Abstract
Biofilms colonizing surfaces inside drinking water distribution networks may provide a habitat and shelter to pathogenic viruses and parasites. If released from biofilms, these pathogens may disseminate in the water distribution system and cause waterborne diseases. Our study aimed to investigate the interactions of protozoan parasites (Cryptosporidium parvum and Giardia lamblia [oo]cysts) and viruses (vaccinal poliovirus type 1, φX174, and MS2) with two contrasting biofilms. First, attachment, persistence, and detachment of the protozoan parasites and the viruses were assessed with a drinking water biofilm. This biofilm was allowed to develop inside a rotating annular reactor fed with tap water for 7 months prior to the inoculation. Our results show that viable parasites and infectious viruses attached to the drinking water biofilm within 1 h and persisted within the biofilm. Indeed, infectious viruses were detected in the drinking water biofilm up to 6 days after the inoculation, while viral genome and viable parasites were still detected at day 34, corresponding to the last day of the monitoring period. Since viral genome was detected much longer than infectious particles, our results raise the question of the significance of detecting viral genomes in biofilms. A transfer of viable parasites and viruses from the biofilm to the water phase was observed after the flow velocity was increased but also with a constant laminar flow rate. Similar results regarding parasite and virus attachment and detachment were obtained using a treated wastewater biofilm, suggesting that our observations might be extrapolated to a wide range of environmental biofilms and confirming that biofilms can be considered a potential secondary source of contamination.
Human enteric viruses and protozoan parasites, such as Cryptosporidium parvum and Giardia lamblia, are known to persist outside their host for a long period of time, and epidemiological studies have shown their ability to cause waterborne outbreaks (17, 25, 36, 38). However, in many cases, the origin and/or the pathogen routes remain undetermined. It has been hypothesized that biofilms may play a role in the accumulation, protection, and dissemination of pathogens through drinking water distribution networks (13, 35, 48). On the one hand, it has been shown that both viruses and protozoan parasites can attach to biofilms (24, 39, 42, 52), and on the other hand, detachment of pieces of biofilm by shearing forces of the moving water has been observed (5, 7, 8, 10, 46, 49). However, a public health risk exists only if biofilm detachment leads to the release of entrapped pathogens that are still infectious. Although many studies on the interactions between drinking water biofilms and pathogenic bacteria have been carried out (3, 6, 9, 12, 37, 53), little is known about the specific interactions of viruses with drinking water biofilms. In a review, Skraber et al. (42) highlighted the lack of quantitative data that precludes concluding whether drinking water biofilms play a significant role or not in the fate of viruses. Since this review, Lehtola et al. (27) have reported the high level of persistence of the feline calicivirus genome in a drinking water biofilm. However, the infectivity was not assessed, which precludes conclusions on the persistence of infectious viruses within a drinking water biofilm. Regarding the specific interactions of Cryptosporidium parvum with drinking water biofilms, few studies have been published. In a lab-scale experiment, Keevil (24) reported the attachment and persistence of high concentrations of Cryptosporidium parvum within a 16-day-old drinking water biofilm 24 h after the inoculation. More recently, Warnecke (52) highlighted the possible role of biofilms in Cryptosporidium dissemination through drinking water networks, but it has to be underlined that the attachment of the parasites was not confirmed by direct examination of the biofilm. Based on this work, Angles et al. (2) proposed to take Cryptosporidium interactions with drinking water biofilm into account for risk assessment. In contrast, no publications could be found on the interactions of Giardia lamblia with drinking water biofilms.
Since pathogen-biofilm interactions are difficult to study in situ (e.g., in the water distribution system), pilot-scale experiments represent an alternative. Different pilots are available for studying pathogen-biofilm interactions, including the rotating annular reactor (RAR). The RAR is a system designed to provide coupon surfaces on which biofilm grows under fluid hydrodynamic conditions representative of water distribution pipelines. This reactor type has been successfully used to study various aspects of biofilms, including structural developments (32, 54), metabolism in biofilms (26), effect of biocides (31, 44), degradation of contaminants (1), and biofilm detachment (7).
