Abstract
Minerotrophic fen peatlands are widely distributed in northern latitudes and, because of their rapid turnover of organic matter, are potentially larger sources of atmospheric methane than bog peatlands per unit area. However, studies of the archaeal community composition in fens are scarce particularly in minerotrophic sites. Several 16S rRNA-based primer sets were used to obtain a broad characterization of the archaeal community in a minerotrophic fen in central New York State. A wide archaeal diversity was observed in the site: 11 euryarchaeal and 2 crenarchaeal groups, most of which were uncultured. The E1 group, a novel cluster in the order Methanomicrobiales, and Methanosaetaceae were the codominant groups in all libraries and results of terminal restriction fragment length polymorphism (T-RFLP) analysis. Given its abundance and potential hydrogenotrophic methane contribution, the E1 group was targeted for culture attempts with a low-ionic-strength medium (PM1). Initial attempts yielded Methanospirillum-dominated cultures. However, by incorporating a T-RFLP analysis as a quick selection tool for treatments and replicates, we were able to select an enrichment dominated by E1. Further dilutions to 10−9 and tracking with T-RFLP yielded a strain named E1-9c. E1-9c is a novel coccoid hydrogenotrophic, mesophilic, slightly acidophilic methanogen and is highly sensitive to Na2S concentrations (requires <0.12 mM for growth). We propose E1-9c as the first representative of a novel genus in the Methanomicrobiales order.
Peatlands are wetlands where the rate of accumulation of organic matter exceeds its rate of decomposition (26), producing organic peat soil. On a global basis, these ecosystems are estimated to store more than 30% of all terrestrial soil carbon (28), and their anoxic and reducing conditions are suitable for CH4 production by methanogenic Archaea (67). It is important to gain a better understanding of the processes and microorganisms involved in CH4 production in peatlands because these ecosystems represent the largest natural sources of CH4 for the atmosphere, levels of which have more than doubled in the past 200 years (17). Moreover, peatlands are not homogeneous ecosystems but include a wide range of sites that can substantially differ in vegetation, hydrology, and chemistry (9, 62). The distinctions between bogs and fens include many of these differences.
Bogs (ombrotrophic or rain-fed sites) are dominated by Sphagnum mosses (27); receive only atmospheric inputs of water, cations, and nutrients; and are nutrient-poor, low-pH (≤4) ecosystems (26). In contrast, fens have a greater plant diversity, are commonly dominated by sedges and woody plants, receive the inputs of groundwater and runoff from surrounding uplands, and exhibit higher alkalinity and pH levels (5.5 to 7 in moderate minerotrophic fens; >7 in calcareous fens) (26, 64). Bogs are abundant in boreal regions and have been the focus of many studies of methanogenic processes, environmental controls, and methanogenic Archaea (2, 3, 9, 11, 36). However, fens have received far less attention, despite their abundance and significant potential for CH4 emissions.
Minerotrophic fens are widely distributed in the subarctic regions (55), the western boreal forests of North America (65), and other northern temperate zones (6, 68). Additionally, minerotrophic fens have been found to have higher rates of CH4 production (7, 70), higher overall CH4 cycling (13, 39, 59), and relatively rapid hydrology and short residence time compared to bogs (59), and their highly abundant vascular plants such as sedges and cattails facilitate the flow of CH4 from deeper layers (50, 71). Thus, fens could have a greater global impact on CH4 emissions than bogs in terms of CH4 equivalents per unit area, in the near future.
Only a few studies have characterized the archaeal community composition in fens from northern peatlands. Studies have been done in oligotrophic or poor fens (21, 22, 47), as well as in mesotrophic ones (20, 36), but minerotrophic fens have remained largely ignored. Nevertheless, the studies of poor fens and bogs (5, 22, 36, 42) showed that Archaea are primarily associated with novel or uncultured groups in addition to some close relatives to known methanogens. Thus, we could expect that minerotrophic fens contain some of these or new uncultured groups.
Cultured representatives of these novel archaeal groups are desirable for understanding their ecology and role in peatlands. Although the cultivation of methanogens from peatlands has proven to be difficult (69), some recent attempts have succeeded, enriching (12, 58) or isolating novel methanogens from bogs, such as “Candidatus Methanoregula boonei” (11) in the order Methanomicrobiales, as well as known methanogens from the order Methanobacteriales (41).
The goal of this study was to characterize the archaeal community of a minerotrophic fen, with particular emphasis on methanogenic Archaea and related groups. The community characterization showed that members of an uncultured group in the order Methanomicrobiales, previously designated E1 (15), were likely to be important in methanogenesis from H2-CO2. Members of E1 were targeted, and only after culturing efforts were coupled to molecular profiling using terminal restriction fragment length polymorphism (T-RFLP) analysis were we able to isolate a methanogen from the group. Phylotypes related to the strain E1-9c have been found in minerotrophic fens, landfills, and anaerobic bioreactors, and the strain is proposed as a representative of a new genus in the order Methanomicrobiales.
MATERIALS AND METHODS
Study site and sampling.
Michigan Hollow (MH; local name) is a 15-ha, minerotrophic fen near the village of Danby in central New York State (42°21′N, 76°28′W) and was initially described by Bernard and MacDonald (8). The peatland is located in the lower part of a small forested watershed and receives surface and subsurface water flow from the surrounding forested uplands (59). The vegetation at the site is currently dominated by Carex lacustris (lake sedge), Typha latifolia (common cattail), and Lythrum salicaria (purple loosestrife). Triplicate peat samples were collected anaerobically at a 20-cm depth in April 2005, using procedures described previously (10). Samples were processed inside of an anaerobic glove box (Coy) no later than 2 h after sampling.
16S rRNA gene amplification, cloning, and phylogenetic analysis.
DNAs from peat samples and enrichment cultures were extracted in duplicate with a Power Soil DNA or Ultraclean Microbial DNA extraction kit (MoBio), respectively, using the manufacturer's protocol. PCR amplifications were performed as previously described (15). Amplifications were done with three archaeal primer combinations and a eubacterial combination with their corresponding annealing temperatures: primer 1Af -1100r (30) at 50°C, primer 1Af-1492r at 50°C, primer 1Af-ArchLSU47 (11) at 53°C, and primer 27f-1492r at 50°C. PCR products were examined by electrophoresis on 1% agarose gels.
Two clone libraries for each of the three archaeal primer combinations were constructed as described previously (5, 15). Sequences were compared against those in the GenBank database to ensure that newly reported relatives were included in the analysis. The alignment of sequences was done with ARB software (44) and exported from it by using a nucleotide base frequency filter that included positions with more than 50% invariance (1,420 valid columns). Trees were constructed by Bayesian analyses using MrBayes software (version 3.0 [53]) and four-chain metropolis-coupled Markov-chain Monte-Carlo (MCMCMC) analysis. Chains commonly converged after 1,000,000 generations. A Bayesian consensus tree was built with a burnout of 300, and nodes with a posterior probability value of ≥0.80 were considered significant. Trees were imported back to ARB, and smaller sequences were added by using the parsimony tool without altering the global tree topology. Tree topology was confirmed by using maximum-likelihood and neighbor-joining methods (implemented in ARB) with Olsen evolutionary distance correction. The tree presented represents the topology most frequently observed across various phylogenetic analyses.
