Abstract
Bioethanol produced by microbial fermentations of plant biomass hydrolysates consisting of hexose and pentose mixtures is an excellent alternative to fossil transportation fuels. However, the yeast Saccharomyces cerevisiae, commonly used in bioethanol production, can utilize pentose sugars like l-arabinose or d-xylose only after heterologous expression of corresponding metabolic pathways from other organisms. Here we report the improvement of a bacterial l-arabinose utilization pathway consisting of l-arabinose isomerase from Bacillus subtilis and l-ribulokinase and l-ribulose-5-P 4-epimerase from Escherichia coli after expression of the corresponding genes in S. cerevisiae. l-Arabinose isomerase from B. subtilis turned out to be the limiting step for growth on l-arabinose as the sole carbon source. The corresponding enzyme could be effectively replaced by the enzyme from Bacillus licheniformis, leading to a considerably decreased lag phase. Subsequently, the codon usage of all the genes involved in the l-arabinose pathway was adapted to that of the highly expressed genes encoding glycolytic enzymes in S. cerevisiae. Yeast transformants expressing the codon-optimized genes showed strongly improved l-arabinose conversion rates. With this rational approach, the ethanol production rate from l-arabinose could be increased more than 2.5-fold from 0.014 g ethanol h−1 (g dry weight)−1 to 0.036 g ethanol h−1 (g dry weight)−1 and the ethanol yield could be increased from 0.24 g ethanol (g consumed l-arabinose)−1 to 0.39 g ethanol (g consumed l-arabinose)−1. These improvements make up a new starting point for the construction of more-efficient industrial l-arabinose-fermenting yeast strains by evolutionary engineering.
Decreasing fossil energy resources and the climate change caused by emissions of CO2 from their burning have led to a growing interest in renewable-energy alternatives. Bioethanol produced by microbial fermentations of plant biomass is an excellent alternative to fossil fuels. However, for economical ethanol production, it is not enough to convert only the starch and sucrose fractions of plant biomass as is mainly done in conventional ethanol plants. The importance of the conversion of the lignocellulosic fraction as well becomes more and more evident. Lignocellulosic hydrolysates consist of easily fermentable hexose sugars but also significant amounts of pentose sugars. Depending on the choice of raw material, the amounts of pentoses found in lignocellulosic hydrolysates range from, for example, 16% xylan and 5% arabinan in grass and 15% arabinan and 19% xylan in wheat bran (14). These numbers clearly indicate that both hexoses and pentoses must be fermented in an efficient ethanol production process. Even small increases in substrate utilization should significantly improve the overall process costs (28).
Although Saccharomyces cerevisiae is the organism most widely used for ethanol production and is able to convert hexoses rapidly and with high ethanol yields, wild-type S. cerevisiae strains are not able to ferment pentose sugars, such as d-xylose and l-arabinose, efficiently. Even though xylose can be slowly metabolized, at least by adapted strains (2), the second most abundant pentose, l-arabinose, cannot be converted at all. Several different genetic-engineering approaches have been used in attempts to enable d-xylose and l-arabinose fermentation in yeast. The most promising approaches for efficient xylose fermentation were the functional expression of a fungal xylose isomerase from Piromyces sp. strain E2 (18, 19) or a combination of xylose reductase and xylitol dehydrogenase from the yeast Pichia stipitis (10, 24, 27).
Even though several yeasts and fungi can utilize L-arabinose as a carbon and energy source, most of them are not able to ferment it into ethanol (8, 17, 20). l-Arabinose utilization in S. cerevisiae has been achieved by expressing fungal or bacterial pathways. However, even though genes for a fungal l-arabinose pathway have been expressed and the resulting strain could grow on l-arabinose medium, it produced only trace amounts of ethanol from this substrate (21). The lack of ethanol production was attributed to a proposed low l-arabinose uptake rate in S. cerevisiae and a cofactor imbalance, which results from the utilization of NADPH in two reduction reactions and the generation of NADH in the oxidation reactions (21).
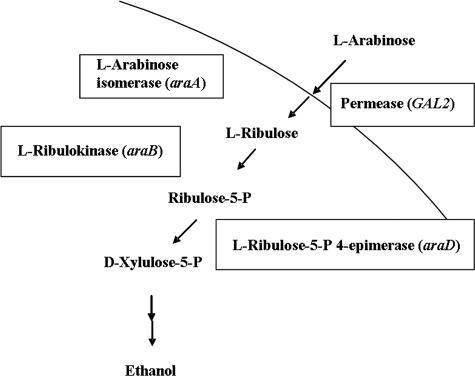
In contrast, the bacterial pathway for l-arabinose utilization does not employ NADPH/NADH-dependent redox reactions and consequently does not cause a cofactor imbalance. Indeed, l-arabinose fermentation in S. cerevisiae could be demonstrated after introducing a bacterial l-arabinose utilization pathway (5) (Fig. 1). There, the araB and araD genes from Escherichia coli, encoding l-ribulokinase and l-ribulose-5-P 4-epimerase, respectively, and the araA gene from Bacillus subtilis, encoding l-arabinose isomerase, have been expressed in a yeast strain, converting l-arabinose into xylulose-5-P as an intermediate of the endogenous pentose-phosphate pathway. Nevertheless, although all enzymes tested as functional, the strain could not grow on l-arabinose medium. Only after prolonged evolutionary engineering was an S. cerevisiae strain obtained which was able to grow on l-arabinose medium with a doubling time of 7.9 h and with an ethanol production rate of 0.06 to 0.08 g ethanol h−1 (g dry weight)−1 under oxygen-limited conditions (5). Molecular analysis of this strain revealed that a crucial prerequisite for efficient l-arabinose utilization was a lowered activity of the l-ribulokinase and an enhanced activity of the endogenous transaldolase enzyme (5).