The aim of our work was to assess the interactions of two parasites (Cryptosporidium parvum oocysts and Giardia lamblia cysts) and three viruses (vaccinal poliovirus type 1, φX174 [a somatic coliphage], and MS2 [an F-specific coliphage]) with two contrasting biofilms: a drinking water biofilm and a wastewater biofilm. These biofilms, exhibiting diverse bacterial communities, might better represent environmental biofilms than monoculture biofilms. The use of both drinking water and wastewater biofilms in our study aimed to assess to what extent attachment and detachment processes are biofilm dependent. Virus and parasite attachment rates were assessed under a laminar regimen, while detachment rates were examined after changing the flow velocity from laminar to turbulent. In addition, during the drinking water experiment, the persistence of infectious viruses and viable parasites as well as their transfer from the biofilm to the water phase under a laminar regimen were assessed during a 34-day monitoring period.
MATERIALS AND METHODS
Virus and parasites.
A stock suspension of vaccinal poliovirus type 1 Sabin (Lsc 2ab) was prepared by inoculating a monolayer of BGM cells. After 24 h of virus multiplication, the cells were frozen and thawed three times and centrifuged at 6,000 × g for 30 min, and the supernatant was filtered through a 0.22-μm polycarbonate membrane. The concentration of the viral stock was about 8.1 log most probable number of cytopathogenic units (MPNCU)·ml−1. The filtered supernatant was divided into aliquots and kept at −80°C until the spiking day.
Stock suspensions of the somatic coliphage φX174 and the F-specific coliphage MS2 were prepared by inoculating Escherichia coli strain WG5 and Salmonella enterica serovar Typhimurium strain WG49, respectively, at 108 CFU·ml−1. After 18 h of virus multiplication, the suspension was centrifuged (6,000 × g for 30 min) and the supernatant was filtered through a 0.22-μm polycarbonate membrane. The concentrations of the viral stocks were about 9.2 and 9.7 log PFU·ml−1 for φX174 and MS2, respectively. The filtered supernatant was divided into aliquots and kept at −80°C until the spiking day.
The Giardia lamblia and Cryptosporidium parvum (oo)cyst suspensions were supplied by Waterborne, Inc. (New Orleans, LA) at a concentration of 6.1 log (oo)cysts·ml−1 and kept at 4°C until the spiking day.
RAR.
The RAR used for this experiment was the 1320 LJ (BioSurface Technologies Corp., Bozeman, MT). With a capacity of 1 liter, the RAR consists of an outer glass cylinder that encompasses an inner rotating drum comprising 20 removable polycarbonate coupons which support biofilm growth. Each coupon exhibits a colonizing surface of 18.75 cm2. A variable-speed motor located above the reactor controls the rotation of the drum, which in turn controls the shear stress on the coupon. An outer glass cylinder allows maintenance of the reactor at a constant temperature using a cooling water bath (Lauda, Koningshoven, Germany). Sanitization of the reactor was done before each experiment by washing and autoclaving the reactor. The polycarbonate coupons were additionally disinfected with ethanol before insertion into the reactor.
Drinking water biofilm experiment.
A drinking water biofilm was allowed to develop on coupons in an RAR fed with drinking water for 7 months in order to share bacterial communities similar to those on biofilms that naturally develop within water distribution networks. Briefly, the reactor was fed with tap water in an open circuit with a flow rate of 100 ml·min−1, and the overflow was discarded to the sewer. The biofilm was allowed to develop for 218 days under a rotational speed of 30 rpm, which results in a laminar regimen in the RAR (the corresponding Reynolds number is 217 based on Brennen's calculation [4]). This rotational speed of 30 rpm corresponds to a flow of approximately 0.1 m·s−1 in a 100-mm-diameter pipe (B. Warwood, personal communication).
