Abstract
Lactococcus lactis subsp. lactis bv. diacetylactis strains are aroma-producing organisms used in starter cultures for the elaboration of dairy products. This species is essentially a fermentative microorganism, which cometabolizes glucose and citrate to yield aroma compounds through the diacetyl/acetoin biosynthetic pathway. Our previous results have shown that under acidic growth Lactococcus bv. diacetylactis CRL264 expresses coordinately the genes responsible for citrate transport and its conversion into pyruvate. In the present work the impact of acidic growth on glucose, citrate, and pyruvate metabolism of Lactococcus bv. diacetylactis CRL264 has been investigated by proteomic analysis. The results indicated that acid growth triggers the conversion of citrate, but not glucose, into α-acetolactate via pyruvate. Moreover, they showed that low pH has no influence on levels of lactate dehydrogenase and pyruvate dehydrogenase. Therefore, the influence of external pH on regulation of the diacetyl/acetoin biosynthetic pathway in Lactococcus bv. diacetylactis CRL264 has been analyzed at the transcriptional level. Expression of the als, aldB, aldC, and butBA genes encoding the enzymes involved in conversion of pyruvate into aroma compounds has been investigated by primer extension, reverse transcription-PCR analysis, and transcriptional fusions. The results support that this biosynthetic pathway is induced at the transcriptional level by acidic growth conditions, presumably contributing to lactococcal pH homeostasis by synthesis of neutral compounds and by decreasing levels of pyruvate.
Lactococcus lactis subsp. lactis bv. diacetylactis is widely used in the food industries because it can convert citrate to aroma (C4) compounds and carbon dioxide. These compounds improve the organoleptic characteristics of fermented foods. Lactococcal dairy strains are divided in two subspecies, cremoris and lactis, and a biovariety of the latter is called diacetylactis, due its ability to convert citrate to diacetyl. The exact mechanisms whereby lactic acid bacteria (LAB) produce diacetyl has been debated, and several metabolic engineering strategies have been proposed to improve diacetyl production (5). L. lactis also generates lactic acid and therefore has to overcome a self-imposed acidic stress. As a consequence, like other LAB, it has evolved various mechanisms to adapt to low pH, including ATPases, the glutamate decarboxylase-GAB antiporter, the arginine deiminase pathway, and general stress proteins (29). The genomes of L. lactis IL1403 and of two L. cremoris strains, MG1363 and SK11, are available in the NCBI database, and recent proteomic (3) and genomic (31) analyses of MG1363 and IL1403 have provided insights into the contribution of metabolic pathways and regulatory circuits to their acid stress response (see below). Previously, we have demonstrated that, in Lactococcus bv. diacetylactis strain CRL264, citrate transport and its conversion into pyruvate are involved in the adaptive response to acid conditions (6, 15, 16).
Citrate transport is mediated by the pH-sensitive citrate permease P (CitP) (6, 13-15, 26), which exchanges extracellular citrate for intracellular lactate. In the cytosol citrate is split into acetate and oxaloacetate by the citrate lyase complex (15, 16). Oxaloacetate is converted to pyruvate by oxaloacetate decarboxylase (CitM); this reaction consumes protons and contributes to the cytoplasmic proton gradient (15, 25). Previously, we reported that in Lactococcus bv. diacetylactis strain CRL264 acid conditions induce the expression of the plasmidic citP gene, and the chromosomal operon citMCDEFXG, which encodes the enzymes that convert citrate into pyruvate (13, 16). During sugar fermentation, lactococcal cells produce and excrete high quantities of lactate. At low pH, the lactate in the medium is protonated and easily reenters the cell, where it dissociates. Thus, the proton-consuming citrate fermentation pathway plays a role in pH homeostasis (6, 15, 16). In aerobic conditions Lactococcus bv. diacetylactis, growing on glucose and citrate, produces high intracellular levels of pyruvate (10); at low culture pH, this is diverted to the production of the C4 compounds (10, 28). The molecular regulation of this rerouting has not been investigated.
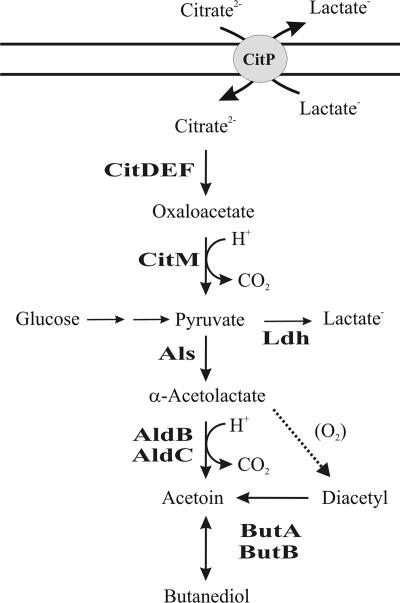
In the C4 compound pathway (see Fig. 6), α-acetolactate synthase (Als) condenses two molecules of pyruvate to yield one molecule of α-acetolactate (AL). Als has a low affinity for pyruvate and an optimal pH of 6.0, so pyruvate excess and acidic pH favor the production of AL and hence of the other C4 compounds (27). Lactococcus bv. diacetylactis also encodes (ilvB and ilvN genes) anabolic Als (acetohydroxyl acid synthetase), which is essential for the biosynthesis of branched-chain amino acids (BCAA) (7). The expression of this enzyme depends on the presence of Leu and Ile in the medium and is not subject to feedback control by BCAAs (8, 9).
FIG. 6.
Acid induction of citrate transport, citrate metabolism, and diacetyl/acetoin pathways in L. lactis. Proposed pathway for citrate fermentation and C4 compound production. CitP, citrate transporter; CitDEF; citrate lyase complex; AldB and AldC, α-acetolactate decarboxylases; ButA and ButB, diacetyl acetoin reductases (also named butanediol dehydrogenases).