Putative chimeras were identified and excluded from phylogenetic analysis, using a Bellerophon server (32). Rarefaction analyses of sequences were done using distance-based operational taxonomic unit and richness (DOTUR) software (57), with the furthest neighbor assignment method.
T-RFLP analysis.
T-RFLP analysis was performed as described previously (15) with the 1Af-1100r primers and the reverse primer fluorescently labeled on its 5′ end with carboxyfluorescein. Fragments were resolved by using an Applied BioSystems GeneScan 500 Liz marker with a 3730 DNA Analyzer (Bio Resources Center, Cornell University, NY). Terminal restriction fragment sequence lengths and peak heights and areas were determined using GeneScan Analysis software (version 3.1.; Applied Biosystems). To exclude potential pseudo-terminal restriction fragments, traces were standardized to an accumulated terminal restriction fragment area of 100 relative fluorescence units, and peaks below the arbitrary value of 0.5 were eliminated from the analysis.
CH4 production by peat slurries.
Peat slurry incubations with or without substrate additions were performed as described previously (10). Substrates were added to anaerobic stock solutions along with (final concentrations) sodium acetate (1 mM) and rifampin (10 mg liter−1), which was added to impede the growth of acetogens in H2-CO2 enrichments (10), and to corresponding controls without H2-CO2. Sterile O2-scrubbed H2-CO2 (80%/20%; 70.7 kPa; Mixed Gas Industries) was added to appropriate tubes. Data represent the averages of triplicate samples.
Chemical analyses.
CH4 content in headspace gas samples was analyzed using a Perkin-Elmer 3920B gas chromatography column with a flame ionization detector (Phoenix Equipment), as previously described (10). Peat slurries were vortexed prior to headspace analysis.
Growth of methanogenic culture.
Culturing efforts were done by using the low-ionic-strength medium PM1, as described previously (11, 12), with the following anaerobic additions to their final concentrations: 1.0 mM titanium(III) nitrilotriacetate (48), 10 mM MES [2-(N-morpholino)ethane sulfonic acid (pKa = 6.2 at 28°C; 1 M stock solution adjusted to pH 7)], 0.5 mM coenzyme-M (2-mercapthoethanesulfonic acid), 0.5 mM sodium acetate, 100 μM fatty acid mixture (isobutyric, valeric, isovaleric, and dl-2-methyl butyric acid), 0.1 g liter−1 yeast extract, 1% (vol/vol) vitamin solution (4), 1 μmol H2S (added as sterile gas), and 10 mg liter−1 rifampin. The final liquid volumes in the tubes were ca. 5 ml, and 70.7 kPa H2-CO2 was added to the headspaces. The cultures were incubated in a shaker (at 200 rpm) at 28°C. After obtaining a culture from a 10−9 dilution, tests were done to determine which additions were strictly required. The yeast extract, the fatty acid mixture, and the rifampin were not required for E1-9c growth and consequently were excluded from further culture transfers and tests.
For experiments on the effect of pH, the pH was adjusted by the addition of 30 mM MES, adjusted to various pH values. Low pH evaluations were also attempted by adding 5 mM of either homopiperazine-N,N′-bis-2(ethanesulfonic acid) (HOMOPIPES) or citric acid instead of MES. The pH of the cultures was assessed at the end of incubations. For the Na2S sensitivity test, Na2S·9H2O filter-sterilized anaerobic solutions were prepared with different concentrations, so the same volume of reagent (0.05-ml to 5-ml cultures) was added to replicate cultures. For experiments on the effect of temperature, cultures were incubated under static conditions. All the tests were performed in triplicate and repeated at least once. Growth rates were calculated from methane production from the exponential parts of the methane accumulation curve.
Bright-field microscopy, fluorescence in situ hybridization (FISH), and electron microscopy.
A Nikon Eclipse E600 microscope equipped with a Hamamatsu charge-coupled device digital camera was used for light and fluorescence microscopy. For the examination of cells by FISH, cells were fixed in 4% paraformaldehyde for 15 to 24 h, filtered onto black 0.2-μm polycarbonate membrane filters, and stored with desiccant at −20°C. FISH was performed as described by Morris et al. (49). The hybridization buffer, without formamide, contained 2 μg ml−1 4′,6′-diamidino-2-phenylindole and an indocarbocyanine dye-labeled probe at a final concentration of 2 ng μl−1 (ARCH 915 or EUB 338) (1).
Negative staining transmission electron microscopy was performed as follows. Five microliters of cells resuspended in deionized water was mixed with a drop of bacitracin (100 μg ml−1) and allowed to settle onto Formvar-coated (100-mesh) copper grids for 5 min. Staining with 2% uranyl acetate (pH 6.5) was done for a few seconds. Micrographs were acquired with a Phillips EM-201 electron microscope and a Gotan Model 780 camera at 100 kV.
Nucleotide sequence accession numbers.
16S rRNA, intergenic transcribed sequences (ITS), and 23S rRNA gene sequences were deposited in the GenBank nucleotide sequence database under the accession numbers EU155896 to EU155999 for environmental clones and EU156000 for strain E1-9c.
RESULTS
Archaeal diversity assessed by 16S rRNA gene analysis.
The 16S rRNA gene clone libraries revealed a diverse archaeal community on the shallow layer of MH fen (Fig. 1 and 2). Some euryarchaeal phylotypes were associated with known methanogen clades such as Methanosaetaceae, Methanosarcinaceae, Methanobacteriaceae, and Methanospirillaceae. Other phylotypes were related to uncultured groups such as “group E1,” “subaqueous cluster” (SC), “group E1′,” marine benthic group D (MBD), and rice cluster II (RC-II) or to recently cultured groups such as “E2/Methanoregula” (E2) (11) and rice cluster I (RC-I) (56). Crenarchaeal phylotypes that belonged to the uncultured groups rice cluster IV (RC-IV) and rice cluster VI (RC-VI) were also recovered (Fig. 2; see Fig. S1 in the supplemental material).
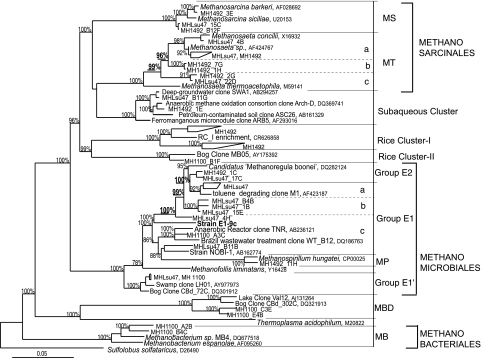
FIG. 1.
