genomes cloned as bacterial artificial chromosomes can be of great help in resistance gene mapping.
Our conclusion that a single nucleotide change in UL27 was necessary and sufficient for resistance to maribavir is in accord with results of a new study by Chou and colleagues (S. Chou, A. E. Senters, R. H. Waldemer, M. G. Davis, and K. K. Biron, 9th Int. Cytomegalovirus Workshop 1st Int. Betaherpesvirus Workshop, abstr. H.03, 2003). Extensive sequencing of >130 kb each of three different isolates of HCMV resistant to maribavir revealed that changes were present only in UL27. Each strain contained a different mutation leading to putative amino acid changes (R233S, A406V/C415stop, and W362R), all of which were different from the L335P mutation reported herein. We note that all four of these mutations are in the middle of this 1,824-nucleotide ORF and speculate that this region either contains a binding site for maribavir or is a binding site for UL97, which is inhibited by maribavir.
Because the strain from which our maribavir-resistant virus was derived was Towne and the sequences of UL26 to UL36, U44, UL57, and UL98 of this HCMV strain were not available in GenBank, we sequenced these ORFs. This afforded us the opportunity to compare the sequences of several genes from the Towne and AD169 HCMV strains. We found numerous sequence differences between the two strains, the list of which is available on our laboratory website (http://sitemaker.umich.edu/drachlab). There were 11 noncoding changes between Towne and AD169 in the 1,824 bp of UL27 and four coding amino acid changes. The greatest number of differences between the two strains was in UL33, where there were 108 noncoding and 24 coding amino acid changes in a gene of 1,170 bp. We also found that approximately one-third of the middle portion of UL36 is missing in the strain of Towne with which we work, which is interesting because the Towne strain used in the study by Patterson and Shenk did not have this deletion (28). Like our strain, a UL36 deletion mutant which they made was not replication deficient.
The discovery that a single amino acid change in ORF UL27 is necessary and sufficient for resistance to maribavir could have clinical implications. Inasmuch as maribavir is a promising drug candidate, knowing where drug resistance mutations might arise will help in monitoring appearance of resistant viral strains in patients receiving the drug. Regardless of ultimate clinical utility, this study shows that UL27 is either directly or indirectly involved in the mechanism of action of maribavir.
Little appears to be known about the function of HCMV UL27. Gene expression analysis by cytomegalovirus microarrays showed that UL27 belongs to an early kinetic class of HCMV gene expression (8). A BLAST search revealed homologues of HCMV UL27 in other herpesviruses: CCMV, rat cytomegalovirus, MCMV, HHV6, HHV7, and THV. Interestingly, alignment of HCMV UL27 with its homologues showed that the amino acid (leucine 335) that mutated to proline in the maribavir-resistant HCMV is conserved in all of these herpesviruses. Based upon what is known about the mechanism of action of maribavir, UL27 could play a role in either HCMV DNA synthesis or nuclear egress of viral particles, since both of these processes are inhibited by maribavir (5, 23). Because HCMV DNA processivity factor UL44 is a substrate of UL97 and is needed for DNA synthesis (K. K. Biron, personal communication), inhibition of UL97 kinase activity by maribavir may be sufficient to explain the action of the drug on DNA synthesis. By analogy, UL27 also could be a substrate for UL97, and its phosphorylation could be involved in the regulation of nuclear egress of HCMV. Alternatively, direct inhibition of UL27 function by maribavir cannot be ruled out. Currently we have no data to support either hypothesis, but experiments are ongoing in our laboratory to test such possibilities.
Acknowledgments
This study was supported by National Institute for Allergy and Infectious Diseases grants U19-AI-31718 and P01-AI-46390 and research funds from the University of Michigan. Use of GCG programs was supported by grant M01RR00042 to the University of Michigan General Clinical Research Center. G.K. gratefully acknowledges fellowship support from the University of Michigan College of Pharmacy. We are particularly grateful to Thomas Shenk and Wolfram Brune, Princeton University, for training G.K. in BAC techniques and to Karen K. Biron, GlaxoSmithKline, for providing unpublished data and helpful discussions. We also thank Sunwen Chou, Oregon Health Sciences University, for discussion after the present study was complete of his unpublished work which identified other UL27 mutations in different isolates of maribavir-resistant HCMV.