FIG. 1.
Scheme of the l-arabinose fermentation pathway engineered in S. cerevisiae. l-Arabinose is transported into the cell by the galactose permease Gal2 and subsequently converted into d-xylulose-5-P via three enzymatic steps. d-Xylulose-5-P is a common intermediate in the metabolic pathways of yeast. See the text for details. The figure is adapted from reference 5.
Recently another recombinant yeast strain has been described that produces high ethanol yields from l-arabinose and can even grow with l-arabinose under anaerobic conditions (31). Via expression of the araA, araB, and araD genes from Lactobacillus plantarum, the overexpression of the genes encoding the enzymes of the nonoxidative pentose-phosphate pathway, a GRE3 deletion, and a long-term selection on l-arabinose medium as the sole carbon source, a strain was constructed which exhibited high rates of l-arabinose consumption (0.70 g h−1 [g dry weight]−1) and of ethanol production (0.29 g h−1 [g dry weight]−1) and a high ethanol yield (0.43 g g−1) (31). Nevertheless, the increase in the growth rate under anaerobic evolutionary selection conditions finally reached a plateau and could not further be optimized to those values obtained for glucose and xylose fermentations, indicating an unknown constraint on l-arabinose consumption.
In this article, we report new efforts to rationally improve l-arabinose fermentation in S. cerevisiae. In our approach, we were able to elucidate B. subtilis l-arabinose isomerase as a limiting step in the l-arabinose utilization pathway. When it was replaced with the corresponding enzyme from Bacillus licheniformis, ethanol production from l-arabinose could be improved. Furthermore, we adapted all genes of the l-arabinose utilization pathway to the codon usage of the highly expressed glycolytic genes of S. cerevisiae. With this approach, we could further accelerate growth on l-arabinose as well as ethanol production and ethanol yield. Importantly, our systematic approach makes transfer of these traits into industrial yeast strains and applications feasible. Moreover, it may provide an improved starting point for evolutionary engineering of anaerobic l-arabinose metabolism.
MATERIALS AND METHODS
Strains and media.
Yeast strains and plasmids used in this work are listed in Table 1. Strain BWY1 was obtained from JBY25 (5) by selecting for improved growth on a pure l-arabinose medium for 17 further subcultivations under oxygen-limiting conditions, essentially as described by Becker and Boles (5). In aerobic batch cultivations, S. cerevisiae was grown in yeast nitrogen base medium (YNB) (6.7 g liter−1 Difco yeast nitrogen base without amino acids) supplemented with 20 g liter−1 glucose or 20 g liter−1 l-arabinose as carbon sources and buffered at pH 5.5 with 20 mM KH2PO4. When the sugar concentration in the medium exceeded 20 g liter−1, the concentration of YNB was doubled. For anaerobic fermentations, defined medium (15) was used, supplemented with 30 g liter−1 l-arabinose and 150 μl liter−1 of silicone antifoam (Sigma), as well as with the anaerobic growth factors ergosterol (0.01 g liter−1) and Tween 80 (0.40 g liter−1) dissolved in ethanol (resulting in 2.5 g liter−1 ethanol in the medium).
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| S. cerevisiae strain or plasmid | Relevant genotype or phenotype | Reference |
|---|---|---|
| Strains | ||
| BWY1 | MATaleu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC; unknown beneficial mutations for l-arabinose utilization | This work |
| BWY1-4M | BWY1 with plasmids YEparaA, YEparaBG361A, YEparaD, and YEpGAL2 | This work |
| BWY1-4S | BWY1 with plasmids YEparaAsynth, YEparaBsynth, YEparaDsynth, and YEpGAL2 | This work |
| Plasmids | ||
| YEparaA | pHXT7-AraA-tCYC1, HIS3 | 5 |
| YEpGAL2 | pHXT7-GAL2-tCYC, URA3 | 5 |
| YEparaB | pHXT7-AraB-tCYC1, TRP1 | 5 |
| YEparaD | pHXT7-AraD-tCYC1, LEU2 | 5 |
| YEparaAsynth | pHXT7-codon-optimized AraAB. liche-tCYC1, HIS3 | This work |
| YEparaBsynth | pHXT7-codon-optimized AraB-tCYC1, TRP1 | This work |
| YEparaDsynth | pHXT7-codon-optimized AraD-tCYC1, LEU2 | This work |
| p423H7 | HIS3 | 13 |
| p424H7 | TRP1 | 13 |
| p425H7 | LEU2 | 13 |
| p426H7 | URA3 | 13 |
| YEparaAB. liche | pHXT7-AraA from B. licheniformis tCYC1, HIS3 | This work |
| YEparaAC. aceto | pHXT7-AraA from C. acetobutylicum tCYC1, HIS3 | This work |
| YEparaAP. pento | pHXT7-AraA from P. pentosaceus tCYC1, HIS3 | This work |
| YEparaAL. planta | pHXT7-AraA from L. plantarum tCYC1, HIS3 | This work |
| YEparaAL. mesent | pHXT7-AraA from L. mesenteroides tCYC1, HIS3 | This work |
| YEparaAB. liche-6HIS | pHXT7-AraA from B. licheniformis tCYC1, HIS3; His6 epitope | This work |
| YEparaAC. aceto-6HIS | pHXT7-AraA from C. acetobutylicum tCYC1, HIS3; His6 epitope | This work |
| YEparaAB. subtilis-6HIS | pHXT7-AraA from B. subtilis tCYC1, HIS3; His6 epitope | This work |
Plasmids were amplified in Escherichia coli strain DH5α (Gibco BRL, Gaithersburg, MD) and strain SURE (Stratagene, La Jolla, CA). E. coli transformations were performed via electroporation according to the methods of Dower et al. (9) and Wirth (30). E. coli was grown on LB (Luria-Bertani) medium with 40 μg ml−1 ampicillin for plasmid selection.
