Abstract
Myxobacteria are very important due to their unique characteristics, such as multicellular social behavior and the production of diverse and novel bioactive secondary metabolites. However, the lack of autonomously replicating plasmids has hindered genetic manipulation of myxobacteria for decades. To determine whether indigenous plasmids are present, we screened about 150 myxobacterial strains, and a circular plasmid designated pMF1 was isolated from Myxococcus fulvus 124B02. Sequence analysis showed that this plasmid was 18,634 bp long and had a G+C content of 68.7%. Twenty-three open reading frames were found in the plasmid, and 14 of them were not homologous to any known sequence. Plasmids containing the gene designated pMF1.14, which encodes a large unknown protein, were shown to transform Myxococcus xanthus DZ1 and DK1622 at high frequencies (∼105 CFU/μg DNA), suggesting that the locus is responsible for the autonomous replication of pMF1. Shuttle vectors were constructed for both M. xanthus and Escherichia coli. The pilA gene, which is essential for pilus formation and social motility in M. xanthus, was cloned into the shuttle vectors and introduced into the pilA-deficient mutant DK10410. The transformants subsequently exhibited the ability to form pili and social motility. Autonomously replicating plasmid pMF1 provides a new tool for genetic manipulation in Myxococcus.
Myxobacteria are gram-negative gliding bacteria that are phylogenetically located in the delta division of the Proteobacteria (29, 34, 41). The two most intriguing characteristics of myxobacteria are their complicated multicellular social behavior, which provides an excellent model for studies of cell-to-cell communication and evolution (6, 18, 39, 47), and their excellent capacity for production of diverse and novel bioactive secondary metabolites. Their production of bioactive secondary metabolites makes myxobacteria an important source of potential new drugs, although this possibility has not been well explored (36). The study and utilization of myxobacteria have been limited by the formidable isolation and culture techniques required (35) and the difficulty of performing genetic manipulations. In the past few decades, genetic studies of myxobacteria were performed mainly with the model species Myxococcus xanthus using transduction (7, 23) and the more efficient electroporation protocols (19). Besides these studies, Sorangium strains were also studied using conjugation protocols (13, 14, 22, 31, 33). Sorangium is a special cellulose degrader among the 17 myxobacterial genera (34, 50) and produces almost one-half of the known secondary metabolites produced by myxobacteria (8). Because no naturally occurring self-replicating plasmid has been discovered previously and no broad-host-range vectors can replicate in myxobacterial cells, all the genetic transfer systems used have been based on integration of introduced plasmids or phages into the recipient chromosomes. Consequently, some genetic manipulations are hard to perform in myxobacteria or are not very efficient (16). Thus, there is an urgent need to develop genetic protocols that are based on self-replicating plasmids in myxobacteria. Discovering indigenous self-replicating plasmids in myxobacteria was a viable approach. During the 1970s and 1980s, there was extensive screening of myxobacteria, but no indigenous plasmids were found, although several phages were discovered (4, 28, 43). To reevaluate whether myxobacterial cells contain plasmids naturally, we screened about 150 strains, mainly belonging to the genus Myxococcus but also belonging to the phylogenetically closely related genus Corallococcus. Using the methods previously used for Streptomyces (20), we discovered a circular plasmid, designated pMF1, in Myxococcus fulvus strain 124B02 and characterized a novel replication gene on this plasmid. Shuttle plasmids based on the pMF1 replicon were constructed. We transferred pilA-containing plasmids into the pilA-deficient mutant M. xanthus DK10410 by electroporation (49). Plasmid-based expression of the pilA gene restored pilus formation and social motility in the transformants.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
We randomly screened strains obtained from our established myxobacterial bank (26). The other bacterial strains and plasmids used in this study are listed in Table 1. The myxobacterial strains were routinely cultivated in VY/2 (35) or CTT (15) medium at 30°C. The Escherichia coli strains were cultivated in LB medium at 37°C. Solid medium was prepared by addition of 1.5% agar. If required, 40 μg/ml kanamycin or 100 μg/ml ampicillin was added for selection.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source |
|---|---|---|
| Strains | ||
| M. fulvus 124B02 | Wild-type strain, containing plasmid pMF1 | This study |
| M. xanthus DK1622 | Wild-type strain | D. Kaiser, Stanford University |
| M. xanthus DZ1 | Nonmotile, nonfruiting, dispersed growth | D. R. Zusman, University of California, Berkeley |
| M. xanthus DK10410 | ΔpilA mutant of DK1622 | M. H. Singer, University of California, Davisb |
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Life Technologies Inc. |
| Plasmids | ||
| pSP72 | Ampr | Life Technologies Inc. |
| ColE I | Kmr | D. Kaiser, Stanford University |
| pMF1 | Cryptic plasmid from M. fulvus 124B02 | This study |
| pZJY1 | Ampr Kmr; pSP72 with insertion of 1.2-kb Kmr gene from ColE I | This study |
| pZJY2 | Ampr Kmr; pZJY1 with insertion of 14.1-kb XbaI fragment (bp 1 to 14132) of pMF1 | This study |
| pZJY7 | Ampr Kmr; pZJY1 with insertion of 3.0-kb SalI fragment (bp 10953 to 13980) of pMF1 | This study |
| pZJY8 | Ampr Kmr; pZJY1 with insertion of 940-bp SalI-SacII fragment of pZJY7 (bp 10953 to 11891) of pMF1 | This study |
| pZJY9 | Ampr Kmr; pZJY1 with insertion of 2.4-kb MluI fragment of pZJY7 (bp 11183 to 13608) of pMF1 | This study |
| pZJY13 | Ampr Kmr; pZJY1 with insertion of 1.4-kb fragment (bp 12527 to 13980) of pMF1 | This study |
| pZJY15 | Ampr Kmr; pZJY1 with insertion of 2.2-kb fragment (bp 11645 to 13853) of pMF1 | This study |
| pZJY41 | Stable shuttle plasmid from a DZ1 transformant of pZJY7 | This study |
| pZJY156 | Stable shuttle plasmid from a DK1622 transformant of pZJY15 | This study |
| pZJY24_30 | Ampr Kmr; pZJY41 containing the pilA gene, parallelc | This study |
| pZJY25_35 | Ampr Kmr; pZJY156 containing the pilA gene, parallelc | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance.