Biochemical (10) and 13C nuclear magnetic resonance data (30) support that diacetyl is produced by the oxidative decarboxylation of AL. Acetoin is biosynthesized by decarboxylation of AL or by reduction of diacetyl in reactions catalyzed, respectively, by α-acetolactate decarboxylase (ALDC) (20) and diacetyl-acetoin reductase (DAR) (1, 22). At acid pH, acetoin can also be produced by chemical decarboxylation of AL (17). In L. lactis subsp. lactis (hereafter referred to as simply L. lactis) NCDO2118, ALDC (AldB) is encoded by the aldB gene located downstream of the BCAA biosynthetic genes. Apart from acetoin biosynthesis, AldB also plays a role in regulating the pool of AL during BCAA metabolism, and therefore its activity is strictly regulated at the transcriptional level (8, 9) and postranslationally by allosteric activation by Leu (20). As mentioned above, diacetyl can be converted to acetoin, which can then be reduced to 2,3-butanediol, with DAR catalyzing both steps. The reduction of diacetyl to acetoin is irreversible, whereas the reduction of acetoin to 2,3-butanediol is reversible. Diacetyl reductase activity is strain dependent in LAB (18). The conversion of 2,3-butanediol into acetoin is catalyzed by 2,3-butanediol dehydrogenase (BDH), and in Lactococcus bv. diacetylactis two proteins seem to possess this activity, as well as DAR activity (4); regulation of their expression has not been investigated.
Given the industrial importance of C4 aroma compound production, there is a need for further knowledge of the molecular regulation of the diacetyl/acetoin route in Lactococcus bv. diacetylactis. Therefore, we describe here work using genetic and proteomic techniques to study the effects of acidic growth conditions on the induction of the transcription of the genes encoding the diacetyl/acetoin pathway and of the genes responsible for the transport and conversion of citrate into pyruvate.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The lactococcal strains and plasmids used in the present study are shown in Table 1. Bacteria were grown in M17 medium, adjusted to the required pH, and supplemented with 1% glucose (M17G) or with 1% glucose and 0.4% citrate (M17GC) without shaking at 30°C. For measurement of β-galactosidase activity, cells were grown in a 50-ml, stirred fermentor in M17 broth containing 0.5% glucose with the pH automatically controlled at 5.5 or 7.0. The fermentor was inoculated with an overnight batch culture in M17 medium containing 0.5% glucose. Cells were harvested during exponential growth phase by centrifugation.
TABLE 1.
Bacterial strains and plasmids used in this study
| L. lactis strain or plasmid | Characteristics and/or genotype | Source or reference |
|---|---|---|
| L. lactis strains | ||
| CRL264 | Lac+ Pro+ Cit+ harboring pCIT264 | 26 |
| CRL30 | Lac+ Pro+ Cit− | 13 |
| IL1403 | Trp+ plasmid-free | 2 |
| Plasmids | ||
| pAK80 | Promoter selection vector containing the promoterless L. mesenteroides β-galactosidase (lacL and lacM) genes | 11 |
| pIL86 | pAK80 derivative containing the lacLM genes under the control of the 457-bp als promoter region from Lactococcus bv. diacetylactis CRL264 | This study |
| pIL91 | pAK80 derivative containing the lacLM genes under the control of the 297-bp aldB promoter region from Lactococcus bv. diacetylactis CRL264 | This study |
| pIL95 | pAK80 derivative containing the lacLM genes under the control of the 280-bp aldC promoter region from Lactococcus bv. diacetylactis CRL264 | This study |
| pIL77 | pAK80 derivative containing the lacLM genes under the control of the 554-bp butBA promoter region from Lactococcus bv. diacetylactis CRL264 | This study |
Transcriptional fusions to the lacLM reporter genes.
In order to clone the promoter (P) regions of the als, aldB, aldC, and butBA genes, the corresponding DNA fragments were amplified by a standard PCR protocol using genomic DNA from Lactococcus bv. diacetylactis strain CRL264 as a template. The oligonucleotide pairs used for the PCR amplification of Pals, PaldB, PaldC, or PbutBA were, respectively, Als-F1 and Als-R1, AldB-F1 and AldB-R1, AldC-F1 and AldC-R1, or But-F1 and But-R1. The DNA sequences of these oligonucleotides, as well as those used for reverse transcription-PCR (RT-PCR) and primer extension assays, are shown in Table S1 in the supplemental material. The amplicons were digested with BamHI and HindIII and ligated into the pAK80 plasmid vector (11) previously digested with the same enzymes. The recombinant plasmids (see the details in Table 1) carrying the above-mentioned promoters fused to the lacLM gene were established by electroporation in L. lactis IL1403, since Lactococcus bv. diacetylactis CRL264 is refractory to the usual techniques of transformation.
β-Galactosidase assay.
Samples of L. lactis IL1403 harboring the indicated plasmids were taken throughout the exponential growth phase, and the enzymatic activity was assayed as described by Israelsen et al. (11). Miller units were calculated as (522·A420)/(t·v·A600), where t is the time (in minutes), v is the volume (in milliliters) of the culture used in the assay, and A600 is the absorbance of the culture at 600 nm.
RT-PCR and primer extension transcriptional analysis.
Lactococcal strains were grown in the indicated media to an A600 of 0.5, and total RNAs were extracted from the cell pellets by using the Fast RNA Pro Blue kit (Q-Biogen).
Semiquantitative analysis of transcript levels was performed by two-step RT-PCR assays. For RT, RNA (500 ng) was added to 20-μl reaction mixtures containing 4 μl of cDNA synthesis buffer, 5 mM dithiothreitol, 40 U of RNaseOUT (Invitrogen), 1 mM deoxynucleoside triphosphate mix, the appropriate gene-specific primer (either Als-R2, AldB-R2, AldC-R2, or ButA-R1) at 10 μM, and 15 U of ThermoScript RT (Invitrogen), followed by incubation for 30 min at 58°C (Als-R2), 57°C (AldB-R2 and AldC-R2), or 62°C (ButA-R1). RT was terminated by incubation at 37°C for 20 min in the presence of 2 U of RNase H (Invitrogen). For the amplification of either cDNA, 2 μl of the RT reaction, 50 pmol of each primer, 500 μM concentrations of each deoxynucleoside triphosphate, 3.5 mM MgCl2, 20 mM Tris-HCl (pH 8.4), and 50 mM KCl were used for each 50-μl PCR, which was performed with 2 U of Taq DNA polymerase (Invitrogen). The primers used to obtain 372-nucleotide (nt) als, 531-nt aldB, 577-nt aldC, or 585-nt butBA amplicons were, respectively, Als-F2 and Als-R2, AldB-F2 and AldB-R2, AldC-F2 and AldC-R2, or ButB-F1 and ButA-R1. The cycling conditions were as follows: 1 cycle of 96°C for 2 min, followed by either (i) 15, 18, 21, 24, and 27 cycles of 94°C for 1 min or 58 or 62°C for 1 min (for the als or butBA genes, respectively), and 72°C for 1 min or (ii) 16, 19, 22, 25, and 28 cycles of 94°C for 1 min or 57°C for 1 min (for the aldB or aldC genes, respectively), and 72°C for 1 min. A total of 6 μl of the RT-PCR was analyzed by 1% agarose gel electrophoresis, and the amplicons were visualized and quantified with the QuantityOne Gel Doc 2000 software (Bio-Rad Laboratories).