A total drinking water volume of approximately 31.4 m3 passed through the reactor prior to the addition of parasites and viruses. The reactor was kept in the dark in order to limit the growth of phototrophic organisms. During the biofilm development process, the temperature of the reactor was not regulated in order to have the same temperature variations as in the distribution system. Tap water characteristics (obtained using a Hydrolab probe Datasonde 4A instrument; Hach Company, Loveland, CO) are described in Table 1. The chlorine concentration, measured twice a day using N-N-diethyl-p-phenylenediamine (Hach), never exceeded 0.02 mg·liter−1, which corresponds to the detection limit of the method (U.S. Environmental Protection Agency [EPA] method 330.5). Two hundred eighteen days (7 months) after the beginning of the experiment, a complex biofilm developed on the coupons inside the reactor as observed using scanning electron microscopy (data not shown). At day 218 (day 0 of the experiment), the water inflow was stopped. The temperature inside the RAR was stabilized at 10°C in order to avoid the possible effects of temperature variations on the attachment/detachment phenomenon, and the reactor was spiked with viruses (φX174, MS2, and poliovirus). The following day (day 1), the reactor was spiked with parasites (Giardia lamblia cysts and Cryptosporidium parvum oocysts) at the concentrations described in Table 2. A tap water control, kept at 10°C under a constant magnetic stirring (30 rpm), was inoculated in the same way in order to assess the evolution of parasite viability and the inactivation of the viruses in the absence of biofilm. One hour after spiking, viruses and parasites were quantified in the biofilms and in the water phase of the reactor, as well as in the water control. The same analyses were performed 24 h (for viruses and parasites) and 48 h (for viruses) after spiking.
TABLE 1.
Tap water characteristics measured for a period of 6 days before virus spikinga
| Value type | Temperature (°C) | pH | Conductivity (μS·cm−1) | DO (%) | DO (mg·liter−1) |
|---|---|---|---|---|---|
| Avg | 15.2 | 7.67 | 385 | 96.8 | 9.83 |
| SD | 2.7 | 0.05 | 18 | 3.3 | 0.54 |
| Min | 10.6 | 7.52 | 345 | 86.3 | 9.02 |
| Max | 19.4 | 7.76 | 420 | 144.6 | 13.40 |
One measurement was taken per half-hour (n = 281). DO, dissolved oxygen; min, minimum; max, maximum.
TABLE 2.
Quantities of parasites and viruses spiked inside the RARa
| Expt | Quantity of organism (units)
|
||||
|---|---|---|---|---|---|
| Infectious vaccinal poliovirus type 1 (log MPNCU) | φX174 (log PFU) | MS2 (log PFU) | G. lamblia cysts (log units) | C. parvum oocysts (log units) | |
| Wastewater | 6.57 | NAb | NA | 6.02 | 6.03 |
| Drinking water | 5.04 | 5.58 | 6.11 | 5.84 | 6.00 |
Total reactor volume is 1 liter.
NA, not applicable, as φX174 and MS2 were not inoculated into the RAR during the wastewater experiment.