See reference 49.
The pilA gene is in the same orientation as the pMF1.14 gene of pMF1 in pZJY41 or pZJY156.
Isolation and detection of plasmids from myxobacteria.
Myxobacterial plasmids were isolated by using the methods previously used for Streptomyces (20), with slight modifications. Cultures of different myxobacterial strains were inoculated into 3 ml of liquid VY/2 medium and cultivated at 30°C with shaking at 200 rpm for 60 to 72 h. The cells were harvested by centrifugation and suspended in 500 μl of TE25S buffer containing 10.3% sucrose, 25 mM Tris-HCl (pH 8.0), 25 mM EDTA (pH 8.0), and 2 mg/ml lysozyme. The suspension was incubated at 37°C for 1 h with periodic gentle inversion of the tubes. Then 250 μl of an alkali digestion solution containing 0.3 M NaOH and 2% sodium dodecyl sulfate was added, and the mixture was incubated at 55°C for 30 min with periodic gentle inversion. The mixture was extracted using 250 μl of water-saturated phenol-chloroform-isoamyl alcohol (25:24:1). After centrifugation, the supernatant was extracted using an equal volume of Tris-saturated phenol-chloroform-isoamyl alcohol (25:24:1) repeatedly until there was no visible layer of proteins. The final supernatant was mixed with 0.1 volume of 3 M sodium acetate and an equal volume of isopropyl alcohol to precipitate the DNA. After centrifugation, the DNA pellet was washed with 70% ethanol, air dried, and dissolved in 20 μl of TE buffer containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA (pH 8.0). The DNA was detected using agarose gel electrophoresis.
Cloning, sequencing, and assembly of pMF1.
Plasmid pMF1 from M. fulvus strain 124B02 was isolated using the protocol described above and was purified using an agarose gel DNA extraction kit (Roche) by following the manufacturer's instructions. After purification, the plasmid was digested with restriction enzymes. It contained no cut sites for BamHI, BglII, XhoI, HindIII, PstI, SacI, and KpnI, but there were multiple sites for XbaI and two sites for EcoRI. The EcoRI-digested segments were incubated in a ligation solution containing EcoRI and alkaline phosphatase-pretreated plasmid pSP72 (Life Technologies). The resulting recombinant plasmids were transferred into E. coli DH5α using the protocol described by Sambrook and Russell (37). Clones containing the expected segment inserts were selected, and the inserts were sequenced with an Applied Biosystems model 377 genetic analyzer at the Chinese Human Genome Center in Shanghai. The sequenced segments were assembled to obtain the complete sequence of pMF1 using ContigExpress software (InforMax Inc). The open reading frames (ORFs) in the pMF1 sequence were located using FramePlot 3.0 (12).
Determination of the replication locus on pMF1.
The kanamycin resistance gene aphII (aminoglycoside 3′-O-phosphotransferase gene) was amplified from plasmid ColE I by PCR using Pfu DNA polymerase (Fermentas) and primers 5′-GGGAAGCTTGTGCTGACCCCGGGTGAATGTCAG-3′ and 5′-GGGAAGCTTATCGAGCCCGGGGTGGGCGAAGAA-3′ (21). The primers contained HindIII sites at their 5′ ends. The 1,269-bp product was digested with HindIII and inserted into pSP72 to form a recombinant plasmid, pZJY1 (see Fig. 2A), which was used to locate the replication region of pMF1. Plasmid pMF1 was digested with different restriction enzymes, and the digested segments were subcloned into pZJY1. The recombinant plasmids containing different segments were electroporated into M. xanthus strain DZ1 or DK1622 by using the protocol described by Kashefi and Hartzell (19). Kanamycin-resistant clones from CTT medium plates containing 40 μg/ml kanamycin were selected and then purified. The presence of plasmids was determined using the method described above, except that liquid CTT medium containing 40 μg/ml kanamycin was used.
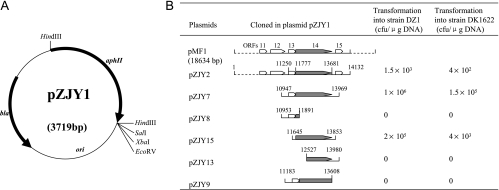
FIG. 2.