Primer extension analysis was performed as previously described (6) with the following modifications. The primers used to detect the start site of the als, aldC, and butBA mRNAs were Als-R3, AldC-R3, and ButA-R2, respectively. A total of 1 pmol of either primer was annealed to 15 μg of total RNA preparation. The primers were previously labeled at their 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase. Primer extension reactions were performed by incubating the annealing mixture with 15 U of avian myeloblastosis virus reverse transcriptase (Promega) at 42°C for 30 min for the als and butBA transcripts and with 15 U of ThermoScript RT (Invitrogen) at 50°C for 30 min for aldC mRNA. The reaction products were analyzed by 8% polyacrylamide gel electrophoresis, and 32P-labeled bands were detected and directly quantified with a PhosphorImager system (Fuji). The length of the extended products was inferred by the use of unrelated DNA sequence ladders. The double-stranded plasmid pFS21 (26), which harbors the lactococcal citQRP operon heterologous sequence (accession no. S77101 in the EMBL data bank), and unlabeled primer R-Pcit (5′-CGGGTATCAAGTCATGG-3′) were used for sequencing reactions. DNA sequencing was performed using a T7 polymerase sequencing kit (Amersham-Pharmacia) and labeled with [α-32P]dCTP.
Proteomics.
Cultures of Lactococcus bv. diacetylactis strains CRL264 and CRL30 were grown in M17G or M17GC media adjusted at pH 7.0 or 5.0 to an A600 of 0.4. After growth, cells were harvested by centrifugation, and protein extracts were prepared as previously described (19). For proteomic analysis, bacterial lysates containing 100 μg of protein were analyzed in individual experiments as previously described (19), with the following modifications. The proteins were fractionated in the first dimension in a Protean IEF cell (Bio-Rad) by using gel strips for pH 4.0 to 7.0 and in the second dimension in a Protean IIxi cell (Bio-Rad) by using 10% Duracryl gels. After staining of the two-dimensional gels with Sypro Ruby, the protein spots were detected and quantified with the PDQuest 2D analysis 7.2.0 program (Bio-Rad). For this quantification, 136 stained spots were matched in all gels and used for normalization of the average intensity.
Mass spectrometric analysis.
Spots of interest were excised by using a Bio-Rad spot cutter and then analyzed at the Centro Nacional de Investigaciones Proteomic Unit (Madrid, Spain) as follows. The spots were digested in gel by using a Propineer DP digestion station (Brucker-Daltonic) according to the protocol of Schevchenko et al. (24). Then, 0.5% trifluoroacetic acid was added for peptide extraction. For peptide mass fingerprinting, the digestion solution described above was mixed with 2,5-dihydroxybenzoic acid in 33% aqueous acetonitrile and 0.1% trifluoroacetic acid and then transferred onto an AnchorChip MALDI probe (Bruker-Daltonics) and allowed to dry at room temperature. Selected samples subjected to LIFT TOF/TOF acquisition were mixed with α-cyano-4-hydroxycinnamic acid in 33% aqueous acetonitrile and 0.1% trifluoroacetic acid, deposited onto the MALDI probe, and allowed to dry at room temperature. Peptide mass fingerprint spectra were measured on a Bruker Ultraflex TOF/TOF MALDI mass spectrometer (Bruker-Daltonics) in positive-ion reflector mode. The measured tryptic peptide masses were transferred through the MS BioTools program (Bruker-Daltonics) as inputs to search the NCBInr database using Mascot software (Matrix Science, London, United Kingdom). When available, tandem mass spectrometry data from LIFT TOF/TOF spectra were combined with mass spectrometric peptide mass fingerprint data for database searching.
RESULTS AND DISCUSSION
Proteomic analysis of citrate and glucose metabolism under acidic growth.