Phylogenetic analysis of archaeal 16S rRNA gene clones from the MH fen. Members of the order Methanosarcinales include Methanosarcinaceae (MS), Methanosaetaceae (MT); subaqueous cluster (SC); rice cluster I (RC-I); rice cluster II (RC-II). Member of the order Methanomicrobiales include group E1, group E1′, group E2, and Methanospirillaceae (MP). Members of the order Methanobacteriales include Methanobacteriaceae (MB), marine benthic group D (MBD). The tree was constructed by Bayesian analysis using MrBayes software version 3 (see Materials and Methods); posterior probabilities greater than 80 are indicated. Clones from MH fen are indicated by the initials MH, followed by the indication of their primer mixture of origin: 1100 (1af-1100r), 1492r (1af-1492r), and LSU47 (1af-LSU47r).
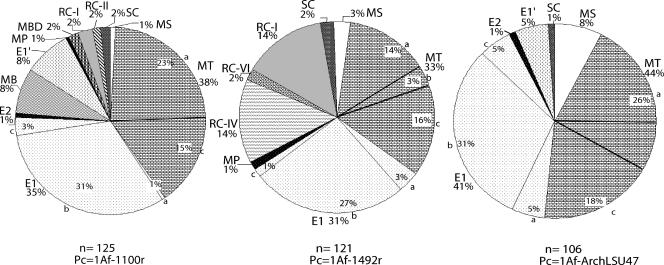
FIG. 2.
Clone distribution from libraries constructed with different primer combinations. The total number of clones (n) and primer combination (Pc) are indicated for each primer set. The recovered groups are Methanosarcinaceae (MS), Methanosaetaceae (MT), subaqueous cluster (SC), rice cluster I (RC-I), rice cluster II (RC-II), group E1 (E1), group E1′ (E1′), group E2 (E2), Methanospirillaceae (MP), Methanobacteriaceae (MB), marine benthic group D (MBD), rice cluster IV (RC-IV), and rice cluster VI (RC-VI).
The clones associated with the E1 and Methanosaetaceae groups were numerically dominant in all libraries, regardless of the primer combination, with average abundances of 35% and 39%, respectively. The other archaeal groups always represented smaller fractions in the libraries (Fig. 2). The dominant E1 and Methanosaetaceae groups were similarly well covered by the different primer combinations. However, differences in coverage were observed in the detection of minority groups. The 1AF-ArchLSU47 primer combination did not recover sequences from the RC-I, RC-II, MBD, Methanospirillaceae, and Methanobacteriaceae groups, although its libraries contained fewer clones than the other two primer sets (Fig. 2). The 1AF-1492r primer set did not recover the RC-II, Methanobacteriaceae, MBD, and E1′ groups. However, this set recovered several crenarchaeal sequences (RC-IV and VI) that made up 16% of these libraries (Fig. 2). The rarefaction analysis of OTUs at 97.5% sequence identity (see Fig. S2 in the supplemental material) indicated that 1Af-ArchLSU47r has a narrow scope and that the 1AF-1492r and 1Af-1100r primers had higher efficiencies. The collective analysis of the libraries showed that the total archaeal diversity is significantly higher than for each primer and far from saturation (see Fig. S2 in the supplemental material).
Additionally, the sequences from groups E1 and Methanosaetaceae showed phylogenetically robust clusters (Fig. 1, underlined bootstrap values) containing three clusters each when using a 97.5% similarity cutoff value. Some phylotypes in cluster E1-c (Fig. 1) did not satisfy the similarity criterion, but because they were represented only by single sequences, they were provisionally classified as part of this single cluster. Similar patterns of clusters were observed when the ITS region, amplified by the 1AF-ArchLSU47 combination, was included in the sequence analysis (not shown). None of the E1 clusters contained a sequence from a cultured organism, whereas only cluster a in Methanosaetaceae contained sequences from cultured Methanosaeta strains (Fig. 1).
CH4 production by peat soil incubated with substrates and T-RFLP analysis.
CH4 production (Fig. 3) values were similar among all treatments over the initial 10 days of incubation, but after 14 days, the acetate and the H2-CO2/rifampin-amended samples produced more CH4. The euryarchaeal community structure of peat samples was analyzed with T-RFLP using the 1Af-1100r primers (Fig. 4). Peak identity was predicted by in silico digestion of the clone libraries, as previously described (15). Methanospirillum was the only member from the E2′ group (15) whose sequences were recovered from MH fen, and thus, Methanospirillaceae was used, instead of E2′, as the identity of the peak. T-RFLP profiles of peat samples before incubation (Fig. 4A and B, panel 1) showed E1 and Methanosaetaceae as the dominant peaks, along with smaller peaks predicted to represent groups such as Methanosarcinaceae, Methanobacteriaceae, E2, Methanospirillaceae, RC-I/SC, and RC-II, in agreement with the clone libraries.
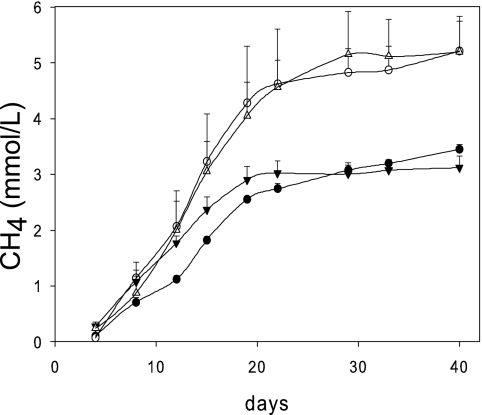
FIG. 3.
CH4 production by peat slurries from MH fen. Samples were nonamended statically grown (white circles) or statically grown and amended with 1 mM acetate (black circles), shaken and amended with rifampin (black triangles), or shaken and amended with H2-CO2 and rifampin (white triangles). Points represent the means ± standard deviations for three samples.
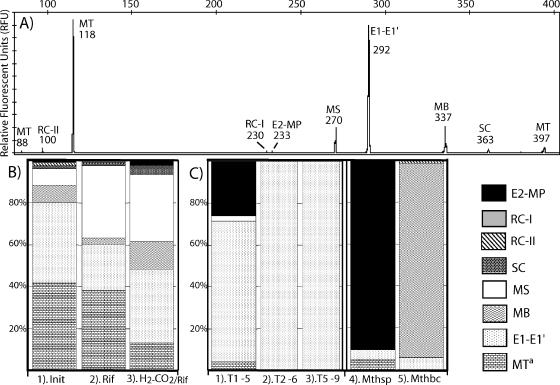
FIG. 4.