Identification of the locus on pMF1 for propagation in M. xanthus DZ1 and DK1622. (A) Schematic map of pZJY1. The antibiotic selection markers bla and aphII are indicated by bold arrows. The cloning sites used in this study are also indicated. (B) Transformation of strains DZ1 and DK1622 by pZJY1 containing various segments of pMF1 (positions are indicated by sequence numbers). Plasmids were constructed in E. coli (see Table 1). Transformation frequencies are shown. Most of the genes on pMF1 are indicated by open boxes; the only exception is the replication gene, which is indicated by a filled box. The direction of transcription is indicated by arrowheads.
Plasmid copy number.
The number of copies of the plasmids was determined by using the methods used for Streptomyces (32). M. xanthus transformants containing plasmids were grown, and total chromosomal DNA and plasmid DNA were isolated and diluted 10-, 100-, and 1,000-fold in TE buffer. The DNA was separated on agarose gels by electrophoresis. By comparing the intensity of the diluted chromosomal DNA band with the intensity of the nondiluted plasmid DNA band using the AlphaEaseFC image analysis software (Alpha Innotech Corp.), we calculated the copy number of the plasmid in the cells as follows:
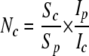 |
where Nc is the copy number of the plasmid, Sc and Sp are the chromosomal DNA size (9,000 kb) and plasmid DNA size, respectively, and Ip and Ic are the intensities of the plasmid and chromosomal DNA bands.
Inheritance of plasmids.
The inheritance of plasmids in Myxococcus was analyzed using the method described by De Mot et al. (5), with small modifications. The Myxococcus transformants were inoculated into liquid CTT medium containing 40 μg/ml kanamycin and grown to the late exponential growth phase. The cells were diluted 25-fold using fresh liquid CTT medium without the antibiotic and grown at 30°C for 24 h. The dilution and cultivation procedure was repeated. After each round, the cells were spread onto CTT medium plates. One hundred random clones were moved to fresh CTT medium plates with and without kanamycin, and the numbers of clones that grew on these plates were counted to determine the survival frequency of the plasmids.
Construction and transformation of the pilA-containing plasmids.
The stable shuttle plasmids pZJY41 and pZJY156 were used for further construction. The pilA gene and its upstream σ54 promoter sequence were amplified from M. xanthus DK1622 using Pfu DNA polymerase (Fermentas) and primers 5′-GGGAGCGCTTCGGATGCGTAGGC-3′ and 5′-CGAGTTACTGGGCCGCGCCGTCG-3′. After purification, the 874-bp product was ligated into EcoRV-digested plasmids pZJY41 and pZJY156 in parallel with the pMF1 replicon, resulting in recombinant plasmids pZJY24_30 and pZJY25_35. The pilA-containing plasmids were transferred into E. coli DH5α and sequenced. Each plasmid was separately transferred into M. xanthus DK10410 (ΔpilA) by electroporation. After 7 days, transformants were selected from CTT medium plates containing 40 μg/ml kanamycin. These transformants were purified twice, and then the plasmids were extracted for confirmation.
Swarming assay.
We used the method described by Shi and Zusman (38) to measure the swarming capacity of the transformants. A 2-μl aliquot of cells (5 × 109 cells/ml) was inoculated onto CTT medium plates containing 1.5 or 0.3% agar. After 3 or 5 days of incubation at 30°C, the size of the swarming colonies was determined.
Examination of the pili by transmission electron microscopy.
To determine whether the transformants produced pili, the method described by Li et al. (25) was used. The cells were harvested and washed with TPM buffer. Then they were moved to carbon-coated microscope grids, stained with 2% phosphotungstic acid, and examined with a JEOL JEM-100CX II electron microscope.
Nucleotide sequence accession numbers.
The complete nucleotide sequence of pMF1 has been deposited in the GenBank database under accession number EU137666. The 16S rRNA gene sequence of strain 124B02 has been deposited in the GenBank database under accession number EU137665. The complete nucleotide sequences of pZJY41 and pZJY156 have been deposited in the GenBank database under accession numbers EU328349 and EU328350.
RESULTS
Detection of the indigenous autonomously replicating plasmid pMF1 in M. fulvus 124B02.
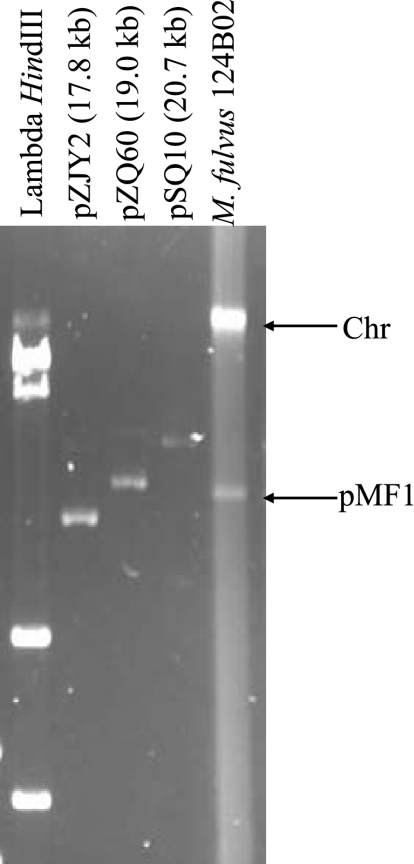
To isolate naturally occurring plasmids, approximate 150 myxobacterial strains were screened. The strains, which were isolated from samples collected in different regions and from different habitats in China (26), are mainly members of the genus Myxococcus; some of them are members of the genus Corallococcus, a genus that is phylogenetically close to Myxococcus (40, 42). The myxobacterial strains produced abundant extracellular slime and matrix (1, 35), which greatly hampered extraction of the plasmid DNA. Using the protocol described previously for Streptomyces, with modifications, we detected a ∼18.5-kb plasmid DNA band (designated pMF1) in strain 124B02 (Fig. 1).