Plasmid construction.
The coding regions of araA from B. licheniformis, Clostridium acetobutylicum, P. pentosaceus, L. plantarum, and Leuconostic mesenteroides were amplified by whole-cell PCR from strains DSM 13, DSM 792, DSM 20336, DSM 20174, and DSM 20343 (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany), respectively, and cloned into the linearized vector p423H7 (HIS3) by recombination cloning, employing the methods described by Wieczorke et al. (29) and omitting the six histidine codons.
Furthermore, araA genes from B. licheniformis, C. acetobutylicum, and B. subtilis were also cloned by recombination cloning into the vector p423H7, fusing six histidine codons at their 3-terminal ends. Molecular techniques were performed according to published procedures (22). Yeast transformations and reisolation of plasmid DNA from yeast cells were carried out as described previously (6, 11).
Codon-optimized gene versions were obtained from Sloning BioTechnology (Puchheim, Germany) by changing the original codons of the respective genes to those used in the genes encoding glycolytic enzymes in S. cerevisiae (Table 2). The coding regions of E. coli araB and araD and B. licheniformis araA were synthesized with the optimized codon sequence and cloned into the vectors p424H7, p425H7, and p423H7, respectively, by recombination cloning as described above. The gene araB from E. coli was synthesized with the Asp→Asn mutation on position 121, which was found in the reisolated araB coding sequence in the work of Becker and Boles (5).
TABLE 2.
Preferred codon usage of glycolytic genes in S. cerevisiae
| Amino acid | Codon usage of codon-optimized genes |
|---|---|
| Ala | GCT |
| Arg | AGA |
| Asn | AAC |
| Asp | GAC |
| Cys | TGT |
| Gln | CAA |
| Glu | GAA |
| Gly | GGT |
| His | CAC |
| Ile | ATT |
| Leu | TTG |
| Lys | AAG |
| Met | ATG |
| Phe | TTC |
| Pro | CCA |
| Ser | TCT |
| Thr | ACC |
| Trp | TGG |
| Tyr | TAC |
| Val | GTT, (GTC)a |
| End | TAA |
GTC was used only twice: synthetic isomerase, position 384; synthetic kinase, position 452.
Growth assays.
Cultures (20 ml) were grown in 200-ml shake flasks (Erlenmeyer flasks) at 30°C in a shaker. Precultures were grown to stationary phase in YNB medium containing 20 g liter−1 l-arabinose. The control preculture was grown in YNB medium with 20 g liter−1 glucose. Cells were washed with sterile water and inoculated to an optical density at 600 nm (OD600) of 0.2. All growth assays were carried out at least in duplicate, with deviations less than 10%.
Anaerobic batch fermentations.
Anaerobic batch fermentations were performed in Minifors bioreactors with a working volume of 2 liters (Infors AG, Bottmingen, Switzerland). Shake flask precultures were grown until late exponential phase in YNB medium with 20 g liter−1 l-arabinose. Cells were washed with sterile water. Cultures were inoculated at an OD600 of about 0.6 and incubated at 30°C with 250-rpm stirring and at pH 5.5, maintained by addition of 4 M KOH. The synthetic medium contained 30 g liter−1 l-arabinose. Cells were grown under aerobic conditions until about 10 g liter−1 l-arabinose was consumed and then were shifted to anaerobic conditions by sparging with nitrogen gas (containing less then 5 ppm of O2; Air Liquide, Düsseldorf, Germany) for 30 min with a flow rate of 1 liter min−1. Ethanol evaporation was not calculated. Evaporation of ethanol was minimized by using a reflux condenser at 4°C. The experiments were performed in duplicate.
Metabolite analysis.
The concentrations of l-arabinose, arabitol, glycerol, acetic acid, and ethanol were determined by high-performance liquid chromatography (Dionex) using a Nugleogel Sugar 810 H exchange column (Macherey-Nagel GmbH & Co, Germany). The column was eluted at the temperature of 65°C with 5 mM H2SO4 as a mobile phase with a flow rate of 0.6 ml min−1. Detection was done by means of a Shodex RI-101 refractive-index detector. Chromeleon software (version 6.50) was used for data evaluation. Rates of l-arabinose consumption and ethanol production were determined in the time frame between 50 and 90 h during the fermentation.
Determination of culture dry weight.
Determinations of dry weight were performed in duplicate by filtering 10 ml of the culture through a preweighed nitrocellulose filter with 0.45-μm pores (Roth, Germany). The filters were washed with demineralized water, dried in a microwave oven for 20 min at 140 W, and weighed again.
Western blot analysis.