T-RFLP analysis of peat samples and enrichments using the 1Af-1100r primer set and HhaI-Sau96 restriction enzymes. T-RFLP profiles (A) were standardized to a total of 100 relative fluorescence units (RFU), and the peaks were matched with their corresponding groups are presented in a single-column format (B and C). Initial (panel 1. Init) or nonincubated samples are grouped with incubated slurries (A and B) amended with rifampin only (panel 2. rif) or H2-CO2/rifampin (panel 3. H2-CO2/rif). Enrichment culture (C) profiling was done with the highest dilution reached for of each E1-targeted culture transfer: transfer 1 dilution, 10−5 (T1-5); transfer 2 dilution, 10−6 (T2-6); transfer 5 dilution, 10−9 (T5-9). Other enrichment attempts that yielded Methanospirillum (Mthsp) and Methanobacterium (Mthbc) and related organisms were also included. MS, Methanosarcinaceae; MT, Methanosaetaceae; SC, subaqueous cluster; RC-I, rice cluster I; RC-II, rice cluster II; E1, group E1; E1′, group E1′; E2, group E2; MP, Methanospirillaceae; MB, Methanobacteriaceae.
Endpoint analysis of peat samples incubated with shaking and amended with rifampin but without H2-CO2 (Fig. 4B, panel 2), showed an increase in the proportion of the peak corresponding to Methanosarcinaceae, with a corresponding reduction in the E1 and Methanobacteriaceae peaks. A smaller increase in the Methanosarcinaceae peak was found in cultures incubated statically without rifampin (data not shown). Meanwhile, in the H2-CO2/rifampin-amended peat, E1 remained as the dominant peak (Fig. 4B, panel 3), while Methanobacteriaceae and Methanosarcinaceae also increased their proportions, and Methanosaetaceae was significantly decreased.
T-RFLP-assisted isolation of a novel methanogen.
Several attempts were made to isolate a member of the E1 group, using the low-ionic-strength medium PM1 (11, 12) adjusted to higher pH (ca. 5.5 to 6) to reflect fen conditions. The initial attempts using fresh or incubated peat and dilution-to-extinction transfers, in PM1 plus H2-CO2, yielded Methanospirillaceae-dominated or Methanospirillaceae-only cultures (data not shown).
In order to target the isolation of a member of the E1 group, a euryarchaeal-specific T-RFLP profiling was used to assess culturing conditions or replicated samples where E1 increased its proportion. The H2-CO2/rifampin-incubated peat (Fig. 4B, panel 3) was used to inoculate replicated sets of PM1 plus H2-CO2 with different buffers (HOMOPIPES or MES), different pH values (4.5 or 5.6), and different incubation temperatures (28 or 34°C). A set of tubes in the treatment with MES, pH 5.6, and 28°C produced CH4 in dilutions as high as 10−5. T-RFLP profiling showed that in these 10−5 tubes, E1 increased its fraction to around 70%, while Methanosarcinaceae and Methanosaetaceae were less than 9%. Methanospirillaceae was also significantly present, making up around 21% of the total profile (Fig. 4C, panel 1). The 10−5 enrichment was transferred to a new round of dilutions, and CH4 production was observed with 10−6 dilutions. T-RFLP analysis of a sample from this dilution indicated that E1 was the only euryarchaeal group detectable by our 1Af-1100r primers (Fig. 4C, panel2). We performed four more dilution rounds, obtaining growth (turbidity) and CH4 production at 10−9 dilutions. T-RFLP analysis indicated that E1 was the only group present in the culture (Fig. 4C, panel 3).
An additional set of PM1 tubes with the same conditions that successfully enriched for E1 were inoculated with fresh peat soil and produced CH4 up to the 10−3 dilution. However, the T-RFLP profiling showed that Methanospirillaceae was the group primarily enriched (Fig. 4C, panel 4). Microscopy observations of this enrichment confirmed the dominance of Methanospirillaceae-like cells (not shown). Other treatments, such as that with HOMOPIPES, pH 4.5, and 34°C incubation, produced CH4 at dilutions only as high as 10−2, and T-RFLP analysis of this dilution indicated a strong enrichment of the Methanobacteriaceae group (Fig. 4C, panel 5). These two treatments were not pursued further, since E1 was not preferentially enriched.
Purity and initial characterization of isolate E1-9c.
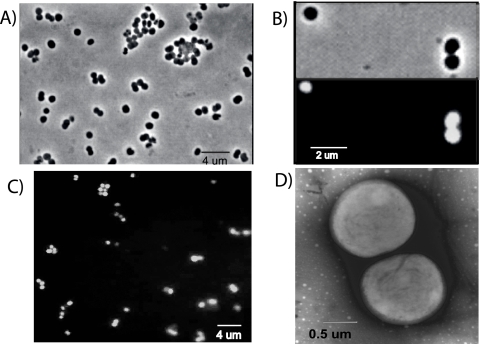
The culture, produced by six sequential dilutions to extinction, was designated strain E1-9c and contained cells with a single morphology (Fig. 5A), and results from several tests indicated its purity. Organic substrates such as yeast extract (0.2 g liter−1), glucose, pyruvate, and lactate (20 mM) were added to the culture in the absence of rifampin to test for heterotrophic contaminants, and no growth or methanogenesis was observed. Attempts at PCR amplification with the universal bacterial 16S rRNA gene primers 27F-1492r were also unsuccessful. FISH with the universal archaeal probe ARCH915 showed hybridization with all the cells in the culture (Fig. 5C) and no hybridization with the bacterial probe EUB 318 (not shown). Additionally, we constructed a clone library with the primers 1AF-ArchLSU47. All 70 clones had the same restriction pattern, and the three sequenced clones had differences of only 3 bases out of 1,784 bases, which was well within the margin of error for PCR amplifications (66). From these observations, the purity of the E1-9c culture was established. The phylogenetic analysis of the full 16S rRNA gene sequence positioned E1-9c in the order Methanomicrobiales, forming a unique cluster closely related to environmental sequences of the group E1-c (Fig. 1). The E1-9c strain's closest described cultured relatives are “Candidatus Methanoregula boonei” and Methanospirillum hungatei (with 93% and 92% identity, respectively).
FIG. 5.
Microscopy examination of E1-9c cells. (A) Phase-contrast microscopy; (B) phase-contrast and fluorescence microscopy showing the autofluorescence of cells illuminated with light near 420 nm; (C) FISH, using the 915-Arch probe; (D) negative stain electron microscopy.
Strain E1-9c cells had a coccoid shape with diameters ranging from 0.5 to 0.8 μm and were often in pairs (Fig. 5). Cells were nonmotile and showed strong blue fluorescence when illuminated with light near 420 nm, indicative of abundant coenzyme factor 420 (F420) in the cells (Fig. 5.B). E1-9c used H2-CO2 as a methanogenic substrate, with a doubling time of ca. 2 days at pH 5.6 and 28°C (standard growth conditions). In addition to mineral nutrients, E1-9c also required a vitamin solution (4), coenzyme M (0.5 mM), and acetate (0.4 mM) as a carbon source in the growth medium.