In order to assess the possible transfer of viruses and parasites from the biofilm to the water phase, the RAR was drained once a week in order to eliminate unadsorbed particles. Briefly, 50 liters of tap water was introduced inside the reactor (from the bottom) at a low flow rate (100 ml·min−1, corresponding to a resident time of 10 min) in order to limit surface tension forces that could have affected cell attachment/detachment. Each draining operation lasted 8 h, and the overflow was collected in a 50-liter carboy. The total volume of collected water containing unadsorbed or detached particles was further filtered on an Envirocheck HV cartridge (Pall Gelman, Ann Arbor, MI) for parasite quantification, giving the amount of parasites present in the reactor prior to the draining operation. During the 8 h of the draining operation, the RAR was maintained at 10°C under a laminar flow rate (30 rpm) in the dark. Biofilms were analyzed once a week, before each draining operation. In total, six draining operations were performed, at days 3, 7, 14, 21, 28, and 35. At day 42, a turbulent regimen (380 rpm) was applied for 1 min inside the reactor in order to assess the detachment of remaining particles from the biofilm to the water phase. A rotational speed of 380 rpm inside the RAR corresponds to a turbulent regimen in the RAR (the corresponding Reynolds number is 2,752, based on Brennen's calculation [4]). This rotational speed of 380 rpm corresponds to a flow of approximately 0.9 m·s−1 in a 100-mm-diameter pipe (B. Warwood, personal communication). After the turbulent regimen, the total volume of the RAR (1 liter) was centrifuged at 1,250 × g for 15 min. One hundred forty milliliters of the supernatant was concentrated using a Centricon filter (Millipore, Billerica, MA) and analyzed for the presence of viruses. The pellet was eluted with 1% pasta beef extract (Difco, Sparks, MD)-0.4% glycine (Bio-Rad, Richmond, CA) at a pH of 9.5 and centrifuged at 1,250 × g for 15 min. The eluate was analyzed for the presence of viruses, whereas the pellet was further analyzed for the presence of parasites.
Wastewater biofilm experiment.
The RAR was fed with 25 liters of biologically treated wastewater with a flow rate of 100 ml·min−1 for 14 days in a closed circuit while the regimen inside the reactor was set to laminar (30 rpm) and the temperature was set to 4°C in order to limit bacterial inactivation. The RAR was maintained at 4°C during the whole experiment. Two hundred fifty milligrams of glucose was added to the wastewater as a source of carbon for the bacteria in order to keep them metabolically active. Fourteen days after the beginning of the experiment, a biofilm had developed on the coupons inside the reactor. The colonization of the coupons was observed using a scanning electron microscope, as for the drinking water biofilm experiment (data not shown). Then, the RAR was rinsed with phosphate-buffered saline (PBS) (pH 7.4) (Sigma, St. Louis, MO), filled with PBS, and kept at a constant laminar flow rate. The reactor was spiked with poliovirus and parasites (Giardia lamblia cysts and Cryptosporidium parvum oocysts) stock suspensions with the quantities described in Table 2. Bacteria, viruses, and parasites were quantified in duplicate in the biofilms and in the water phase of the reactor 1, 24, and 48 h after the reactor was spiked. In order to assess the detachment of particles when the flow velocity inside the reactor was increasing, samples were taken in triplicate at day 2 before and after applying a turbulent regimen (380 rpm for 1 min).
Biofilm and water sampling.
The biofilm was scraped from the 18.75-cm2 polycarbonate coupon using a cell scraper (Sarstedt, Germany) and resuspended in 5 ml (in the case of the drinking water biofilm) or 10 ml (in the case of the wastewater biofilm) of eluate (1% pasta beef extract [Difco, Sparks, MD]-0.4% glycine [Bio-Rad, Richmond, CA] at pH 9.5). The resulting biofilm suspension was vortexed at 2,600 rpm for 2 min. For the experiment with drinking water biofilm, the total volume of eluate (5 ml) was centrifuged at 1,250 × g for 15 min. Giardia lamblia and Cryptosporidium parvum (oo)cysts were quantified in the pellet, while the supernatant was processed for virus detection after the pH was neutralized with 1 M HCl. In the case of the wastewater biofilm, 2 ml was analyzed for total and culturable bacteria and 5 ml was processed for parasite detection. The remaining 3 ml was centrifuged at 3,000 × g for 5 min, and the pH of the supernatant was neutralized with 1 M HCl before it was frozen and saved for poliovirus detection. The water phase of the reactor was analyzed directly, but during the drinking water experiment, 70 ml of water was additionally concentrated into 350 μl using a 100-kDa-cutoff Centricon centrifugal filter for virus quantification, and the total volume after draining (50 liters) was concentrated into 10 ml using an Envirocheck HV cartridge (Pall Gelman, Ann Arbor, MI) for parasite quantification.