REFERENCES
- 1.Abenes, G., M. Lee, E. Haghjoo, T. Tong, X. Zhan, and F. Liu. 2001. Murine cytomegalovirus open reading frame M27 plays an important role in growth and virulence in mice. J. Virol. 75:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin, H. I., E. Ai, H. R. McDonald, and R. N. Johnson. 2000. Retinal toxic effects associated with intravitreal fomivirsen. Arch. Ophthalmol. 118:426-427. [PubMed] [Google Scholar]
- 3.Baek, M. C., P. M. Krosky, and D. M. Coen. 2002. Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. J. Virol. 76:11943-11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek, M. C., P. M. Krosky, Z. He, and D. M. Coen. 2002. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J. Biol. Chem. 277:29593-29599. [DOI] [PubMed] [Google Scholar]
- 5.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94: a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt, W. J., R. F. Pass, S. Stagno, and C. A. Alford. 1991. Pediatric cytomegalovirus infection. Transplant Proc. 23:115-117. [PubMed] [Google Scholar]
- 8.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrisp, P., and S. P. Clissold. 1991. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs 41:104-129. [DOI] [PubMed] [Google Scholar]
- 10.Crumpacker, C. S. 1996. Ganciclovir. Drug Ther. 335:721-729. [DOI] [PubMed] [Google Scholar]
- 11.Curran, M., and S. Noble. 2001. Valganciclovir. Drugs 61:1145-1152. [DOI] [PubMed] [Google Scholar]
- 12.Deray, G., F. Martinez, C. Katlama, B. Levaltier, H. Beaufils, M. Danis, M. Rozenheim, A. Baumelou, E. Dohin, M. Gentilini, and C. Jacobs. 1989. Foscarnet nephrotoxicity—mechanism, incidence and prevention. Am. J. Nephrol. 9:316-321. [DOI] [PubMed] [Google Scholar]
- 13.Drocourt, D., T. Calmels, J. P. Reynes, M. Baron, and G. Tiraby. 1990. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 18:4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erice, A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field, A. K., and K. K. Biron. 1994. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin. Microbiol. Rev. 7:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good, S. S., B. S. Owens, L. B. Townsend, and J. C. Drach. 1994. The disposition in rats and monkeys of 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole (BDCRB) and its 2,5,6-trichloro congener (TCRB). Antivir. Res. 23:103. [Google Scholar]
- 17.Grattan, M. T., C. E. Moreno-Cabral, V. A. Starnes, E. B. Stinson, and N. E. Shumway. 1989. Cytomegalovirus infection is associated with cardiac allograph rejection and atherosclerosis. JAMA 261:3561-3566. [PubMed] [Google Scholar]
- 18.Hitchcock, M. J. M., H. S. Jaffe, J. C. Martin, and R. J. Stagg. 1996. Cidofovir, a new agent with potent anti-herpesvirus activity. Antimicrob. Agents Chemother. 7:115-127. [Google Scholar]
- 19.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, R. J. 1975. Isolation of herpes simplex virus clones and drug resistant mutants in microculture. Arch. Virol. 49:73-80. [DOI] [PubMed] [Google Scholar]
- 21.Koszalka, G. W., N. W. Johnson, S. S. Good, L. Boyd, S. D. Chamberlain, L. B. Townsend, J. C. Drach, and K. K. Biron. 2002. Preclinical and toxicology studies of 1263W94, a potent and selective inhibitor of human cytomegalovirus replication. Antimicrob. Agents Chemother. 46:2373-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koszalka, G. W., S. D. Chamberlain, R. J. Harvey, L. W. Frick, S. S. Good, M. L. Davis, A. Smith, K. K. Biron, J. C. Drach, and L. B. Townsend. 1996. Benzimidazoles For The Treatment Of Hum. Cytomegalovirus. Antivir. Res. 30:A43. [Google Scholar]