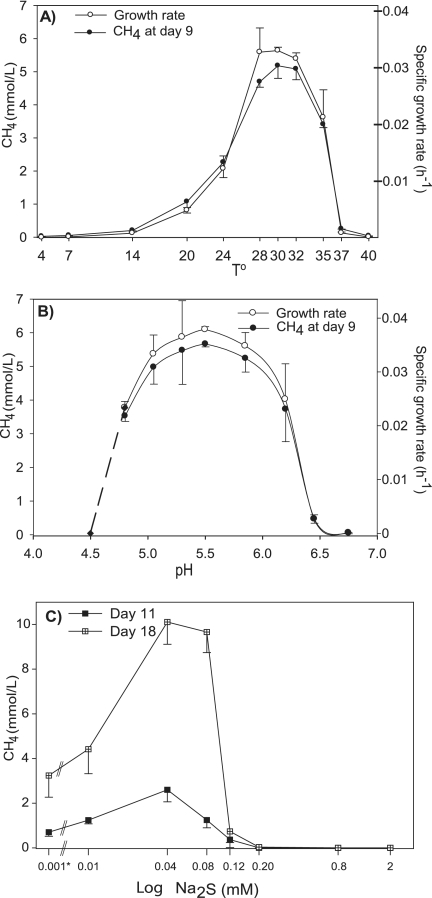
CH4 production by E1-9c was observed from 7°C to 37°C, with an optimum near 30°C (Fig. 6A). CH4 production above half of its maximum rate occurred between 24°C and 35°C, indicating the mesophilic nature of the isolate. E1-9c grew at a pH range of pH 4.8 to 6.5, with an optimum range of 5.3 to 5.5 (Fig. 6B). Growth at pH values lower than 4.8 could not be obtained using MES buffer. We attempted to use HOMOPIPES and citric acid as low-end pH buffers, but no growth was observed even at similar pH values where growth was observed using MES, which suggests some detrimental effects of these buffers on the growth of strain E1-9c.
FIG. 6.
Physiological characterization of E1-9c. Effect of temperature on growth and methanogenesis (A); effect of pH on growth and methanogenesis (B); effect of sulfide (Na2S) on methanogenesis (C). The asterisk represents a set of tubes with H2S gas addition instead of Na2S (as H2S gas was initially used for the isolation of E1-9c). Points represent the means ± standard deviations for three samples.
The effects of H2S or Na2S·9H2O, a common reducing agent and sulfur source in anaerobic growth medium, on the growth of strain E1-9c was examined (Fig. 6C). Very low concentrations of Na2S (0.01 to 0.08 mM) improved the growth of E1-9c compared to that of our standard H2S gas addition (∼0.001 mmol). The optimum Na2S addition was around 0.04 mM, and additions above 0.12 mM completely inhibited the growth of strain E1-9c.
DISCUSSION
MH is a minerotrophic fen where multiyear observations of CH4 flux have shown significant CH4 emissions into the atmosphere, particularly in rainy years (59). Minerotrophic fens such as MH are abundant in the boreal forest biome, particularly in North America (6, 62); nevertheless, the methanogenic communities inhabiting these fens have scarcely been assessed. To better characterize the euryarchaeal community from the MH fen site, multiple clone libraries were constructed using different primer combinations targeting the 16S rRNA gene (Fig. 1 and 2). Individually, each primer combination achieved near sampling saturation but with only a 58 to 26% coverage of all OTUs at 97.5% sequence identity (see Fig. S2 in the supplemental material), suggesting that a multiple primer approach is required for thorough community characterization. The libraries showed a diverse community where the majority of phylotypes are associated with uncultured groups (Fig. 1 and 2). The recovered phylotypes were associated with a total of 11 euryarchaeal and 2 crenarchaeal groups, as follows: Methanosarcinaceae, Methanosaetaceae, SC, RC-I, RC-II, group E2, group E1, Methanospirillaceae, group E1′, Methanobacteriaceae, MBD, RC-IV, and RC-VI (Fig. 1 and 2). Although this study had a more extensive number of screened clones (352 total in Fig. 2) and used different primers from other studies, the euryarchaeal composition in the fen had a diversity that was similar or greater (see Fig. S3 in the supplemental material) than that in some other bogs, oligotrophic fens, or mesotrophic fens (5, 21, 42, 54).
In terms of primers, the 1Af-1100r combination (30) amplified all euryarchaeal groups detected by the different mixes, making this primer set optimal for the euryarchaeal coverage in MH fen (Fig. 2). The 1Af-1492r and 1Af-ArchLSU47 primers missed sequences of ca. five minor groups each, and the 1Af-1492r mixture was the only one that amplified crenarchaeal sequences in addition to euryarchaeal ones (Fig. 2 and see Fig. S1 in the supplemental material). The 1AF-ArchLSU47 primer mixture allowed the recovery of full 16S and ITS rRNA gene sequences from many euryarchaeal groups in MH fen, although RC-I sequences were not recovered. rRNA gene sequences from the RC-I genome (19) had perfect matches with the 1Af-ArchLSU47 primers, but the size of the ITS regions, 582 bases in two and 344 bases in one (19), would lead to fragments larger than 2 kb, which are less efficient to amplify or clone with standard techniques. Fragment size could represent a limiting factor in the coverage of the 1Af-ArchLSU47 primers. Nevertheless, the retrieval of a large fragment of the rRNA operon (∼1.75 kb, average size) allowed the first examination of the variability (size and sequence) of the ITS regions of uncultured Euryarchaea, including the presence or absence of tRNA (see Table S1 in the supplemental material), in addition to providing full 16S rRNA sequences for phylogenetic analysis and primer or probe design to study novel groups such as the SC, E1, or E1′ group. A previous study has examined the ITS variability of uncultured Archaea, and it examined marine Crenarchaea (24).
Multiple phylotypes associated with several uncultured euryarchaeal groups were also recovered from MH fen. Groups such as RC-I, RC-II, RC-IV, RC-VI (30), and MBD (63) have also been found in bogs (5, 15, 42, 58), poor fens (22, 36), and lake sediments (25, 37). The abundance of these groups varies in peatlands, from modest to significant fractions in clone libraries, but information about their role or physiology is limited by the lack of cultures. A member of the RC-I group was recently isolated (56), confirming the group as an H2-CO2-utilizing methanogen, as observed with previous enrichments (58). The RC-I group is particularly abundant in rice paddies, where it can make up 20 to 50% of the total number of methanogens (43); however, in the MH fen, this group represented only 2 to 14% of the clone libraries. RC-I has been suggested to be well adapted to low H2 habitats (56) and relatively oxic conditions (19) such as the rice rhizosphere where RC-I phylotypes have been commonly found (29). RC-I could play a role in the rhizosphere of sedges in minerotrophic fens; however, this would require further investigation, since our study focused on bulk peat.