FIG. 1.
Detection of plasmid pMF1 in M. fulvus 124B02. Plasmid DNA was isolated and separated by gel electrophoresis in a 0.6% agarose gel at 42 V for 12 h. HindIII-digested lambda DNA and other sequenced circular plasmids were used as size markers. Chr, chromosome.
Strain 124B02 is a gram-negative bacterium that produces vegetative cells that are 6 to 10 by 1.2 μm. This strain produces typical Myxococcus-type fruiting bodies containing spherical myxospores that are 1.2 to 1.5 μm in diameter. The 16S rRNA gene sequence of strain 124B02 closely resembles the 16S rRNA gene sequence of M. fulvus strain ATCC 25199 (99% identity). Thus, the strain was designated M. fulvus 124B02 and has been deposited in the China Center for Type Culture Collection under accession number M 206081.
Characterization of the pMF1 sequence.
pMF1 DNA was digested with EcoRI and cloned into E. coli plasmid pSP72 for sequencing. The complete nucleotide sequence of the plasmid consisted of 18,634 bp, had a G+C content of 68.7%, and resembled the genome sequence of M. xanthus DK1622 (G+C content, 68.9% [9]). The ORFs of pMF1 were predicted using FramePlot 3.0, a program for sequences with high G+C contents (12). There were 23 predicted ORFs in pMF1; 21 were on the sense strand, and 2 (pMF1.19c and pMF1.20c) were on the complementary strand (Table 2). Nine ORFs resembled known genes in the GenBank database, especially pMF1.19c and pMF1.20c, which have homology to two hypothetical genes (MXAN_6992 and MXAN_6330) of M. xanthus DK1622. The remaining 14 ORFs were largely unknown. About 86% of the plasmid sequence consisted of protein coding sequences resembling those of the Myxococcus genome (90%) (9). However, bioinformatic analysis did not reveal the replicon region of pMF1, suggesting that it is a new locus.
TABLE 2.
Predicted ORFs in the complete sequence of pMF1
| ORF | Position (bp) | Size of product (amino acids) | E value | Similarity or function (organism) |
|---|---|---|---|---|
| pMF1.1 | 803-1753 | 316 | 2 × 10−8 | APEG precursor protein (Xenopus laevis) |
| pMF1.2 | 1750-2502 | 250 | Unknown | |
| pMF1.3 | 2516-2692 | 58 | Unknown | |
| pMF1.4 | 2757-4220 | 487 | Unknown | |
| pMF1.5 | 4664-5530 | 288 | 3 × 10−10 | RTX toxins and related Ca2+-binding protein (Magnetospirillum) |
| pMF1.6 | 6107-6571 | 154 | 3 × 10−8 | Mucin-associated surface protein (Jannaschia) |
| pMF1.7 | 6574-7332 | 252 | Unknown | |
| pMF1.8 | 7329-7964 | 211 | 6 × 10−38 | Glycoside hydrolase (Stigmatella) |
| pMF1.9 | 7961-8263 | 100 | Unknown | |
| pMF1.10 | 8275-9225 | 316 | Unknown | |
| pMF1.11 | 9264-9704 | 146 | Unknown | |
| pMF1.12 | 10005-11045 | 346 | 3 × 10−10 | Protein kinase (Stigmatella) |
| pMF1.13 | 11250-11780 | 176 | Unknown | |
| pMF1.14 | 11777-13681 | 634 | Unknown | |
| pMF1.15 | 13678-14010 | 110 | Unknown | |
| pMF1.16 | 14029-14853 | 274 | 1 × 10−28 | Resolvase (Anaeromyxobacter) |
| pMF1.17 | 14974-15453 | 159 | Unknown | |
| pMF1.18 | 15443-15826 | 127 | Unknown | |
| pMF1.19ca | 16590-15823 | 255 | 6 × 10−42 | Hypothetical MXAN_6992 protein (M. xanthus DK1622) |
| pMF1.20ca | 17309-16587 | 240 | 2 × 10−19 | Hypothetical MXAN_6330 protein (M. xanthus DK1622) |
| pMF1.21 | 17391-17654 | 87 | Unknown | |
| pMF1.22 | 17651-18334 | 227 | 4 × 10−33 | Cobyrinic acid a,c-diamide synthase (Pseudomonas) |
| pMF1.23 | 18336-18596 | 86 | Unknown |
c indicates that the ORF is on the complementary strand.
Identification of a novel replication locus in pMF1.