Yeast transformants expressing C-terminal His6 epitope-tagged variants of araA from C. acetobutylicum, B. subtilis, and B. licheniformis (carried on multicopy vectors) were cultivated until early exponential growth phase in selective medium. Cells were harvested and disrupted with glass beads (Φ = 0.45 mm) using a Vibrax cell disrupter (Vibrax VXR; Janke & Kunkel). The protein content was determined according to the method of Bradford (7) and adjusted for equal loading on a sodium dodecyl sulfate (SDS)-polyacrylamide gel. Twenty micrograms of total protein was applied in each lane. For Western blot analysis, protein was transferred from the SDS gel to a nitrocellulose membrane by submerse electroblotting. AraA-His6 proteins were detected with mouse anti-His5 antibody (Roche) and rabbit anti-mouse immunoglobulin G coupled to peroxidase (Roche).
RESULTS
Screen for improved l-arabinose isomerase.
The functional expression of a bacterial l-arabinose pathway leading to an l-arabinose-fermenting yeast strain has previously been shown (5). From physiological analyses, it became apparent that the first enzyme in the pathway, l-arabinose isomerase, seems to be limiting for efficient l-arabinose conversion in recombinant yeast cells (5, 23). Moreover, we observed that in a yeast strain containing three different plasmids expressing the individual genes of the l-arabinose pathway and growing on an l-arabinose medium, the copy number of the plasmid containing the l-arabinose isomerase gene from B. subtilis was significantly increased (data not shown), indicating an increase in gene dosage that compensates for a low enzyme activity.
Therefore, we aimed to screen for l-arabinose isomerases from various organisms of different phylogenetic affiliations with improved activity after expression in yeast cells.
We chose the l-arabinose isomerases from Bacillus licheniformis, Pediococcus pentosaceus, Lactobacillus plantarum, Leuconostoc mesenteroides, and Clostridium acetobutylicum. To test the performance of the different l-arabinose isomerases, we amplified the coding sequences of the genes by PCR and cloned them into the high-copy-number yeast vector p423H7 (13) under the control of the strong and constitutive HXT7 promoter and the CYC1 terminator via homologous recombination. To test the performance of the various isomerases within the l-arabinose utilization pathway, yeast strains expressing the whole pathway including the different isomerases were constructed. Growth of these strains in synthetic medium with 20 g liter−1 l-arabinose as the only carbon source was compared.
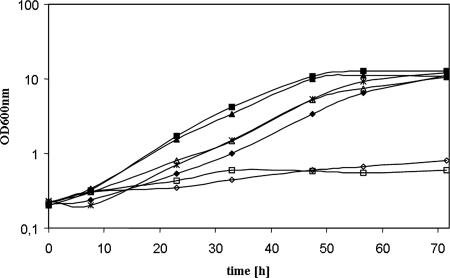
It turned out that, in particular, yeast transformants containing the l-arabinose isomerases from B. licheniformis and from C. acetobutylicum showed highly accelerated growth on l-arabinose medium (Fig. 2). Notably, they reached their maximal specific growth rate much faster than the other strains. After 33 h, the strains containing the isomerase from C. acetobutylicum or B. licheniformis had reached an OD600 of 4 whereas the strain containing the l-arabinose isomerase from B. subtilis had reached only an OD600 of 1. The latter strain needed an additional 20 h to reach an OD600 of 4. Nevertheless, comparison of the maximal specific growth rates of the transformed strains showed nearly identical values. Except for the recombinant strain containing the l-arabinose isomerase from P. pentosaceus (growth rate of 0.02 h−1), yeast strains containing the other l-arabinose isomerases all exhibited a maximal specific growth rate of about 0.08 h−1 (Fig. 2).
FIG. 2.
Growth of recombinant S. cerevisiae strains expressing different l-arabinose isomerases. YNB medium contained 20 g liter−1 l-arabinose as the sole carbon source. Yeast strains were grown aerobically as shake flask cultures at 30°C. The yeast strains were derived from strain BWY1 and contained the l-arabinose utilization pathway genes, including one of the various l-arabinose isomerases, from the following organisms: ▴, B. licheniformis; ⧫, B. subtilis; ▪, C. acetobutylicum; ⋄, P. pentosaceus; ▵, L. plantarum; ×, L. mesenteroides; □, vector control.
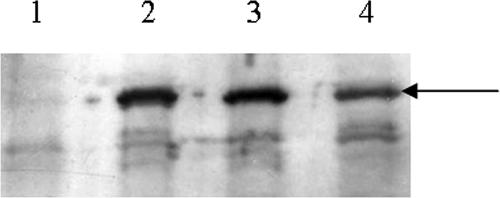
Since determination of the enzyme activity of l-arabinose isomerase in yeast is difficult to perform and is not possible in a quantitative way (5), the quantities of the enzymes were determined by Western blot analysis (Fig. 3). The l-arabinose isomerase genes of B. licheniformis, C. acetobutylicum, and B. subtilis were tagged with a His6 epitope and expressed in yeast cells. Crude extracts were prepared after growth in maltose medium, and the same amounts of protein were used in SDS-polyacrylamide gel electrophoresis. As a negative control, yeast cells containing the empty vector p426H7 were used. We observed that nearly the same amounts of isomerase protein were present in the strains containing the isomerase from B. subtilis or from C. acetobutylicum (Fig. 3). However, the amount of the isomerase in the strain producing the enzyme from B. licheniformis seemed to be lower. This indicates that the improvement in growth performance on l-arabinose medium was not caused by a higher protein level of the B. licheniformis and C. acetobutylicum l-arabinose isomerases but obviously by higher activities of the two enzymes in yeast cells.
FIG. 3.