- 23.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole nucleoside analogs maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalezari, J. P., J. A. Aberg, L. H. Wang, M. B. Wire, R. Miner, W. Snowden, C. L. Talarico, S. Shaw, M. A. Jacobson, and W. L. Drew. 2002. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 46:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie, R., M. W. D. Travis, S. A. Dolan, S. Pittaluga, I. M. Feuerstein, J. Shelhamer, R. Yarchoan, and H. Masur. 1991. The cause of death in patients with human immunodeficiency virus infection: a clinical and pathological study with emphasis on the role of pulmonary disease. Medicine 70:326-343. [DOI] [PubMed] [Google Scholar]
- 28.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry, C. M., and J. A. Balfour. 1999. Fomivirsen. Drugs 57:375-381. [DOI] [PubMed] [Google Scholar]
- 30.Prichard, M. N., S. R. Turk, L. A. Coleman, S. L. Englehardt, C. J. Shipman, and J. C. Drach. 1990. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus. J. Virol. Methods 28:101-106. [DOI] [PubMed] [Google Scholar]
- 31.Shipman, C. J. 1969. Evaluation of 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) as a tissue culture buffer. Proc. Soc. Exp. Biol. 130:305-310. [DOI] [PubMed] [Google Scholar]
- 32.Sia, I. G., and R. Patel. 2000. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin. Microbiol. Rev. 13:83-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan, V., K. K. Biron, C. Talarico, S. Stanat, M. Davis, L. M. Pozzi, and D. M. Coen. 1993. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob. Agents Chemother. 37:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend, L. B., R. V. Devivar, S. R. Turk, M. R. Nassiri, and J. C. Drach. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(β-D-ribofuranosyl)benzimidazoles. J. Med. Chem. 38:4098-4105. [DOI] [PubMed] [Google Scholar]
- 35.Townsend, L. B., K. S. Gudmundsson, S. M. Daluge, J. J. Chen, Z. Zhu, G. W. Koszalka, L. Boyd, S. D. Chamberlain, G. A. Freeman, K. K. Biron, and J. C. Drach. 1999. Studies designed to increase the stability and antiviral activity (HCMV) of the active benzimidazole nucleoside, TCRB. Nucleosides Nucleotides 18:509-519. [DOI] [PubMed] [Google Scholar]
- 36.Turk, S. R., C. Shipman, Jr., R. Nassiri, G. Genzlinger, S. H. Krawczyk, L. B. Townsend, and J. C. Drach. 1987. Pyrrolo[2,3-d]pyrimidine nucleosides as inhibitors of human cytomegalovirus. Antimicrob. Agents Chemother. 31:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams, S. L., C. B. Hartline, N. L. Kushner, E. A. Harden, D. J. Bidanset, J. C. Drach, L. B. Townsend, M. R. Underwood, K. K. Biron, and E. R. Kern. 2003. In vitro activities of benzimidazole d- and l-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wingard, J. R., S. Piantadosi, W. H. Burns, M. L. Zahurak, G. W. Santos, and R. Saral. 1990. Cytomegalovirus infections in bone marrow transplant recipients given intensive cytoreductive therapy. Rev. Infect. Dis. 12(Suppl. 7):S793-S804. [DOI] [PubMed] [Google Scholar]
- 40.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacny, V. L., E. Gershburg, M. G. Davis, K. K. Biron, and J. S. Pagano. 1999. Inhibition of Epstein-Barr virus replication by a benzimidazole l-riboside: novel antiviral mechanism of 5,6-dichloro-2-(isopropylamino)-1-beta-l-ribofuranosyl-1H-benzimidazole. J. Virol. 73:7271-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]