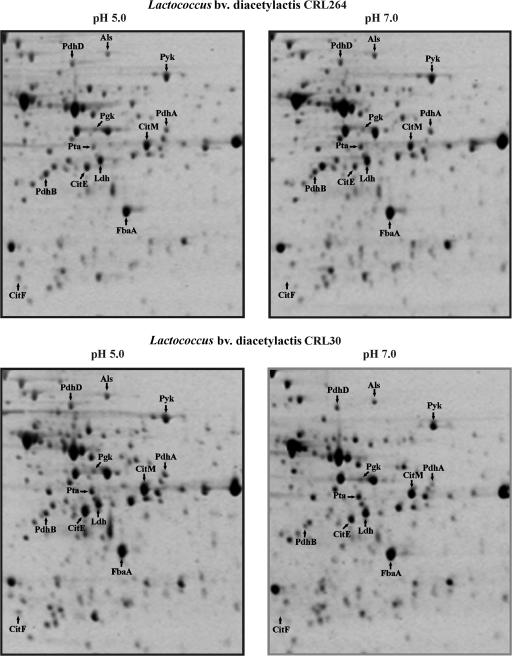
The previously reported transcriptional induction of the first steps of citrate metabolism in Lactococcus bv. diacetylactis at low pH (6, 14-16) prompted us to perform a proteomic analysis of cells grown under acidic conditions in the presence of citrate. Two strains were investigated: the citrate-metabolizing Lactococcus bv. diacetylactis strain CRL264, which carries the pCIT264 plasmid encoding the CitP transporter, and its isogenic strain, CRL30, cured of pCIT264 and consequently unable to take up citrate. Cultures were grown in M17GC media adjusted at pH 7.0 or 5.0 to an A600 of 0.4. These conditions did not affect the growth rate of the cultures nor the initial external pH values by cellular metabolism (results not shown). Protein extracts were prepared from three cultures of each strain at each condition and analyzed on two-dimensional gels (examples shown in Fig. 1). The proteome of L. lactis IL1403 has been determined (2), which allowed us to identify 12 polypeptides, corresponding to nine proteins, involved in glucose and/or citrate metabolism (Fig. 1). Their identity was confirmed by excision from the gel, digestion with trypsin, and matrix-assisted laser desorption ionization-time of flight analysis. Quantification of the spots showed the same level of expression of these polypeptides in both strains (Table 2 and Table S2 in the supplemental material). When grown at acidic pH, increased levels of the α (CitF) and β (CitE) subunits of the citrate lyase, CitM, and Als, were detected, whereas the levels of fructose-biphosphate aldolase (FbaA), phosphoglycerate kinase (Pgk), and pyruvate kinase (Pyk) were not affected. Similarly, the shift to pH 5.0 did not increase the levels of lactate dehydrogenase (Ldh), the PdhA and PdhB subunits, or the PdhD component of pyruvate dehydrogenase (Pdh) or phosphate acetyltransferase. The increased levels of the CitE and CitF subunits of citrate lyase and of CitM in CRL264, under acidic growth, correlated with the previously demonstrated transcriptional induction of their coding genes (cit operon) in CRL264 (14) and with the detection, by microarray analysis, of increased levels of CitE and CitF mRNAs in the L. lactis IL1403 strain (31). The overall proteomic analysis of the Lactococcus bv. diacetylactis CRL264 and CRL30 strains indicated that the conversion of citrate and not of glucose into C4 compounds via pyruvate is activated under acidic growth, independently of the presence of intracellular citrate. Moreover, the analysis suggested that only the C4 compound biosynthetic pathway, and not some other pyruvate metabolism, was induced at the transcriptional level under our experimental conditions.
FIG. 1.
Proteomic analysis of influence of external pH on L. lactis CRL264 and CRL30 strains. Protein extracts of exponential-phase cultures grown in M17GC at the indicated pHs. Cultures were analyzed by two-dimensional gel electrophoresis as described in the text. Arrows indicate polypeptides identified by matrix-assisted laser desorption ionization-time of flight analysis after tryptic digest. The names correspond to those used for the proteins listed in Table 2.
TABLE 2.
Influence of acidic growth in levels of proteins involved in citrate and glucose metabolisms
| Protein
|
Fold inductiona
|
pI | Mr | Scoreb | Coveragec (%) | ||
|---|---|---|---|---|---|---|---|
| Function | Name | CRL30 | CRL264 | ||||
| Citrate metabolism | |||||||
| Citrate lyase α subunitd | CitF | 1.36 | 1.45 | 5.20 | 55,440 | 106 | 12 |
| Citrate lyase β subunit | CitE | 1.81 | 2.06 | 4.84 | 33,307 | 252 | 55 |
| Oxalacetate decarboxylase | CitM | 1.82 | 2.16 | 5.21 | 40,475 | 137 | 26 |
| Pyruvate metabolism | |||||||
| α-Acetolactate synthase | Als | 1.40 | 1.24 | 4.80 | 60,863 | 255 | 34 |
| l-Lactate dehydrogenase | Ldh | 0.94 | 1.06 | 4.90 | 35,050 | 176 | 25 |
| Pdh E1 component α subunit | PdhA | 1.18 | 1.16 | 5.10 | 41,325 | 99 | 20 |
| Pdh E1 component β subunit | PdhB | 1.04 | 1.05 | 4.73 | 35,210 | 197 | 41 |
| Pdh lipoamide dehydrogenase component | PdhD | 1.18 | 1.17 | 4.85 | 59,867 | 149 | 24 |
| Phosphate acetyltransferase | Pta | 0.89 | 1.07 | 4.91 | 35,256 | 126 | 28 |
| Glucolysis | |||||||
| Fructose-bisphosphate aldolase | FbaA | 0.95 | 1.01 | 5.04 | 31,989 | 260 | 41 |
| Phosphoglycerate kinase | Pgk | 1.03 | 1.14 | 5.00 | 42,070 | 222 | 48 |
| Pyruvate kinase | Pyk | 1.19 | 0.87 | 5.21 | 54,255 | 110 | 19 |
The fold induction at acidic pH values were calculated by dividing the mean values of the spot intensities (from three independent experiments) obtained from cultures grown at pH 5.0 or 7.0. The mean values and the standard deviation of the spot intensities are depicted in Table S1 in the supplemental material.
The score is −10·log(P), where P is the probability that the observed match is a random event. Protein scores greater than 76 are significant (P < 0.05).
Coverage is the ratio of amino acids (identified in peptides/in theoretical peptides from sequence data) expressed as a percentage.
Detected a proteolytic C-terminal fragment of CitF with an Mr of 22,000 and a pI of 4.6.
L. cremoris MG1363 has also been investigated by proteomic analysis, after a challenge for a short period at pH 3.5 (3). The lack of a functional cit operon in this strain prevented detection of the citrate lyase subunits and the CitM. However, an increase in Als levels was observed after exposure to acid. This effect was detected in cultures grown in a defined medium but not in M17 rich medium. In contrast to CRL264, a low pH triggers increased levels of PdhC subunit of the Pdh and Pta in MG1363 grown in M17 and elevated levels of Ldh in cultures grown in defined medium. This indicates a different pyruvate metabolism in the citrate-fermenting Lactococcus bv. diacetylactis with respect to the non-citrate-fermenting L. cremoris under acidic growth conditions.
Effect of acidic pH on expression of the acetoin/diacetyl biosynthetic pathway genes.
From the published genome of the Lactococcus bv. diacetylactis-derived L. lactis IL1403 (2) we identified the genes als, aldB, aldC, butB, and butA, which could encode proteins that convert pyruvate to C4 compounds (see Fig. 6). The upstream and coding regions of these five genes of Lactococcus bv. diacetylactis strain CRL264 were PCR amplified using oligonucleotides inferred from the IL1403 genome sequence. There was a high (99.5%) identity between genes from both strains (results not shown), and we therefore investigated the expression of the Lactococcus bv. diacetylactis strain CRL264 genes at the transcriptional level.