Quantification of parasites, viruses, and bacteria. (i) Quantification and viability assessment of Giardia lamblia and Cryptosporidium parvum (oo)cysts.
In biofilm eluates or water samples, parasite quantification was carried out by epifluorescence microscopic observation (DMRB; Leica, Mannheim, Germany) according to EPA method 1623 (50) using a fluorescein-labeled double monoclonal antibody solution for the simultaneous detection of Giardia lamblia cysts and Cryptosporidium parvum oocysts and 4′,6′-diamidino-2-phenylindole (DAPI) staining (Waterborne, Inc., New Orleans, LA). In the case of the wastewater experiment, the biofilm eluate and 10 ml of the water phase (PBS) were analyzed after immunomagnetic separation (Dynabeads; Invitrogen, Oslo, Norway), and the concentrate obtained (100 μl) was deposited on a well slide for microscopic observation. In the case of the drinking water experiment, the pellet of the eluate, resuspended in 1 ml of PBS, was recentrifuged at 1,250 × g for 5 min. The supernatant was discarded and the pellet resuspended in 200 μl of PBS, and 100 μl was deposited on a well slide for microscopic observation. In addition, viability assessment was carried out using a 0.1-mg·ml−1 propidium iodide solution (Invitrogen-Molecular Probes, Eugene, OR). For the water phase, 10 ml or the total volume of the eluate after concentration of the 50 liters after the draining operation on an Envirochek cartridge (Pall Gelman, Ann Arbor, MI) was centrifuged at 1,250 × g for 15 min or 30 min, respectively. The supernatant was discarded and the pellet resuspended in 200 μl of PBS, and 100 μl was used for microscopic observation. For the water phase, the theoretical limits of parasite detection assuming 100% recovery were 2.0 log units·liter−1 when analyzing 10 ml directly and 0.0 log units·liter−1 when analyzing the totality of the reactor volume after each draining. For biofilm analysis, the limit of detection was −0.97 log units·cm−2.
(ii) Quantification of bacteriophages φX174 and MS2.
Infectious φX174 and MS2 were quantified in the drinking water biofilm eluate and in the water phase using the bacterial host strains Escherichia coli WG5 and Salmonella enterica serovar Typhimurium WG49, respectively, according to the standardized methods described in ISO/FDIS 10705-2 (22) and ISO/FDIS 10705-1 (21), respectively. Concerning the water phase, the theoretical limits of phage detection, assuming 100% recovery, were 2.3 log PFU·liter−1 when analyzing 5 ml directly and 1.7 log PFU·liter−1 when analyzing 100 μl from the 350-μl concentrate. For biofilm analysis, the limit of detection was −0.5 log PFU·cm−2.
(iii) Quantification of infectious poliovirus by cell culture.
Ten milliliters of water samples and a 1-ml aliquot of the biofilm eluate were treated with 10% antibiotic and antimycotic solution (Sigma, St. Louis, MO) for 2 h at 37°C. Samples were then log diluted in Ca2+- and Mg2+- free Dulbecco's phosphate-buffered saline (Gibco, Grand Island, NY). Quantification of infectious poliovirus type 1 was performed in 96-well microplates using BGM cells. Fifty microliters of three successive log-diluted suspensions for each analyzed sample was added to 200 μl of 7.5 × 104 cells·ml−1 in minimum essential medium solution (Gibco, Paisley, United Kingdom) supplemented with 2% fetal calf serum (Gibco, Paisley, United Kingdom). After 6 days' incubation at 37°C under a 5%-CO2 atmosphere, the plates were examined under the microscope and the cytopathogenic effects were quantified, allowing calculation of the MPNCU (29). The theoretical limits of detection in water and biofilm, assuming 100% recovery, were 2.0 log MPNCU·liter−1 and −0.5 log MPNCU·cm−2, respectively.
(iv) Quantification of poliovirus genome by reverse transcription and qPCR.