Groups E1′ and SC have recently been identified in peatlands (15), and some associated phylotypes have been observed for other ecosystems. Group E1′, which belongs to the order Methanomicrobiales, has until now been found only in forested and acidic peatlands in New York State (5, 15). At MH fen, E1′ made up only 5 to 8% of the clones, suggesting a small population size, which could make its detection difficult with low coverage libraries. In contrast, phylotypes related to the SC group have been found in lake sediments, contaminated soils, and nitrate-rich canal sediments (38, 51, 61). Interestingly, a sequence affiliated with SC (Fig. 1, Arch-D) was shown to be the unique archaeal partner in an enrichment from nitrate-rich sediments, performing anaerobic oxidation of methane coupled to denitrification (51). Whether members of the SC group play a role in anaerobic oxidation of methane in freshwater sites like the minerotrophic MH fen is unknown.
CH4 production among H2-CO2- or acetate-amended and nonamended samples (stimulated and endogenous methanogenesis, respectively) was similar in the initial days of incubation (Fig. 3), suggesting that both aceticlastic and hydrogenotrophic methanogens were active and not substrate limited in unamended peat slurries, whether grown shaken or static. Although incubation of peat slurries provides homogeneous conditions and reduces environmental variables, the processing of the samples by diluting the peat soil and shaking the incubations (18) can perturb them (sometimes called the “vial effect”), often releasing substrates or disrupting syntrophic interactions. The shift in nonamended samples toward members of Methanosarcinaceae, metabolically versatile and relatively fast-growing methanogens (72), is consistent with greater substrate availability and suggests that the endogenous rates of methanogenesis in these samples are overestimates of those in situ.
Several studies have reported that hydrogenotrophic methanogenesis is dominant in bogs, while aceticlastic methanogenesis is rated from important to dominant in minerotrophic fens (2, 3, 15, 16, 20, 31, 39, 50). In minerotrophic fens, the relative contribution of hydrogenotrophic methanogenesis has been found to range from 30 to 55% (20, 39). Thus, both aceticlastic and hydrogenotrophic methanogenesis can be significantly important in minerotrophic fens such as MH. The importance of both methanogenic pathways in MH fen is supported by the abundance of the two dominant members of the euryarchaeal community, Methanosaetaceae, presumably aceticlastic methanogens, and E1, presumably hydrogenotrophic methanogens (Fig. 1 and 4).
All of the Methanosaeta isolates described are known to use only acetate for CH4 production (23) and have a lower minimum threshold for acetate (5 to 70 μM) than the other known aceticlastic methanogenic genus Methanosarcina (0.2 to 1.2 mM) (34). Methanosaeta can outcompete Methanosarcina in sites with low acetate concentrations such as some minerotrophic fens (20), which is in agreement with the clone library and T-RFLP results observed for MH fen. MT was significantly more abundant than Methanosarcinaceae in the amplified portion of the archaeal community (Fig. 2 and 4), and this has also been observed for a Finnish fen (20). The Methanosaeta clusters observed in this study (with a 97.5% similarity threshold) were similarly abundant in the different libraries, but only one cluster (Methanosaetaceae a in Fig. 1 and 2) had associated isolates.
The E1 group was recently identified by phylogenetic analysis and a common terminal restriction site in our T-RFLP analyses (15) and represented a minor fraction of the methanogenic community in nearby bogs. The results of this study suggest that this group can be numerically significant and diverse in the MH fen, as indicated by its abundance in clone libraries, T-RFLP profiles, and the existence of several sub-clusters when applying a 97.5% similarity cutoff to clone libraries (Fig. 1, 2, and 4). In addition, sequences phylogenetically associated with E1 have also been detected in bogs, tundra wetland soil, anaerobic bioreactors, and landfill sites (see Fig. S1 in the supplemental material), suggesting a broad ecological and geographical distribution of its members.
Group E1 belongs to the order Methanomicrobiales and does not have a reported isolated representative, and its closest cultured relatives are the hydrogenotrophic methanogens Methanospirillum hungatei and “Candidatus Methanoregula boonei” (Fig. 1). E1 was likely to be made up of hydrogenotrophic methanogens, as indicated by the increase in the E1 peak in T-RFLP traces from H2-CO2-amended peat slurries (Fig. 4B, panel 3). However, Methanospirillaceae spp., a group of relatively fast-growing H2-CO2-utilizing methanogens that were poorly represented in the MH fen clone libraries, commonly outgrew E1 in MH fen enrichments. Fortunately, the use of T-RFLP analysis made it possible to overcome this interference by identifying a sample, among several treatments, where a member of the E1 group was preferentially enriched (Fig. 4C, panel 1). Several dilutions to extinction were subsequently performed, and the selective enrichment of E1 was verified by T-RFLP (Fig. 4C). Strain E1-9c was obtained by this process, and the purity of the isolate was tested and confirmed by several analyses (Fig. 5).
E1-9c is a novel isolate in the Methanomicrobiales order. It is associated with the E1 group and is closest to members of the E1-c subgroup, with 95% similarity (Fig. 1). Group E1-c made up a small fraction in our libraries (Fig. 2), but given that some sequences within this group showed a similarity lower than 97.5%, this group could contain other clusters which could be resolved as more sequences become available in the future. In addition, very recently the isolation of strain NOBI-1 was reported (56). The NOBI-1 16S rRNA gene sequence has 94% identity with that of E1-9c, and the predicted terminal restriction pattern for its rRNA gene places NOBI-1 in the E1 group, although its relationship with E1-9c and other members of E1 is unclear because of poor resolution in the part of the tree where it is located (Fig. 1). In terms of uncultured phylotypes, the closest relatives to E1-9c were environmental sequences recovered from anaerobic bioreactors and landfills (Fig. 1 and see Fig. S1 in the supplemental material).
Strain E1-9c is a mesophilic and mildly acidophilic methanogen (optimum growth at 30°C and pH 5.5). Interestingly, this strain required the presence of H2S or Na2S but at concentrations below 0.1 mM (Fig. 6C), which was far lower than the 1 to 2 mM commonly added as a reducing agent in growth media for the culture of many other methanogens (40, 45, 60). H2S and Na2S have shown toxicity in anaerobic cultures (14, 46), with a wide variability among methanogenic strains in terms of optimal and inhibitory concentrations (33). Na2S toxicity in an anaerobic medium can be overcome by the addition of alternative reducing agents such as titanium(III) citrate or ferrous sulfide (14, 35) and minimal amounts of Na2S to satisfy the sulfur requirements for growth, reported to be 0.02 to 0.03 mM for some methanogenic strains (52). “Candidatus Methanoregula boonei” strain 6A8, a close relative of E1-9c, is also inhibited by sulfide (12), and it is likely that high sulfide concentrations in many standard growth media for methanogens are inhibitory to some presently uncultured methanogens. Based on its phylogenetic position, morphology, and physiology, we suggest that strain E1-9c (ATCC no. BAA1556; DSM no. 19958) is a member of a novel genus in the order Methanomicrobiales and propose the name “Candidatus Methanosphaerula palustris” (H. Cadillo-Quiroz, J. B. Yavitt, and S. H. Zinder, submitted for publication).