To locate the replication locus in pMF1, Kmr plasmid pZJY1 was constructed (Fig. 2A). When this plasmid was transferred into M. xanthus DZ1 or DK1622 by electroporation, no resistant clones appeared on kanamycin selection plates, indicating that pZJY1 was unable to self-replicate or integrate into the genome of M. xanthus. Plasmid pZJY2 was obtained by ligating a large XbaI fragment (bp 1 to 14132) from pMF1 into pZJY1. Plasmid pZJY2 was transferred into M. xanthus strains DZ1 and DK1622 by electroporation. Kanamycin-resistant transformants were obtained at frequencies of 1.5 × 103 CFU/μg DNA for DZ1 and 4 × 102 CFU/μg DNA for DK1622, suggesting that the plasmid contained an autonomously replicating locus. The transformants were analyzed for the presence of plasmids, and all of the individual clones produced bands of the autonomously replicating plasmid. However, compared with the original pZJY2 plasmid from E. coli, which was 17.8 kb long, almost all of the plasmids extracted from the Myxococcus transformants were smaller and generally 5 to 8 kb long. These results suggested that there was unstable propagation of this E. coli-Myxococcus shuttle vector in M. xanthus. A plasmid (∼6 kb) from one DZ1 transformant, designated pDZ9, was analyzed further to locate the replication region. Digestion with restriction enzymes and PCR sequencing revealed that pDZ9 contained the pMF1.12, pMF1.13, pMF1.14, and pMF1.15 sequences from pMF1. To define the replication region, different segments of the pMF1 sequence in pDZ9 were subcloned into pZJY1 to produce pZJY7, pZJY8, pZJY9, pZJY13, and pZJY15(Fig. 2B). These plasmids were introduced into strains DZ1 and DK1622 by electroporation. Only pZJY7 and pZJY15 produced transformants in M. xanthus cells with very high frequencies (about 1 × 105 and 1 × 104 CFU/μg DNA for pZJY7 and pZJY15, respectively). Plasmid pZJY15 contained only the minimal locus of intact pMF1.14, which is a large gene that was not homologous to any gene in the GenBank database. Thus, pMF1.14 was required for propagation of the plasmid in M. xanthus.
Structurally stable E. coli-Myxococcus shuttle vectors in M. xanthus.
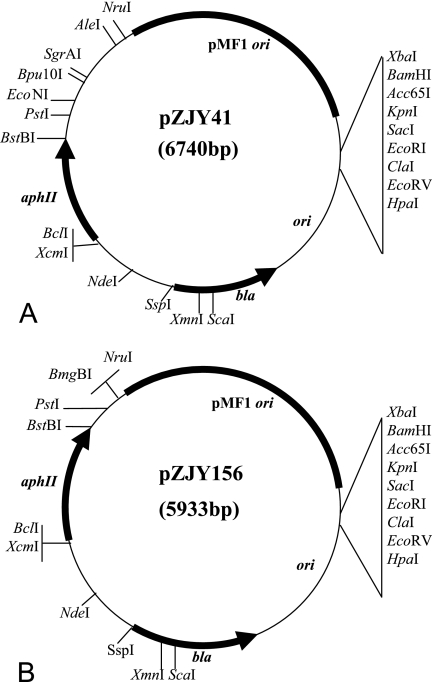
Although the reconstructed plasmids were able to replicate autonomously in M. xanthus cells, almost all the plasmids isolated from the individual transformants were unstable and smaller than the original plasmids from E. coli. While the plasmids extracted from Myxococcus colonies were able to retransform Myxococcus cells, most of them were unable to transform E. coli cells again, suggesting that there were deletions of the E. coli portion of these plasmids. To identify the deleted regions, plasmids from about 50 DZ1 transformants containing pZJY7 or pZJY15 were analyzed by restriction enzyme digestion, and some of them were analyzed by sequencing. All of these plasmids lacked the E. coli origin region of pSP72 and the bla sequence of the ampicillin resistance gene. To produce stable E. coli-Myxococcus shuttle plasmids, we screened the pZJY7 and pZJY15 transformants of Myxococcus and found two plasmids, designated pZJY41 (from pZJY7) (Fig. 3A) and pZJY156 (from pZJY15) (Fig. 3B), which had maintained their original sizes from E. coli. These two plasmids were both able to retransform both M. xanthus and E. coli cells. After several rounds of subculture in the presence of 40 μg/ml kanamycin, about 100 individual clones each of pZJY41 and pZJY156 were randomly selected and analyzed to determine the presence and size of the plasmid. Each clone contained a band that was the same size, indicating that plasmids pZJY41 and pZJY156 were stable E. coli-Myxococcus shuttle plasmids in M. xanthus DZ1. Similar results were obtained for the two plasmids in M. xanthus DK1622 (data not shown). Compared with the sequences of pZJY7 and pZJY15, there was one base deletion in the pMF1 insert in the sequences of pZJY41 and pZJY156, respectively, and there was also one base replacement in the aphII insert in the sequence of pZJY156.
FIG. 3.
Comprehensive restriction maps of stable Myxococcus-E. coli shuttle plasmids pZJY41 (A) and pZJY156 (B), which were derived from pMF1.
Inheritance and copy numbers of plasmids in M. xanthus.