Immunological detection and quantification of different l-arabinose isomerases in crude extracts of recombinant S. cerevisiae strains. Strains carrying the genes for the different l-arabinose isomerases on multicopy vectors were grown as shake flask cultures at 30°C to exponential growth phase in synthetic medium with 10 g liter−1 maltose and without histidine. Crude extracts were prepared, and 20 μg of total protein was applied in each lane. Western blotting was performed as described in Materials and Methods. Lanes: 1, empty vector (p423H7); 2, YEparaAB. subtilis-6HIS; 3, YEparaAC. aceto-6HIS; 4, YEparaAB. liche-6HIS. The arrow indicates the respective His-tagged isomerases cross-reacting with the antibodies.
Comparison of the codon adaptation index (CAI) of the B. licheniformis araA (CAI, 0.063) and C. acetobutylicum araA (CAI, 0.12) gene using the CODONW software program (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=codonw) indicated that the C. acetobutylicum gene matched the codon usage of S. cerevisiae more closely. Therefore, we expected that the B. licheniformis enzyme might improve l-arabinose conversion in yeast even more profoundly after adaptation of the corresponding codons to those used frequently in S. cerevisiae (see below). Therefore, we decided to continue our work with the isomerase from B. licheniformis.
Heterologous expression of codon-optimized gene versions for improved l-arabinose conversion.
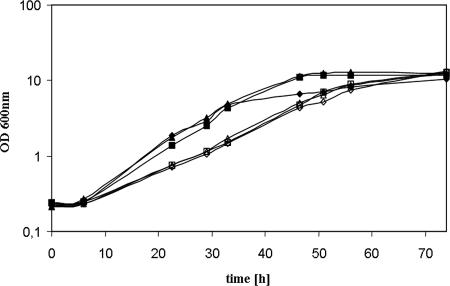
To further improve l-arabinose utilization in recombinant yeast cells, the codon usage of the genes araA from B. licheniformis and araB and araD from E. coli was adapted to that of the genes encoding glycolytic enzymes in S. cerevisiae (Table 2) but without changing the amino acid sequence. All codons of the bacterial genes were changed to those shown in Table 2 (identities of the codon-optimized genes compared to the original genes are as follows: l-arabinose isomerase, 73%; l-ribulokinase, 73%; l-ribulose-5-P 4-epimerase, 75%). Glycolytic genes in S. cerevisiae are highly expressed and have a strongly biased codon usage. To test the effect on l-arabinose conversion, recombinant yeast strains were constructed containing the original genes and the codon-optimized genes in different combinations on different plasmids (Table 3). First, growth on l-arabinose medium was tested under aerobic conditions using shake-flask cultures (Fig. 4). It turned out that growth performance of strain BWY1-SK and strain BWY1-SE, containing the codon-optimized araB and araD genes, respectively, was not improved. The same specific growth rates (0.08 h−1) compared to that of the control strain, BWY1-4M, were observed. However, strain BWY1-SI, expressing the codon-optimized araA gene, showed an increased maximal specific growth rate, up to 0.11 h−1. Strain BWY1-SEK, with optimized araB and araD, and strain BWY1-4S, with all three optimized genes, showed similar growth behavior to that of strain BWY1-SI and grew with a maximal rate of 0.11 h−1. These strains reached their final ODs in less than 50 h, whereas strain BWY1-4M needed an additional 25 h to grow to the same OD.
TABLE 3.
Yeast strains containing different combinations of original and synthetic genes for l-arabinose utilization on four plasmids
| Strain | Relevant enzymes | Plasmids carried | Specific growth rate (h−1)a |
|---|---|---|---|
| BWY1-SI | Synthetic isomerase, mutated kinase, original epimerase, Gal2 transporter | YEparaAsynth, YEparaBG361A, YEparaD, YEpGAL2 | 0.112 ± 0.005 |
| BWY1-SK | Original isomerase, synthetic kinase, original epimerase, Gal2 transporter | YEparaA, YEparaBsynth, YEparaD, YEpGAL2 | 0.081 ± 0.005 |
| BWY1-SE | Original isomerase, mutated kinase, synthetic epimerase, Gal2 transporter | YEparaA, YEparaBG361A, YEparaDsynth, YEpGAL2 | 0.083 ± 0.002 |
| BWY1-SEK | Original isomerase, synthetic kinase, synthetic epimerase, Gal2 transporter | YEparaA, YEparaBsynth, YEparaDsynth, YEpGAL2 | 0.109 ± 0.006 |
| BWY1-4S | Synthetic isomerase, synthetic kinase, synthetic epimerase, Gal2 transporter | YEparaAsynth, YEparaBsynth, YEparaDsynth, YEpGAL2 | 0.111 ± 0.004 |
| BWY1-4M | Original isomerase, mutated kinase, original epimerase, Gal2 transporter | YEparaA, YEparaBG361A, YEparaD, YEpGAL2 | 0.081 ± 0.003 |
The specific growth rates and the standard deviations are indicated.
FIG. 4.
Growth of S. cerevisiae strains expressing codon-optimized genes of the l-arabinose utilization pathway. Growth of recombinant yeast strains with different combinations of codon-optimized genes and original genes were tested in shake flask cultures at 30°C on YNB medium with 20 g liter−1 l-arabinose as the sole carbon source. ⧫, BWY1-SI; ▵, BWY1-SE; □, BWY1-SK; ▴, BWY1-SEK; ▪, BWY1-4S; ⋄, BWY1-4M.