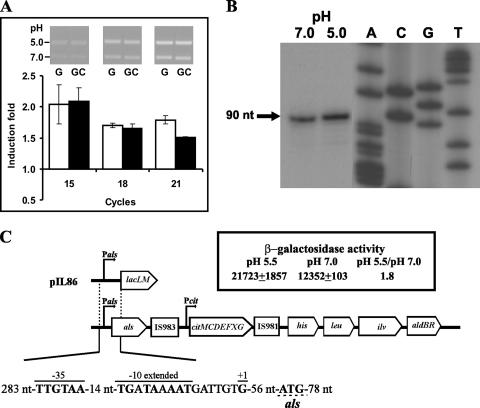
The Als encoded by gene als is located in the IL1403 genome upstream of the citMCDEFXG operon (cit operon), which in turn is located 52 kb upstream of aldB and is flanked by an inactive segment of IS983 and a copy of IS981 (IS981E), which could be a remnant (i.e., the remaining trace) of an integration process (Fig. 2C). The als of Lactococcus bv. diacetylactis DSM20384 has been expressed previously in multicopy in the L. cremoris MG1363 background (17). Detection of the als monocistronic transcript during exponential growth led Marugg et al. (17) to postulate that the gene is expressed constitutively, although these researchers did not analyze the levels of als mRNA in different growth or physiological conditions. Therefore, we tested the influence of acidic growth on als expression. Total RNA was extracted from CRL264 cells grown in the presence or absence of citrate at pH 7.0 or 5.0, and RT-PCR was used to assess the transcription of als. Quantification of DNA fragments after 15, 18, and 21 PCR cycles (see inset in Fig. 2A) revealed that, as predicted from the proteomic results, cultivation at pH 5.0 in M17G and M17GC media resulted in similar (1.8 ± 0.1)-fold and (1.7 ± 0.1)-fold increases of the als mRNA levels (Fig. 2A). The 5′ end of the als transcript in CRL264 grown in M17G at neutral or acidic pH was detected by primer extension assays, and the levels of the extended product were 1.3-fold higher in RNA preparations of cultures grown at pH 5.0 (Fig. 2B). In a previous study, an increase in als mRNA at acid pH was not detected by microarray analysis of IL1403 in M17 medium (31). This difference may be due to the fact that in the IL1403 study the cells were cultivated at pH 5.5, as opposed to pH 5.0 in the CRL264 study. Furthermore, an induction of <2-fold may not have been detected in the microarray analysis. The transcription start site was located at a guanine residue 57 nt upstream of the ATG translational start codon of the CRL264 als gene (Fig. 2C), as has also been reported for the DSM20384 als mRNA (17). In both Lactococcus bv. diacetylactis strains can be found the Pals promoter with −35 (TTGtaA) and −10 extended (TGNTAaAAT) boxes separated by a correct 14-nt spacing for RNA polymerase sigma factor binding and located at the appropriate distance upstream of the transcriptional start site (Fig. 2C) (17). To test whether Pals is a promoter and is susceptible to acidic activation, the plasmid pIL86 (Fig. 2C) was generated by cloning Pals upstream of the lacLM reporter genes of Leuconostoc mesenteroides within the pAK80 vector. The IL1403 strain was chosen as host since its lack of the citrate transport plasmid ensures that any residual citrate in the medium cannot enter into the cell and also because the CRL30 and CRL264 strains are refractory to transformation. However, IL1403 is more sensitive to acidic pH than CRL264 (results not shown), and consequently a pH of 5.5 (rather than 5.0) was chosen to challenge the cells. Thus, the IL1403(pIL86) strain, carrying the Pals-lacLM fusion in multicopy, was grown in M17G at pH 7.0 or 5.5, and samples taken during exponential growth were assayed for β-galactosidase activity (Fig. 2C). High levels of activity were detected at both pHs, supporting that Pals is a strong promoter and correlating with RT-PCR experiments, in which the amplicon was detectable after 15 PCR cycles (Fig. 2A). A significant 1.8-fold-higher enzymatic activity was observed in the culture grown at pH 5.5, confirming that expression from Pals is activated under acidic growth.
FIG. 2.
Transcriptional analysis of als expression. (A) RT-PCR analysis of als mRNA levels in Lactococcus bv. diacetylactis strain CRL264 grown in M17G (G) or M17GC (GC) medium at pH 5.0 or 7.0 (see details in Materials and Methods). The inset presents an example of analysis of the RT-PCRs in 1% agarose gel; amplicons obtained with RNA from cultures grown at pH 5.0 and at 7.0 are shown in each lane. The bands obtained from three determinations after the indicated PCR cycles were quantified, and the ratio of values obtained at pH 5.0 and 7 were calculated. The mean values of the ratios (fold induction), as well as the standard deviation versus PCR cycles, are depicted. (B) Primer extension analysis of the als transcriptional initiation site in CRL264 grown in M17G at pH 7.0 and 5.0. The extension product is indicated by the arrow (+1). (C) Organization of the Lactococcus bv. diacetylactis als gene and downstream regions, with a schematic representation of pIL86 and the β-galactosidase activity encoded by IL1403(pIL86) grown in M17G at the indicated pHs. In the sequence of the als promoter region, the upper lines indicate the −35 box, the −10 extended box, and the transcriptional start site (+1). The translational start site is underlined. The location and size of the fragment fused to the lacLM reporter gene of L. mesenteroides in pIL86 are also shown.