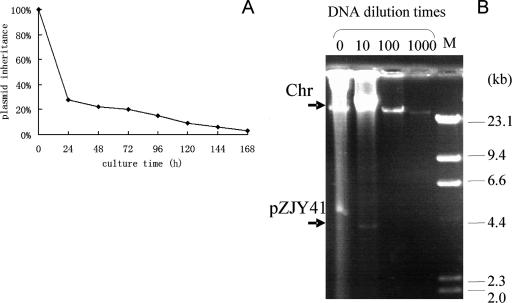
Using the method of De Mot et al. (5), the inheritance of pZJY41 and pZJY156 in M. xanthus cells was investigated. In the absence of antibiotic selection, 72% of the DZ1 transformed cells had lost pZJY41 after six generations. After 24 generations 85% of the cells had lost the plasmid, and after 42 generations 97% of the cells had lost the plasmid (Fig. 4A). The plasmid-containing cells grew somewhat slower, and the generation time was found to be about 4 h. The inheritance of pZJY156 in DZ1 or the inheritance of pZJY41 and pZJY156 in DK1622 was also unstable in the absence of antibiotic selection (data not shown). These results suggested that plasmids pZJY41 and pZJY156 could replicate autonomously in Myxococcus cells but were easily lost without antibiotic selection.
FIG. 4.
Inheritance and copy number of plasmid pZJY41 in M. xanthus DZ1. (A) Inheritance of plasmid pZJY41 in M. xanthus DZ1 in the absence of antibiotic selection. (B) Genomic DNA, including the chromosome and plasmid, were isolated and then diluted 10-, 100-, and 1,000-fold and electrophoresed in a 0.7% agarose gel at 35 V/37 cm for 12 h. Lambda HindIII DNA was used as the size marker. Chr, chromosome.
To determine the copy number of the plasmid, a series of dilutions of the chromosomal and plasmid DNA were prepared and run on agarose gels. We compared the fluorescence intensities of the bands under UV light, and the copy number of pZJY41 was estimated to be 10 to ∼20 in M. xanthus DZ1 (Fig. 4B). The number of copies of plasmid pZJY156 was lower than the number of copies of pZJY41 in DZ1, and the copy numbers of both plasmids were lower in DK1622 than in DZ1 (data not shown).
Restoration of pilA in pilA-deficient Myxococcus cells.
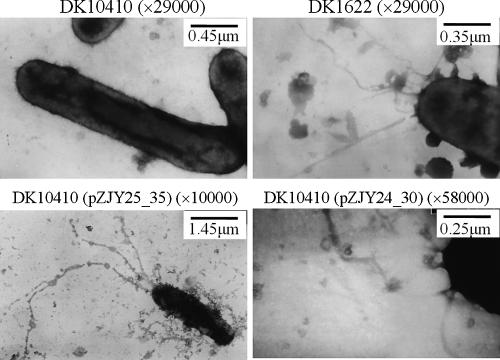
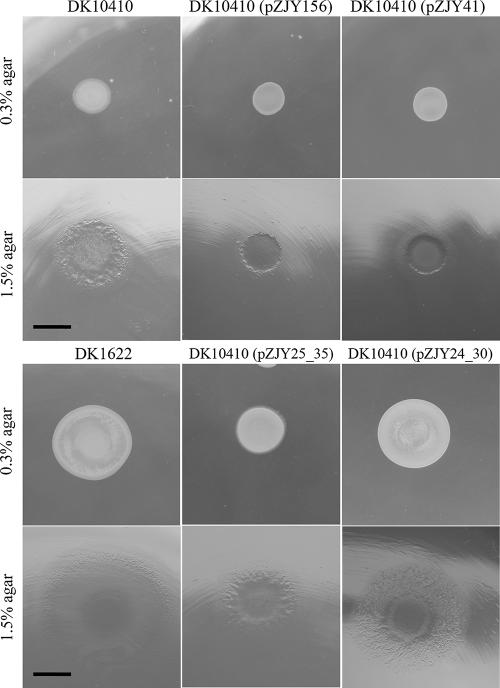
Myxobacterial gliding is controlled by two distinct motility systems, the adventurous system and the social system (10, 11). In M. xanthus, three cell surface components, type IV pili, fibrils, and lipopolysaccharide O antigen, are required for social motility (3, 27). Type IV pili pull the cells forward by pilus extension, attachment, and retraction (17, 25, 44, 45). The proteins for type IV pilus function are encoded mainly in a gene cluster (pil gene cluster) (46). DK10410 is a pilA null mutant which lacks pili and consequently social motility (48). In this study, plasmid-based expression of the pilA gene in the DK10410 mutant with pilA deleted was used to confirm the function of the shuttle plasmids pZJY41 and pZJY156. The pilA gene, together with its upstream σ54 promoter sequence, was inserted into the shuttle vectors in the same direction as the pMF1 replicon (Table 1). Plasmids pZJY24_30 and pZJY25_35 were transferred, and transformants were selected on CTT medium plates containing 40 μg/ml kanamycin. Transformation with either pZJY24_30 or pZJY25_35 produced resistant colonies. Approximately 10 colonies were randomly selected from each transformation to determine whether the cells contained the plasmids, and full-sized plasmids were detected on an agarose gel (data not shown). The isolated plasmids were able to transform both E. coli cells and M. xanthus cells. After two rounds of selection with kanamycin, the transformant cells were examined for the formation of pili by transmission electron microscopy (25), and the motilities on soft and hard agar were determined using the method described by Shi and Zusman (38). The results revealed that the transformants had the pilus structure (Fig. 5) and consequently exhibited social motility on solid medium (Fig. 6).
FIG. 5.