Fermentation characteristics of yeast strains containing codon-optimized l-arabinose utilization genes.
To analyze l-arabinose consumption and ethanol production of yeast transformants containing the codon-optimized genes, anaerobic batch fermenter cultivations were performed in synthetic medium with 30 g liter−1 of l-arabinose as the sole carbon and energy source. Precultures of strain BWY1-4S, containing the codon-optimized genes, and strain BWY1-4M, containing the original genes, were pregrown aerobically in shaking flasks containing 50 ml of YNB medium with 20 g liter−1 l-arabinose. Cells were harvested, washed, and inoculated into batch fermenters. To generate enough biomass, cells were first grown under aerobic conditions until 10 g liter−1 of l-arabinose was consumed (Fig. 5). Anaerobic conditions were maintained by sparging with nitrogen gas until no oxygen was left in the medium (see Materials and Methods).
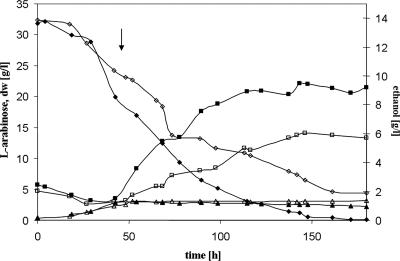
FIG. 5.
Anaerobic batch fermentations by recombinant S. cerevisiae. Shown are typical graphs of anaerobic batch fermentations of strains BWY1-4M and BWY1-4S. Strains were grown in mineral medium with 30 g liter−1 l-arabinose as the sole carbon source. Strains were pregrown in the fermenter under aerobic conditions until about 10 g liter−1 l-arabinose was consumed and then shifted to anaerobic conditions (indicated by the arrow). Symbols for BWY1-4M: ⋄, l-arabinose; ▵, biomass; □, ethanol. Symbols for BWY1-4S: ⧫, l-arabinose; ▪, ethanol; ▴, biomass. dw, dry weight.
l-Arabinose consumption and ethanol production rates were determined in the anaerobic phase of the fermentation, i.e., starting with approximately 20 g liter−1 l-arabinose in the medium. Strain BWY1-4M, with the original genes, consumed l-arabinose at a rate of 0.08 g l-arabinose h−1 (g dry weight)−1, and a residual of 3 g liter−1 l-arabinose was still left in the medium after 200 h. During fermentation by strain BWY1-4S, with the synthetic genes, l-arabinose was utilized at a rate of 0.11 g l-arabinose h−1 (g dry weight)−1 and was consumed completely after 150 h (Fig. 5). The ethanol production rates were 0.014 g ethanol h−1 (g dry weight)−1 for strain BWY1-4M and 0.036 g ethanol h−1 (g dry weight)−1 for strain BWY1-4S. Ethanol yields were 0.24 g ethanol (g consumed l-arabinose)−1 (BWY1-4M) and 0.39 g ethanol (g consumed l-arabinose)−1 (BWY1-4S). As by-products, glycerol and acetate were produced in similar amounts by the two strains (0.06 g glycerol (g consumed l-arabinose)−1; 0.015 g acetate (g consumed l-arabinose)−1), whereas arabitol production was significantly reduced in strain BWY1-4S (0.11 g arabitol [g consumed l-arabinose]−1) to half of the amount with BWY1-4M (0.21 g arabitol [g consumed l-arabinose]−1).
DISCUSSION
Successful l-arabinose fermentation in S. cerevisiae was previously accomplished after heterologous expression of bacterial l-arabinose utilization pathways consisting of three enzymes, l-arabinose isomerase, l-ribulokinase, and l-ribulose-5-phosphate 4-epimerase (5, 31). These efforts resulted in strains capable of growing on l-arabinose and fermenting it into ethanol. Nevertheless, growth of the cells with l-arabinose and its fermentation were still much slower than the utilization of glucose or even xylose in recombinant yeast cells (3, 4, 19), indicating some kind of constraint. Several observations indicated that the l-arabinose isomerase reaction is a limiting step for l-arabinose catabolism in recombinant S. cerevisiae cells. In the first approach, to express a functional l-arabinose catabolic pathway in yeast, Sedlak and Ho (23) found that the activity of the l-arabinose isomerase of E. coli was not sufficient to support any growth of the yeast cells with l-arabinose. In our previous work (5), we were able to confirm this observation, but we found that the l-arabinose isomerases from Mycobacterium smegmatis and B. subtilis could be expressed in S. cerevisiae in a functional form and supported growth of the cells with l-arabinose. Nevertheless, only very low activities of the enzymes could be measured (5). Moreover, an increased dosage of the araA gene was shown to be beneficial for l-arabinose utilization after chromosomal integration of the gene (16). Furthermore, we observed that in a yeast strain containing three different plasmids carrying the individual genes of the l-arabinose pathway and growing on an l-arabinose medium, the copy number of the plasmid containing the l-arabinose isomerase gene was highly increased compared to those of the other two plasmids (data not shown). This indicated that the activity of the enzyme was still limiting for efficient conversion of l-arabinose and was compensated by an increase in the gene dose.
To overcome this limitation, in the present work we investigated the performance of recombinant yeast cells after replacement of the l-arabinose isomerase of B. subtilis with the corresponding enzymes of a variety of different bacterial species. Indeed, we found that the l-arabinose isomerases of some of them, and especially the enzymes from B. licheniformis and C. acetobutylicum, improved growth of the cells on l-arabinose medium. The amounts of the different isomerases, as evidenced by immunodetection of His6-tagged versions, were found to be lowest in the strain expressing the isomerase from B. licheniformis. This finding indicates that the enzymes from B. licheniformis and C. acetobutylicum apparently exhibit higher activities in yeast cells than the enzyme from B. subtilis. Unfortunately, we were not able to directly prove this hypothesis by enzyme activity measurements.