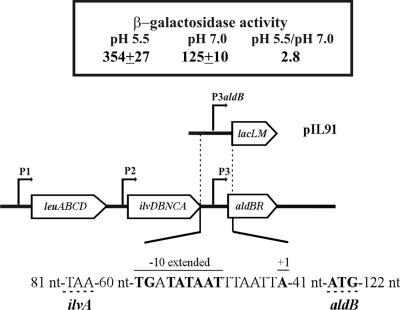
The aldB and aldC genes of Lactococcus bv. diacetylactis could encode α-acetolactate decarboxylase activities. The aldB expression has been transcriptionally characterized in L. lactis NCDO2118 in connection with BCAA biosynthesis (9). Three promoters—P1, P2, and P3—drive its transcription coupled to the aldR regulatory gene, and it has been shown that leucine and isoleucine are effectors of expression from P1 and P2 but not from P3 (9). We analyzed the effect of acidic growth on the expression of CRL264 aldB by RT-PCR as described above for als. The levels of aldB mRNAs in cultures grown at pH 7.0 or 5.0 were not significantly different in either M17G or M17GC medium (results not shown). This could be due to transcription from P1 and P2, unaffected by acidic growth, masking induced expression from P3. The location of P3 (Fig. 3), downstream of P1 and P2, invalidated the analysis of its expression by primer extension in the CRL264 background. Therefore, a P3aldB-lacLM fusion was constructed (plasmid pIL91, Fig. 3), and the influence of acid growth was analyzed in the IL1403 background, where a 2.8-fold-higher level of expression was detected in cultures grown at pH 5.5 compared to those grown at neutral pH (Fig. 3).
FIG. 3.
Transcriptional analysis of aldB expression. (A) RT-PCR analysis of aldB mRNA levels in CRL264 grown in M17G or M17GC medium at pH 5.0 or 7.0. The results are depicted as in Fig. 2. (B) Organization of the Lactococcus bv. diacetylactis leu-ilv-ald cluster, with a schematic representation of pIL91 and the β-galactosidase activity encoded by IL1403(pIL91) grown in M17G at the indicated pHs. In the sequence of P3 promoter region, the upper lines indicate the −10 extended box and the transcriptional start site (+1) of aldB mRNA identified by Goupil-Feuillerat (8). The translational start site is underlined with a discontinuous line. The sequence TAA located 60 nt upstream of −10 extended box indicates the stop codon of ilvA gene. The location and size of the fragment fused to lacLM in pIL91 are also shown.
Although, our RT-PCR experiments suggest that aldB transcription is driven primarily from P1 and P2 promoters, P3 may nevertheless play a role in controlling AldB levels. This assumption is based on the results of Goupil-Feuillerat et al. (9), which showed that expression of aldB is controlled at the posttranscriptional level by a secondary structure, which blocks the ribosomal binding site of the gene and interferes with AldB synthesis. This structure only exists in the aldB transcripts synthesized from P1 and P2. Thus, the transcriptional activation of P3 at acid pH may be designed to increase the levels of AldB, when promotion from P1 and P2 is hindered and when cells need to detoxify pyruvate.
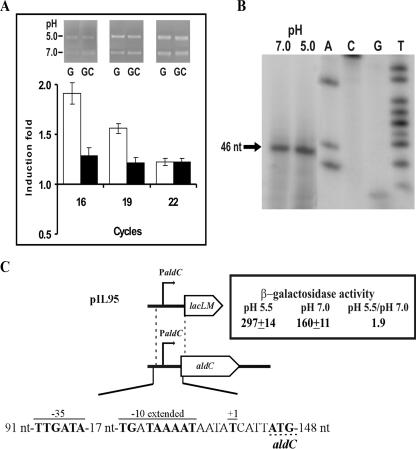
The aldC gene of the L. lactis IL1403 chromosome has 60% similarity to the protein encoded by the paralogous aldB gene, and its expression has not been investigated. Thus, the influence of external acidic pH on the expression of CRL264 aldC was studied in its natural background, as described above for als, by RT-PCR (Fig. 4A) and primer extension (Fig. 4B) analysis. RT-PCR of an internal region of aldC showed that the levels of its mRNA were (1.6 ± 0.4)-fold higher at pH 5.0 than at pH 7.0, when cells were grown in medium lacking citrate. Primer extension analysis located the 5′ end of aldC transcript in a thymidine residue just 4 nt upstream of the first ATG codon of the aldC open reading frame. A promoter region, designated PaldC, was identified preceding the start site of transcription (Fig. 4C). PaldC had a −35 (TTGAtA) and a −10 extended (TGNTAAaAT) region, each deviating by only 1 nt from the consensus sequences separated by an anomalous 17-nt spacing. The response to acid growth detected by RT-PCR was barely observed by primer extension assays, with only a 1.2-fold-higher level of transcript being detected at pH 5.0. Therefore, we carried out the transcriptional analysis in the IL1403 background by generating a PaldC-lacLM fusion in plasmid pIL95 (Fig. 4C). When IL1403(pIL95) was grown in M17G medium at pH 7.0 or 5.5. the production of β-galactosidase confirmed that PaldC, like P3aldB, is a promoter of intermediate strength (Fig. 4C), which is utilized more efficiently by the RNA polymerase at acidic pH.
FIG. 4.
Transcriptional analysis of aldC expression. (A) RT-PCR analysis of aldC mRNA expression levels in CRL264 grown in M17G or M17GC medium at pH 5.0 or 7.0. The results are depicted as in Fig. 2. (B) Analysis of the initiation of transcription from aldC promoter determined by primer extension in CRL264 grown in M17G at pH 7.0 and 5.0. The extension product is indicated by an arrow (+1). (C) Schematic representation of the Lactococcus bv. diacetylactis aldC gene, with representation of pIL77 and the β-galactosidase activity encoded by IL-1403(pIL77) grown in M17G at the indicated pHs. The location and size of the fragment fused with the lacLM reporter gene from L. mesenteroides is also shown. In the sequence of the aldC promoter region, the upper lines indicate the −35 box, the −10 extended box, and the transcriptional start site (+1). The translational start site is underlined with the discontinuous line.
Characterization of the butBA operon in Lactococcus bv. diacetylactis CRL264.