Supplementary Material
Acknowledgments
We thank R. Ward for help with electron microscopy, S. Bräuer and K. Kota for help with culturing efforts, J. P. Euzéby for assistance with the taxonomy and name derivation for group E1-9c, and L. Salzberg for useful comments on the manuscript.
This work was supported by an NSF Microbial Observatories program grant (0132049).
Footnotes
Published ahead of print on 15 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, G. B., R. D. Shannon, J. R. White, C. S. Martens, and M. J. Alperin. 1999. Effect of seasonal changes in the pathways of methanogenesis on the delta C-13 values of pore water methane in a Michigan peatland. Global Biogeochem. Cycles 13:475-484. [Google Scholar]
- 3.Avery, G. B., R. D. Shannon, J. R. White, C. S. Martens, and M. J. Alperin. 2003. Controls on methane production in a tidal freshwater estuary and a peatland: methane production via acetate fermentation and CO2 reduction. Biogeochemistry 62:19-37. [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Mol. Biol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basiliko, N., J. B. Yavitt, P. M. Dees, and S. Merkel. 2003. Methane biogeochemistry and methanogen communities in two Northern peatland ecosystems, New York State. Geomicrobiol. J. 20:563-577. [Google Scholar]
- 6.Bedford, B., and K. Godwin. 2003. Fens of the United States: distribution, characteristics, and scientific connection versus legal isolation. Wetlands 23:608-629. [Google Scholar]
- 7.Bergman, I., B. H. Svensson, and M. Nilsson. 1998. Regulation of methane production in a Swedish acid mire by pH, temperature and substrate. Soil Biol. Biochem. 30:729-741. [Google Scholar]
- 8.Bernard, J. M., and J. G. Macdonald. 1974. Primary production and life history of Carex lacustris. Can. J. Bot. 52:117-123. [Google Scholar]
- 9.Blodau, C. 2002. Carbon cycling in peatlands: a review of processes and controls. Environ. Rev. 10:111-134. [Google Scholar]
- 10.Bräuer, S., J. B. Yavitt, and S. H. Zinder. 2004. Methanogenesis in McLean bog, an acidic peat bog in upstate New York: stimulation by H2/CO2 in the presence of rifampicin, or by low concentrations of acetate. Geomicrobiol. J. 21:433-443. [Google Scholar]
- 11.Bräuer, S. L., H. Cadillo-Quiroz, E. Yashiro, J. B. Yavitt, and S. H. Zinder. 2006. Isolation of a novel acidophilic methanogen from an acidic peat bog. Nature 442:192-194. [DOI] [PubMed] [Google Scholar]
- 12.Bräuer, S. L., E. Yashiro, N. G. Ueno, J. B. Yavitt, and S. H. Zinder. 2006. Characterization of acid-tolerant H2/CO2-utilizing methanogenic enrichment cultures from an acidic peat bog in New York State. FEMS Microbiol. Ecol. 57:206-216. [DOI] [PubMed] [Google Scholar]
- 13.Bridgham, S. D., K. Updegraff, and J. Pastor. 1998. Carbon, nitrogen, and phosphorous mineralization in Northern wetlands. Ecology 79:1545-1561. [Google Scholar]
- 14.Brock, T. D., and K. O'Dea. 1977. Amorphous ferrous sulfide as a reducing agent for culture of anaerobes. Appl. Environ. Microbiol. 33:254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadillo-Quiroz, H., S. Bräuer, E. Yashiro, C. Sun, J. Yavitt, and S. Zinder. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ. Microbiol. 8:1428-1440. [DOI] [PubMed] [Google Scholar]
- 16.Chasar, L. S., J. P. Chanton, P. H. Glaser, and D. I. Siegel. 2000. Methane concentration and stable isotope distribution as evidence of rhizospheric processes: comparison of a fen and bog in the glacial lake Agassiz peatland complex. Ann. Bot. 86:655-663. [Google Scholar]
- 17.Cicerone, R. J., and R. S. Oremland. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 2:299-327. [Google Scholar]
- 18.Dannenberg, S., J. Wudler, and R. Conrad. 1997. Agitation of anoxic paddy soil slurries affects the performance of the methanogenic microbial community. FEMS Microbiol. Ecol. 22:257-263. [Google Scholar]
- 19.Erkel, C., M. Kube, R. Reinhardt, and W. Liesack. 2006. Genome of rice cluster I Archaea: the key methane producers in the rice rhizosphere. Science 313:370-372. [DOI] [PubMed] [Google Scholar]
- 20.Galand, P. E., H. Fritze, R. Conrad, and K. Yrjala. 2005. Pathways for methanogenesis and diversity of methanogenic Archaea in three boreal peatland ecosystems. Appl. Environ. Microbiol. 71:2195-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galand, P. E., H. Fritze, and K. Yrjala. 2003. Microsite-dependent changes in methanogenic populations in a boreal oligotrophic fen. Environ. Microbiol. 5:1133-1143. [DOI] [PubMed] [Google Scholar]
- 22.Galand, P. E., S. Saarnio, H. Fritze, and K. Yrjala. 2002. Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42:441-449. [DOI] [PubMed] [Google Scholar]
- 23.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Martinez, J., and F. Rodriguez-Valera. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of group I. Mol. Ecol. 9:935-948. [DOI] [PubMed] [Google Scholar]
- 25.Glissman, K., J. Chin, P. Casper, and R. Conrad. 2004. Methanogenic pathway and archaeal community structure in the sediment of eutrophic lake Dagow: effect of temperature. Microb. Ecol. 48:389-399. [DOI] [PubMed] [Google Scholar]
- 26.Gore, A. J. P. 1983. Ecosystems of the world 4B. Mires: swamp, bog, fen, and moor. Regional studies. Elsevier Scientific Publishing, Amsterdam, The Netherlands.