Notably, the improvement in growth behavior did not mainly result from an increased maximal growth rate but rather from the cells reaching their maximal specific growth rate much faster than those of the original strain. A likely explanation for this effect might be related to variations in the copy number of the plasmid carrying the araA gene. It is reasonable to assume that if l-arabinose isomerase activity is limiting for growth of the cells on l-arabinose, there must be a continuous selection for cells with a higher copy number of the plasmid expressing l-arabinose isomerase. Therefore, growth of the cells will be slower at the beginning of a culture, until an increasing part of the culture will consist of cells with higher plasmid numbers. If, however, the activity of the enzyme is high and is not limiting for growth, as in the case of cells expressing the enzymes from B. licheniformis and C. acetobutylicum, all cells of the population will start growing rapidly. In accordance with this explanation is the strongly increased copy number of the plasmid containing the B. subtilis araA gene (see above).
The successful elimination of this apparent “bottleneck” in l-arabinose utilization in yeast is especially important because such adaptations of the copy number of plasmids are impossible after chromosomal integration of the l-arabinose utilization genes. Chromosomal integration, in turn, is necessary for transfer of these traits to industrial yeast strains and for commercial applications.
After the initial optimization of l-arabinose utilization by introduction of l-arabinose isomerase from B. licheniformis, all genes of the l-arabinose utilization pathway were adapted to the codon usage of S. cerevisiae. Analysis of the codon usage showed that the l-arabinose isomerase gene from B. licheniformis contains 21 rare codons, which are used by S. cerevisiae to translate the corresponding amino acids at a frequency of only 6% or less (relative to the number in the genome of codons representing a particular amino acid). The l-ribulose-5-P 4-epimerase and the l-ribulokinase from E. coli each contain 10 rare codons (data source, NCBI-GenBank Flat File, release 156.0 [October 15 2006]). It was shown that codon usage plays an important role in the expression levels of genes and the functionality of the corresponding enzymes (1, 25, 26). For example, the gene cyt2Aa1 from Bacillus thuringiensis, important in the development of antitumor agents, could be heterologously expressed in E. coli only with a very low yield of functional product. Subsequently, the gene could successfully be expressed in the yeast Pichia pastoris after adaptation of the codon use to that of P. pastoris. The synthetic gene was effectively used for recombinant production of Cyt2Aa1 (12).
Moreover, in particular, the strongly expressed genes in S. cerevisiae have adapted a highly biased codon usage with a strong preference for the most abundant tRNAs. For example, glycolytic proteins can make up more than 50% of the proteins in a yeast cell, and the carbon flux through this pathway is very high. For most amino acids, the glycolytic genes are restricted to only one of the corresponding synonymous codons (Table 2). Therefore, we thought that it might be beneficial to adapt the bacterial genes not only to the general codon usage of yeast but more specifically to the codon usage of glycolytic genes in yeast.
Indeed, by turning the heterologous genes from the l-arabinose utilization pathway into highly expressed “endogenous” genes, we could further improve, in a rational way, growth on l-arabinose, as well as ethanol production from l-arabinose. This improvement in l-arabinose growth and l-arabinose fermentation resulted in a 2.5-fold increase in ethanol production, from 0.014 g ethanol h−1 (g dry weight)−1 to 0.036 g ethanol h−1 (g dry weight)−1 and also an increase in the ethanol yield from 0.24 g ethanol (g consumed l-arabinose)−1 to 0.39 g ethanol (g consumed l-arabinose)−1. To our knowledge, this is the first report about successful pathway engineering in S. cerevisiae by expression of codon-adapted heterologous genes.
In the growth experiments, it turned out that neither the synthetic kinase nor the synthetic epimerase alone could improve growth. However, expression of both synthetic genes together improved growth, indicating that both activities were limiting for growth on l-arabinose. On the other hand, expression of the synthetic isomerase alone could also increase the specific growth rate, indicating that this enzyme is limiting for growth. Overexpression of all three synthetic genes together did not further improve growth. These results are difficult to explain. It seems that there is some kind of interaction between the three enzymes or their activities are somehow regulated by changing concentrations of intermediate metabolites. Interestingly, in our previous work (5), we also observed that a reduction of the activity of the second enzyme in the pathway, the kinase, was crucial for the catabolism of l-arabinose, although the activity of the first enzyme, the isomerase, obviously was the most limiting factor.
Recently a combined “engineering and evolution” approach resulted in a strain with a strongly increased growth rate on l-arabinose and very efficient l-arabinose fermentation (31). Also, in this adapted strain, it was found that expression of araA, araB, and araD had increased 6-, 52-, and 90-fold, respectively, indicating that the activities of all three enzymes were limiting. In this strain, the complete nonoxidative part of the pentose-phosphate pathway was overexpressed. In a similar approach, it has been shown that such an overexpression together with the heterologous expression of a xylose isomerase led to dramatically improved growth on d-xylose and its fermentation to ethanol (19). Both d-xylose metabolism and that of l-arabinose converge into the pentose-phosphate pathway at the level of xylulose-5-phosphate. However, with the same approach, the l-arabinose-fermenting strain did not reach comparable growth rates on l-arabinose even after extensive adaptive selection, but the growth rates finally reached a plateau value. These results show that there is still a limiting effect upstream of the pentose-phosphate pathway in the bacterial l-arabinose utilization enzymes or their expression levels. We propose that this limitation can be overcome by using the codon-adapted bacterial genes. Therefore, our strain might provide an advanced starting point for even more-improved l-arabinose fermentation by use of evolutionary engineering approaches. This might become especially important when the heterologous genes are stably integrated into the yeast genome as is necessary for industrial applications. Importantly, our systematic approach makes the transfer of these features into industrial yeast strains possible.