Transmission electron micrographs of pili. Cells were harvested from 1-day liquid CTT medium cultures and then negatively stained and viewed with a JEOL JEM-100CX II electron microscope.
FIG. 6.
Gliding behaviors of M. xanthus pilA-deficient mutant DK10410 and transformants with different plasmids on CTT medium containing either 0.3 or 1.5% agar. The images are images of 5-day cultures. The plasmids could be detected in the resistant DK10410 mutants. Bars = 6.5 mm.
DISCUSSION
Previously, myxobacteria were thought to contain no naturally occurring plasmid since analyses performed over many decades had failed to isolate any plasmids. The abundant extracellular slime and polysaccharides produced by myxobacteria (1, 35) certainly hamper the isolation and detection of plasmids, especially low-copy-number plasmids. Using the method for plasmid preparation developed for the mycelium-producing organism Streptomyces (20) to screen a large population of myxobacterial strains (∼150 strains), we discovered a cryptic autonomously replicating plasmid designated pMF1 in one strain, M. fulvus 124B02. Successful transformation with recombinant plasmids containing the pMF1 replicon in M. xanthus DZ1 and DK1622 suggested that more indigenous autonomously replicating plasmids may be present in myxobacteria.
Most of the predicted ORFs (14 of 23 ORFs) in pMF1 were not homologous to sequences in the GenBank database. One of these ORFs, pMF1.14, encoding a large 634-amino-acid protein, was shown to be the essential locus for self-replication in M. xanthus. No single-stranded DNA intermediate was detected, and no typical iterons were predicted at the replication locus (our unpublished data), suggesting that a novel theta-type mechanism is used for plasmid replication.
Some shuttle vectors, including the Streptomyces-E. coli plasmid derived from pIJ350 (30) and pIJ702 (24), are structurally unstable, and there are deletions of the E. coli portion in Streptomyces. Similar phenomena were observed for the pMF1-derived Myxococcus-E. coli shuttle plasmid. After screening a population of Myxococcus transformants, we found that two plasmids, pZJY41 and pZJY156, were propagated with no visible deletions, as determined by gel electrophoresis. The copy numbers of shuttle plasmids pZJY41 and pZJY156 in Myxococcus cells were low, and these plasmids were lost at a high frequency under nonselective conditions, which was probably related to the lack of the ParA-ParB system in the plasmids. The ParA-ParB system is encoded by many low-copy-number plasmids (2). In pMF1, bioinformatic analysis revealed that the PMF1.22 gene was partially homologous to the ParA gene, but no ORF was homologous to the ParB gene. This finding is similar to observations for plasmid pFAJ2600 in Rhodococcus erythropolis (5). We constructed plasmids by inserting the potential parA gene pMF1.22 (together with its neighbor, pMF1.21) and/or the potential recombinase gene pMF1.16 of pMF1 into pZJY41 and pZJY156, but there was no improvement in the stability of the plasmids (data not shown). However, plasmid pMF1 replicates stably in the strain in which it occurs naturally. A more stable shuttle plasmid is still under construction.
The discovery of self-replicating plasmids in Myxococcus and the construction of shuttle plasmids should greatly facilitate genetic manipulations in myxobacteria. In this study, we demonstrated that shuttle plasmid pZJY41 and pZJY156 derivatives, containing the motility gene pilA, restored expression of pilA in pilA-deficient strain DK10410. Cloning of a large DNA fragment, such as the myxobacterial antibiotic biosynthetic gene clusters on pMF1-derived vectors, and expression in M. xanthus will be investigated in other studies.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (grant 30325003) and a Chinese Academy of Sciences project (grants KSCX2-SW-329-3 and KSCX2-YW-G-014) to Z. Qin and from the National Natural Science Foundation of China (grant 30671192) and the Chinese 863 programs (grant 2006AA02Z171) to Y. Li.