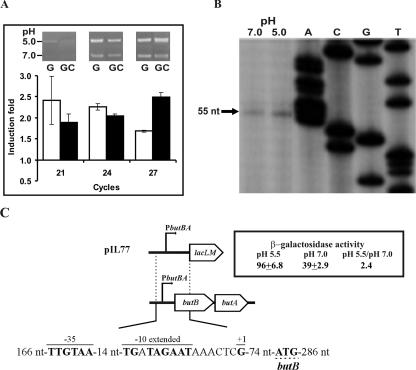
The genes butB and butA (also called dar) are linked in both the Lactococcus bv. diacetylactis and the L. cremoris chromosomes. However, in other species of LAB, such as Leuconostoc pseudomesenteroides (23), butA is carried on plasmids. Inactivation of L. pseudomesenteroides butA (23) and L. cremoris dar (20) resulted, in both cases, in a decrease in DAR activity, confirming the enzymatic identity of the gene products. Bioinformatic analysis on the butA gene from Leuconostoc (23) allowed us to identify the butA gene in the IL1403 genome, whose product shares 81% homology with the ButA from Leuconostoc. Preceding the IL1403 butA is the butB gene, whose product is annotated as BDH. To verify the bicistronic structure of butBA, a DNA fragment containing the 3′ end of butB and the 5′ end of butA was RT-PCR amplified. The detection of the expected amplicon by use of RNA preparation from CRL264 revealed that indeed butBA constitutes an operon. Moreover, the analysis showed that acidic growth resulted in a significant increase of expression of these genes of (2.1 ± 0.2)-fold and (2.1 ± 0.1)-fold in cultures grown, respectively, in M17G and M17GC (Fig. 5A). For detection of the amplified fragment in agarose gel a long (21, 24, and 27 cycles) PCR cycling was needed, suggesting low levels of the transcript. In fact, we were unable to detect the 5′ end of the transcript synthesized by CRL264 by primer extension, even when using a 5′-end 32P-labeled primer and [32P]dCTP-labeled substrates for the primer extension assay (results not shown). Nevertheless, inspection upstream of butB revealed a putative promoter, which was designated PbutBA, located 82 nt from its translational start codon. This promoter possesses both −35 and −10 extended boxes. The first (TTGttA) deviates in 2 nt from the consensus sequence, and the second (TGNTAgAAT) has a substitution of G by T, which would make PbutBA behave as a weak promoter. Therefore, the promoter region of the CRL264 butBA operon was fused to lacLM in the pAK80 vector, generating pIL77 (Fig. 5C), with the aim of testing PbutBA in multicopy state in the L. lactis IL1403 host. Primer extension analysis of IL1403(pIL77) (Fig. 5B) detected the 5′ end of the butBA mRNA at a guanine located exactly at the expected position if transcription is driven from PbutBA. In addition, this analysis revealed a 1.8-fold increase of the transcript at acidic pH. This result was substantiated by detection of 2.4-fold-higher levels of the β-galactosidase activity in cultures subjected to acidic growth (Fig. 5C).
FIG. 5.
Transcriptional analysis of butBA expression. (A) RT-PCR analysis of butBA mRNA levels in Lactococcus bv. diacetylactis strain CRL264 grown in M17G or M17GC medium at pH 5.0 or 7.0. The RT-PCRs were performed as described in Materials and Methods, and the results are depicted as in Fig. 2. (B) Analysis of the initiation of transcription from butBA promoter determined by primer extension in Lactococcus bv. diacetylactis strain CRL264 grown in M17G at pH 7.0 and 5.0. The extension product is indicated by an arrow (+1). (C) Organization of the Lactococcus bv. diacetylactis butBA genes, with a schematic representation of pIL77 and the β-galactosidase activity encoded by IL1403(pIL77) grown in M17G at the indicated pHs. The location and size of the fragment fused to lacLM in pIL77 are also shown. In the sequence of the butBA promoter region, the upper lines indicate the −35 box, the −10 extended box, and the transcriptional start site (+1). The ButB translational start site is underlined with a discontinuous line.
Thus, we have demonstrated that butB and butA in Lactococcus bv. diacetylactis are organized within an operon, whose transcription is driven from the weak PbutBA promoter and activated by acidic growth. Two DARs have been purified from lactococcal cells that could be encoded by the butBA operon; each has less affinity for 2,3-butanediol than for diacetyl and acetoin as substrate, and each has an optimal alkaline and acidic pH, respectively, for its BDH and DAR activities (4). Therefore, at acidic pH a synchronized, moderate increase of the two DARs transcribed from the weak PbutBA promoter would result in an increase of 2,3-butanediol, although still yielding significant amounts of acetoin, as has been observed in Lactococcus bv. diacetylactis strains (21).
Conclusions.
The transcriptional analysis presented here indicates that in Lactococcus bv. diacetylactis CRL264 expression of the als, aldB, and aldC genes and the butBA operon from the Pals, P3aldB, PaldC, and PbutAB promoters is induced at low pH. Functional analysis of the levels of expression from the CRL264 promoters revealed that for all of them, in the IL1403 background, a shift from pH 7.0 to 5.5 resulted in a moderate but significant induction. The mechanism of their low pH induction remains unknown, since they lack the ACID-boxes detected in other pH-inducible promoters, which seem to be the operators of the RcfB regulator (12). All promoters possessed a −10 extended box, and all except P3aldB have a −35 box. In addition, Pals and PbutBA show the appropriate spacing of 14 nt between the boxes for binding of the RNA polymerase vegetative sigma factor. The results showed differences in the strength of the promoters probably due to deviation from the consensus sequence in the boxes and the anomalous 17-nt spacing in PaldC (Fig. 2C, 3B, 4C, and 5C). Pals was the strongest promoter, with transcriptional levels ∼70-fold higher than those for PaldB and PaldC (Fig. 2B versus Fig. 3B and 4C).
Overall, it is therefore logical that under acidic growth conditions the conversion of citrate to C4 compounds (Fig. 6) would be favored. The acidic induction of the C4 biosynthetic pathway would switch the metabolism from pyruvate to lactate into the production of less-acidic and neutral compounds, thus contributing to the pH homeostasis within the cell. This mechanism is not essential for Lactococcus but confers to Lactococcus bv. diacetylactis higher tolerance to, and better survival at, low pH (6, 15).
Supplementary Material
Acknowledgments
We thank M. A. Corrales for technical assistance. We thank Stephen Elson for the critical reading of the manuscript.
This study was supported by European Union grants QLK1-CT-2002-02388 and KBBE-CT-2007-211441, Spanish Ministry of Education grant AGL2006-1193-C05-01, and Agencia Nacional de Promoción Científica y Tecnológica grant PICT 15-38025. The work at the CIB and IBR was performed, respectively, under the auspices of the Consejo Superior de Investigaciones Científicas and Consejo Nacional de Investigaciones Cientifícas y Tecnólogicas (CONICET).