- 27.Gorham, E., and J. A. Jannsens. 1992. Concepts of fen and bog re-examined in relation to bryophyte cover and the acidity of surface waters. Acta Soc. Bot. Pol. 61:7-20. [Google Scholar]
- 28.Gorham, E. 1991. Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1:182-195. [DOI] [PubMed] [Google Scholar]
- 29.Grobkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 33.Jarrel, K. F., and M. L. Kalmokoff. 1988. Nutritional requirements of the methanogenic archaebacteria. Can. J. Microbiol. 34:557-576. [Google Scholar]
- 34.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Lett. 88:181-198. [Google Scholar]
- 35.Jones, G. A., and M. D. Pickard. 1980. Effect of titanium(III) citrate as reducing agent on growth of rumen bacteria. Appl. Environ. Microbiol. 39:1144-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juottonen, H., P. E. Galand, E. S. Tuittila, J. Laine, H. Fritze, and K. Yrjala. 2005. Methanogen communities and Bacteria along an ecohydrological gradient in a northern raised bog complex. Environ. Microbiol. 7:1547-1557. [DOI] [PubMed] [Google Scholar]
- 37.Jurgens, G., F. O. Glöckner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Münster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 38.Kasai, Y., Y. Takahata, T. Hoaki, and K. Watanabe. 2005. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ. Microbiol. 7:806-818. [DOI] [PubMed] [Google Scholar]
- 39.Keller, J. K., and S. D. Bridgham. 2007. Pathways of anaerobic carbon cycling across an ombrotrophic-minerotrophic peatland gradient. Limnol. Oceanogr. 52:96-107. [Google Scholar]
- 40.Kendall, M. M., Y. Liu, M. Sieprawska-Lupa, K. O. Stetter, W. B. Whitman, and D. R. Boone. 2006. Methanococcus aeolicus sp. nov., a mesophilic, methanogenic archaeon from shallow and deep marine sediments. Int. J. Syst. Evol. Microbiol. 56:1525-1529. [DOI] [PubMed] [Google Scholar]
- 41.Kotsyurbenko, O. R., M. W. Friedrich, M. V. Simankova, A. N. Nozhevnikova, P. N. Golyshin, K. N. Timmis, and R. Conrad. 2007. Shift from acetoclastic to H2-dependent methanogenesis in a West Siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl. Environ. Microbiol. 73:2344-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotsyurbenko, O. R., K. J. Chin, M. V. Glagolev, S. Stubner, M. V. Simankova, A. N. Nozhevnikova, and R. Conrad. 2004. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6:1159-1173. [DOI] [PubMed] [Google Scholar]
- 43.Kruger, M., P. Frenzel, D. Kemnitz, and R. Conrad. 2005. Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51:323-331. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma, K., X. Liu, and X. Dong. 2005. Methanobacterium beijingense sp. nov., a novel methanogen isolated from anaerobic digesters. Int. J. Syst. Evol. Microbiol. 55:325-329. [DOI] [PubMed] [Google Scholar]
- 46.Mariotto, C., P. Loubière, G. Goma, and N. D. Lindley. 1989. Influence of various reducing agents on methylotrophic growth and organic acid production of Eubacterium limosum. Appl. Microbiol. Biol. 32:193-198. [Google Scholar]
- 47.Merila, P., P. E. Galand, H. Fritze, E. S. Tuittila, K. Kukko-oja, J. Laine, and K. Yrjala. 2006. Methanogen communities along a primary succession transect of mire ecosystems. FEMS Microbiol. Ecol. 55:221-229. [DOI] [PubMed] [Google Scholar]
- 48.Moench, T. T., and J. G. Zeikus. 1983. An improved preparation method for a titanium (III) media reductant. J. Microbiol. Methods 1:199-202. [Google Scholar]
- 49.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 50.Popp, T. J., J. P. Chanton, G. J. Whiting, and N. Grant. 1999. Methane stable isotope distribution at a Carex dominated fen in North Central Alberta. Global Biogeochem. Cycles 13:1063-1077. [Google Scholar]
- 51.Raghoebarsing, A. A., A. Pol, K. van de Pas-Schoonen, A. J. P. Smolders, K. F. Ettwig, W. I. Rijpstra, S. Schouten, J. S. S. Damste, H. J. M. Op den Camp, M. S. M. Jetten, and M. Strous. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918-921. [DOI] [PubMed] [Google Scholar]
- 52.Rajagopal, B. S., and L. Daniels. 1986. Investigation of mercaptans, organic sulfides, and inorganic sulfur compounds as sulfur sources for the growth of methanogenic bacteria. Curr. Microbiol. 14:137-144. [Google Scholar]
- 53.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 54.Rooney-Varga, J. N., M. W. Giewat, K. N. Duddleston, J. P. Chanton, and M. E. Hines. 2007. Links between archaeal community structure, vegetation type and methanogenic pathway in Alaskan peatlands. FEMS Microbiol. Ecol. 60:240-251. [DOI] [PubMed] [Google Scholar]
- 55.Roulet, N., T. Moore, J. Bubier, and P. Lafleur. 1992. Northern fens: methane flux and climatic change. Tellus B 44:100-105. [Google Scholar]
- 56.Sakai, S., H. Imachi, Y. Sekiguchi, A. Ohashi, H. Harada, and Y. Kamagata. 2007. Isolation of key methanogens for global methane emission from rice paddy field: a novel isolate affiliated with the clone cluster rice cluster I. Appl. Environ. Microbiol. 73:4326-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 59.Smemo, K., and J. B. Yavitt. 2006. A multi-year perspective on methane cycling in a shallow peat fen in central New York State, USA. Wetlands 26:20-29. [Google Scholar]
- 60.Sowers, K. R., and K. M. Noll. 1995. Techniques for anaerobic growth, p. 15-48. In F. T. Robb, A. R. Place, K. R. Sowers, H. J. Schreier, S. DasSarma, and E. M. Fleischmann (ed.), Archaea: a laboratory manual. Methanogens. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 61.Stein, L. Y., G. Jones, B. Alexander, K. Elmund, C. Wright-Jones, and K. H. Nealson. 2002. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol. 42:431-440. [DOI] [PubMed] [Google Scholar]
- 62.Turetsky, M. R., and S. Ripley. 2005. Decomposition in extreme-rich fens of boreal Alberta, Canada. Soil Sci. Soc. Am. J. 69:1856-1860. [Google Scholar]
- 63.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vitt, D. H., and W. L. Chee. 1990. The relationships of vegetation to surface water chemistry and peat chemistry in fens of Alberta, Canada. Plant Ecol. 89:87-106. [Google Scholar]
- 65.Vitt, D. H., L. A. Halsey, I. E. Bauer, and C. Campbell. 2000. Spatial and temporal trends in carbon storage of peatlands of continental western Canada through the Holocene. Can. J. Earth Sci. 37:683-693. [Google Scholar]
- 66.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 67.Wahlen, M. 1993. The global methane cycle. Annu. Rev. Earth Planet. Sci. 21:407-426. [Google Scholar]
- 68.Wang, Z., D. Zeng, and W. H. Patrick. 1996. Methane emissions from natural wetlands. Environ. Monit. Assess. 42:143-161. [DOI] [PubMed] [Google Scholar]
- 69.Williams, R. T., and R. L. Crawford. 1985. Methanogenic bacteria, including an acid-tolerant strain, from peatlands. Appl. Environ. Microbiol. 50:1542-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yavitt, J., G. E. Lang, and A. J. Sextone. 1990. Methane fluxes in wetland and forest soils, beaver ponds, and low-order streams of a temperate forest ecosystem. J. Geophys. Res. 95:22463-22474. [Google Scholar]
- 71.Yavitt, J. B., and A. Knapp. 1995. Methane emission to the atmosphere through emergent cattail (Typha latifolia L.) plants. Tellus B 47: 521-534. [Google Scholar]
- 72.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 536. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapmann and Hall, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.