Acknowledgments
This work was financed by the European Commission through contract no. 019882 (New Improvements for Lignocellulosic Ethanol).
We thank C. Essl for her expert technical assistance.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Akashi, H. 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11:660-666. [DOI] [PubMed] [Google Scholar]
- 2.Attfield, P. V., and P. J. Bell. 2006. Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEMS Yeast Res. 6:862-868. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125-234. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, J. A., R. W. Payne, and D. Yarrow. 1990. Yeasts: characterisation and identification. Cambridge University Press, Cambridge, United Kingdom.
- 5.Becker, J., and E. Boles. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl. Environ. Microbiol. 69:4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boles, E., and F. K. Zimmermann. 1993. Saccharomyces cerevisiae phospho-glucose isomerase and fructose bisphosphate aldolase can be functionally replaced by the corresponding enzymes of Escherichia coli and Drosophila melanogaster. Curr. Genet. 23:187-191. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Dien, B. S., C. P. Kurtzman, B. C. Saha, and R. J. Bothast. 1996. Screening for L-arabinose fermenting yeasts. Appl. Biochem. Biotechnol. 57-58:233-242. [PubMed]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hagerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 12.Gurkan, C., and D. J. Ellar. 2003. Expression of the Bacillus thuringiensis Cyt2Aa1 toxin in Pichia pastoris using a synthetic gene construct. Biotechnol. Appl. Biochem. 38:25-33. [DOI] [PubMed] [Google Scholar]
- 13.Hamacher, T., J. Becker, M. Gardonyi, B. Hahn-Hagerdal, and E. Boles. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783-2788. [DOI] [PubMed] [Google Scholar]
- 14.Hayn, M., W. Steiner, R. Klinger, H. Steinmüller, M. Sinner, and H. Esterbauer. 1993. Basic research and pilot studies on the enzymatic conversion of lignocellulosics, p. 33-72. In J. N. Saddler (ed.), Biocenversion of forest and agricultural plant residues. CAB International, Wallingford, United Kingdom.
- 15.Jeppsson, M., O. Bengtsson, K. Franke, H. Lee, B. Hahn-Hagerdal, and M. F. Gorwa-Grauslund. 2005. The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:665-673. [DOI] [PubMed] [Google Scholar]
- 16.Karhumaa, K., B. Wiedemann, B. Hahn-Hagerdal, E. Boles, and M. F. Gorwa-Grauslund. 2006. Co-utilization of L-arabinose and D-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb. Cell Fact. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurtzman, C. P., and B. S. Dien. 1998. Candida arabinofermentans, a new L-arabinose fermenting yeast. Antonie van Leeuwenhoek 74:237-243. [DOI] [PubMed] [Google Scholar]
- 18.Kuyper, M., H. R. Harhangi, A. K. Stave, A. A. Winkler, M. S. Jetten, W. T. de Laat, J. J. den Ridder, H. J. Op den Camp, J. P. van Dijken, and J. T. Pronk. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 4:69-78. [DOI] [PubMed] [Google Scholar]
- 19.Kuyper, M., M. M. Hartog, M. J. Toirkens, M. J. Almering, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399-409. [DOI] [PubMed] [Google Scholar]
- 20.McMillan, J. D., and B. L. Boynton. 1994. Arabinose utilization by xylose-fermenting yeasts and fungi. Appl. Biochem. Biotechnol. 45-46:569-584. [DOI] [PubMed]
- 21.Richard, P., M. Putkonen, R. Vaananen, J. Londesborough, and M. Penttila. 2002. The missing link in the fungal L-arabinose catabolic pathway, identification of the L-xylulose reductase gene. Biochemistry 41:6432-6437. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. M. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Sedlak, M., and N. W. Ho. 2001. Expression of E. coli araBAD operon encoding enzymes for metabolizing L-arabinose in Saccharomyces cerevisiae. Enzyme Microb. Technol. 28:16-24. [DOI] [PubMed] [Google Scholar]
- 24.Sedlak, M., and N. W. Ho. 2004. Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. Appl. Biochem. Biotechnol. 113-116:403-416. [DOI] [PubMed]
- 25.Sharp, P. M., and E. Cowe. 1991. Synonymous codon usage in Saccharomyces cerevisiae. Yeast 7:657-678. [DOI] [PubMed] [Google Scholar]
- 26.Sharp, P. M., and W. H. Li. 1987. The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonderegger, M., and U. Sauer. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 69:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Sivers, M., and G. Zacchi. 1996. Ethanol from lignocellulosics: a review of the economy. Bioresour. Technol. 56:131-140. [Google Scholar]
- 29.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 30.Wirth, R. 1989. Elektroporation: eine alternative Methode zur Transformation von Bakterien mit Plasmid-DNA. Forum Mikrobiol. 11:507-515. [Google Scholar]
- 31.Wisselink, H. W., M. J. Toirkens, M. Del Rosario Franco Berriel, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]