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aungpraphapornchai, P., H. G. Griffin, and M. J. Gasson. 1999. Cloning, DNA sequence analysis, and deletion of a gene encoding diacetyl-acetoin reductase from Lactococcus lactis. DNA Seq. 10:163-172. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budin-Verneuil, A., V. Pichereau, Y. Auffray, D. S. Ehrlich, and E. Maguin. 2005. Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics 5:4794-4807. [DOI] [PubMed] [Google Scholar]
- 4.Crow, V. L. 1990. Properties of 2,3-butanediol dehydrogenases from Lactococcus lactis subsp. lactis in relation to citrate fermentation. Appl. Environ. Microbiol. 56:1656-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vos, W. M., and J. Hugenholtz. 2004. Engineering metabolic highways in lactococci and other lactic acid bacteria. Trends Biotechnol. 22:72-79. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Quintáns, N., C. Magni, D. de Mendoza, and P. López. 1998. The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 64:850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godon, J. J., M. C. Chopin, and S. D. Ehrlich. 1992. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 174:6580-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goupil-Feuillerat, N., M. Cocaign-Bousquet, J. J. Godon, S. D. Ehrlich, and P. Renault. 1997. Dual role of α-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J. Bacteriol. 179:6285-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goupil-Feuillerat, N., G. Corthier, J. J. Godon, S. D. Ehrlich, and P. Renault. 2000. Transcriptional and translational regulation of α-acetolactate decarboxylase of Lactococcus lactis subsp. lactis. J. Bacteriol. 182:5399-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz, J. 1993. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 12:165-178. [Google Scholar]
- 11.Israelsen, H., S. M. Madsen, A. Vrang, E. B., Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madsen, S. M., T. Hindré, J. P. Le Pennec, H. Israelsen, and A. Dufour. 2005. Two acid-inducible promoters from Lactococcus lactis require the cis-acting ACID-box and the transcription regulator RcfB. Mol. Microbiol. 56:735-746. [DOI] [PubMed] [Google Scholar]
- 13.Magni, C., F. L. López de Felipe, F. Sesma, P. López, and D. de Mendoza. 1994. Citrate transport in Lactococcus lactis subsp. lactis biovar diacetylactis: expression of the citrate permease P. FEMS Microbiol. Lett. 118:75-82. [Google Scholar]
- 14.Magni, C., F. López de Felipe, P. López, and D. de Mendoza. 1996. Characterization of an insertion sequence-like element identified in plasmid pCIT264 from Lactococcus lactis subsp. lactis biovar diacetylactis. FEMS Microbiol. Lett. 136:289-295. [DOI] [PubMed] [Google Scholar]
- 15.Magni, C., D. de Mendoza, W. N. Konings, and J. S. Lolkema. 1999. Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J. Bacteriol. 181:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín, M. G., P. D. Sender, S. Peirú, D. de Mendoza, and C. Magni. 2004. Acid-inducible transcription of the operon encoding the citrate lyase complex of Lactococcus lactis biovar diacetylactis CRL264. J. Bacteriol. 186:5649-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marugg, J. D., D. Goelling, U. Stahl, A. M. Ledeboer, M. Y. Toonen, W. M. Verhue, and C. T. Verrips. 1994. Identification and characterization of the α-acetolactate synthase gene from Lactococcus lactis subsp. lactis biovar diacetylactis. Appl. Environ. Microbiol. 60:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellerick, D., and T. M. Cogan. 1981. Induction of some enzymes of citrate metabolism in Leuconostoc lactis and other heterofermentative lactic acid bacteria. J. Dairy Res. 48:497-502. [Google Scholar]
- 19.Mohedano, M. L., K. Overweg, A. de la fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. López, and J. Wells. 2005. Inducible expression of the essential response regulator YycF in Streptococcus pneumoniae modulates expression of genes involved in fatty acid biosynthesis and affects membrane composition. J. Bacteriol. 178:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phalip, V., C. Monnet, P. Schmitt, P. Renault, J. J. Godon, and C. Divies. 1994. Purification and properties of the α-acetolactate decarboxylase from Lactococcus lactis subsp. lactis NCDO 2118. FEBS Lett. 351:95-99. [DOI] [PubMed] [Google Scholar]
- 21.Ramos, A., K. N. Jordan, T. M. Cogan, and H. Santos. 1994. 13C nuclear magnetic resonance studies of citrate and glucose cometabolism by Lactococcus lactis. Appl. Environ. Microbiol. 60:1739-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattray, F. P., M. Walfridsson, and D. Nilsson. 2000. Purification and characterization of a diacetyl reductase from Leuconostoc pseudomesenteroides. Int. Dairy J. 10:781-789. [Google Scholar]
- 23.Rattray, F. P., D. Myling-Petersen, D. Larsen, and D. Nilsson. 2003. Plasmid-encoded diacetyl (acetoin) reductase in Leuconostoc pseudomesenteroides. Appl. Environ. Microbiol. 69:304-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 25.Sender, P. D., M. G. Martin, S. Peiru, and C. Magni. 2004. Characterization of an oxaloacetate decarboxylase that belongs to the malic enzyme family. FEBS Lett. 570:217-222. [DOI] [PubMed] [Google Scholar]
- 26.Sesma, F., D. Gardiol, A. P. de Ruiz Holgado, and D. de Mendoza. 1990. Cloning and expression of the citrate permease gene of Lactococcus lactis subsp. lactis biovar diacetylactis in Escherichia coli. Appl. Environ. Microbiol. 56:2099-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snoep, J. L., M. J. Teixeira de Mattos, M. J. Starrenburg, and J. Hugenholtz. 1992. Isolation, characterization, and physiological role of the pyruvate dehydrogenase complex and α-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 174:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starrenburg, M. J., and J. Hugenholtz. 1991. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl. Environ. Microbiol. 57:3535-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van de Gutchte, M., P. Serror, C. Chervaus, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 30.Verhue, W. M., and F. S. B. Tjan. 1991. Study of the citrate metabolism of Lactococcus lactis subsp. lactis biovar diacetylactis by means of 13C nuclear magnetic resonance. Appl. Environ. Microbiol. 57:3371-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie, Y., L. Chou, A. Cutler, and B. Weimer. 2004. DNA microarray of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stress. Appl. Environ. Microbiol. 70:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.