We thank Roberta Greenwood for editing the manuscript.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Behmlander, R. M., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, M. G., and H. B. Kaplan. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30:275-284. [DOI] [PubMed] [Google Scholar]
- 4.Campos, J. M., J. Geisselsoder, and D. R. Zusman. 1978. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119:167-178. [DOI] [PubMed] [Google Scholar]
- 5.De Mot, R., I. Nagy, A. De Schrijver, P. Pattanapipitpaisal, G. Schoofs, and J. Vanderleyden. 1997. Structural analysis of the 6 kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcus shuttle vectors. Microbiology 143:3137-3147. [DOI] [PubMed] [Google Scholar]
- 6.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisselsoder, J., J. M. Campos, and D. R. Zusman. 1978. Physical characterization of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119:179-189. [DOI] [PubMed] [Google Scholar]
- 8.Gerth, K., S. Pradella, O. Perlova, S. Beyer, and R. Müller. 2003. Myxobacteria: proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J. Biotechnol. 106:233-253. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 11.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 12.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 13.Jaoua, S., S. Neff, and T. Schupp. 1992. Transfer of mobilizable plasmids to Sorangium cellulosum and evidence for their integration into the chromosome. Plasmid 28:157-165. [DOI] [PubMed] [Google Scholar]
- 14.Julien, B., and R. Fehd. 2003. Development of a mariner-based transposon for use in Sorangium cellulosum. Appl. Environ. Microbiol. 69:6299-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser, D. 1991. Genetic systems in myxobacteria. Methods Enzymol. 204:357-372. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser, D. 2006. A microbial genetic journey. Annu. Rev. Microbiol. 60:1-25. [DOI] [PubMed] [Google Scholar]
- 19.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 20.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, Y., H. Nakano, H. Terasaka, and K. Takegawa. 2001. Myxococcus xanthus mokA encodes a histidine kinase-response regulator hybrid sensor required for development and osmotic tolerance. J. Bacteriol. 183:1140-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp, M., H. Irschik, F. Gross, O. Perlova, A. Sandmann, K. Gerth, and R. Müller. 2004. Critical variations of conjugational DNA transfer into secondary metabolite multiproducing Sorangium cellulosum strains So ce12 and So ce56: development of a mariner-based transposon mutagenesis system. J. Biotechnol. 107:29-40. [DOI] [PubMed] [Google Scholar]
- 23.Kuner, J. M., and D. Kaiser. 1981. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc. Natl. Acad. Sci. USA 78:425-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, Y. H. W., Z. Y. Tzecheng, S. C. Wang, W. L. Cheng, and C. W. Chen. 1986. Structural stability of heterologous genes cloned in Streptomyces plasmid pIJ702. Biochem. Biophys. Res. Commun. 140:372-378. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. Z., J. Li, L. Zhou, Y. Zhang, W. Hu, and Q. Chen. 2000. Isolation and identification of myxobacterial sources in China. Acta Microbiol. Sin. 40:652-656. (In Chinese.) [PubMed] [Google Scholar]
- 27.Lu, A., K. Cho, W. P. Black, X. Duan, R. Lux, Z. Yang, H. B. Kaplan, D. R. Zusman, and W. Shi. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55:206-220. [DOI] [PubMed] [Google Scholar]
- 28.Martin, S., E. Sodergren, T. Masuda, and D. Kaiser. 1978. Systematic isolation of transducing phages for Myxococcus xanthus. Virology 88:44-53. [DOI] [PubMed] [Google Scholar]
- 29.Oyaizu, H., and C. R. Woese. 1985. Phylogenetic relationships among the sulfate respiring bacteria, myxobacteria and purple bacteria. Syst. Appl. Microbiol. 6:257-263. [Google Scholar]
- 30.Pigac, J., D. Vujaklija, Z. Toman, V. Gamulin, and H. Schrempf. 1988. Structural instability of a bifunctional plasmid pZG1 and single-stranded DNA formation in Streptomyces. Plasmid 19:222-230. [DOI] [PubMed] [Google Scholar]
- 31.Pradella, S., A. Hans, C. Spröer, H. Reichenbach, K. Gerth, and S. Beyer. 2002. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch. Microbiol. 178:484-492. [DOI] [PubMed] [Google Scholar]
- 32.Qin, Z., M. Shen, and S. N. Cohen. 2003. Identification and characterization of a pSLA2 plasmid locus required for linear DNA replication and circular plasmid stable inheritance in Streptomyces lividans. J. Bacteriol. 185:6575-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rachid, S., K. Gerth, I. Kochems, and R. Müller. 2007. Deciphering regulatory mechanisms for secondary metabolite production in the myxobacterium Sorangium cellulosum So ce56. Mol. Microbiol. 63:1783-1796. [DOI] [PubMed] [Google Scholar]
- 34.Reichenbach, H. 2004. Order VIII. Myxococcales, p. 1059-1143. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 35.Reichenbach, H., and M. Dworkin. 1992. The myxobacteria, p. 3416-3487. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, NY.
- 36.Reichenbach, H., and G. Höfle. 1993. Production of bioactive secondary metabolites, p. 347-397. In M. Dworkin and D. Kaiser (ed.), Myxobacteria. American Society for Microbiology, Washington, DC.
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spröer, C., H. Reichenbach, and E. Stackebrandt. 1999. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 49:1255-1262. [DOI] [PubMed] [Google Scholar]
- 41.Stackebrandt, E., R. G. E. Murray, and H. G. Trüper. 1988. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the purple bacteria and their relatives. Int. J. Syst. Bacteriol. 38:321-325. [Google Scholar]
- 42.Stackebrandt, E., and O. Päuker. 2005. Gene sequence heterogeneity of Corallococcus coralloides strains isolated from geographically diverse locations. Environ. Microbiol. 7:1017-1023. [DOI] [PubMed] [Google Scholar]
- 43.Starich, T., and J. Zissler. 1989. Movement of multiple DNA units between Myxococcus xanthus cells. J. Bacteriol. 171:2323-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 45.Vlamakis, H., J. R. Kirby, and D. R. Zusman. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol. Microbiol. 52:1799-1811. [DOI] [PubMed] [Google Scholar]
- 46.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 47.Ward, M. J., and D. R. Zusman. 1999. Motility in Myxococcus xanthus and its role in developmental aggregation. Curr. Opin. Microbiol. 2:624-629. [DOI] [PubMed] [Google Scholar]
- 48.Wu, S. S., and D. Kaiser. 1996. Markerless deletion of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, Z. C., B. Wang, Y. Z. Li, X. Gong, H. Q. Zhang, and P. J. Gao. 2003. Morphologies and phylogenetic classification of cellulolytic myxobacteria. Syst. Appl. Microbiol. 26:104-109. [DOI] [PubMed] [Google Scholar]