One hundred forty microliters of sample (water, concentrate, or biofilm eluate) was mixed with 560 μl of AVL lysis buffer (Qiamp viral RNA kit; Qiagen, Hilden, Germany). The mix was pulse-vortexed and left for 10 min at room temperature. The total volume was then homogenized through a QIAshredder column (Qiagen, Hilden, Germany) by centrifuging at 6,000 × g for 1 min. The filtrate was transferred to a new 2-ml tube, and 560 μl of ethanol was added. Viral RNA was further extracted using the Qiamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Hence, 60 μl of final extracted RNA solution was obtained. Each sample was analyzed undiluted and log diluted (10 μl in 90 μl of TE buffer, consisting of 10 mM Tris-HCl at pH 8 and 1 mM EDTA). The reverse transcription step was applied to 5 μl of extracted RNA (or log-diluted extracted RNA) by adding it to a mix (TaqMan reverse transcription reagents; Applied Biosystems, Branchburg, NJ) containing 1× TaqMan RT buffer, 5.5 mM of MgCl2, 500 μM of each deoxynucleoside triphosphate, 2.5 μM of random hexamers, 0.4 U·μl−1 of RNase inhibitor, 1.25 U·μl−1 of Multiscribe reverse transcriptase, and 0.1 μg·μl−1 of T4 gene 32. Reverse transcription was performed in a Whatman Biometra thermocycler at 20°C for 10 min followed by 42°C for 50 min. RNA-DNA hybrids were denatured, and reverse transcriptase was inactivated by heating to 95°C for 5 min. The resulting cDNA of the poliovirus genome was quantified by quantitative PCR (qPCR) using the primers and probe described by Monpoeho et al. (30). The primers and probe, unchanged from the original published method (except for the BHQ quencher), were as follows: reverse primer Ev1 (5′-GATTGTCACCATAAGCAGC-3′), forward primer Ev2 (5′-CCCCTGAATGCGGCTAATC-3′), and Ev-probe (5′-FAM-CGGAACCGACTACTTTGGGTGTCCGT-BHQ-3′), all synthesized by Eurogentec (San Diego, CA). For the qPCR step, 5 μl of cDNA was added to 20 μl of a mixture (Eurogentec, San Diego, CA) containing 1× reaction buffer, 500 nM of Ev1 primer, 400 nM of Ev2 primer, and 120 nM of Ev-P probe. The PCR assay was performed in an ABI Prism 7500Fast real-time PCR system (Applied Biosystems). The activity of the HotGoldStar DNA polymerase was released by heating it to 95°C for 10 min. Samples were then submitted to 45 cycles (10 s at 95°C, 30 s at 55°C, and 15 s at 72°C). Real-time fluorescence was measured during the extension step, every cycle at 55°C, and analyzed using the SDS v1.3.1 software (Applied Biosystems). TE dilution buffer and water were used as negative controls. A linear relationship was observed between the CT values (y) and the numbers of infectious poliovirus over a 6-log concentration range (101 to 106 MPNCU·ml−1). The calculated linear regression (y = −3.49 x + 39.39; r2 = 0.998) was used to transform each CT value into PCR units.
(v) Quantification of total and culturable bacteria in wastewater biofilm.
Total bacteria were quantified after SYBR green staining. Briefly, 1 ml of log-diluted biofilm suspension was mixed with 10 μl of 10× SYBR green I (Molecular Probes, Eugene, OR). After 10 min in the dark, 100 μl of the suspension was filtered on a 0.22-μm-pore-size polycarbonate filter (Millipore, Tullagreen, Ireland). After the filter was rinsed twice, the total bacteria were enumerated by counting at least 30 random fields or 300 bacteria under blue excitation light at magnification ×1,000 using an epifluorescence microscope (DMRB; Leica, Mannheim, Germany) equipped with an ocular grid. Heterotrophic plate count bacteria and culturable thermotolerant coliforms were quantified in diluted wastewater biofilm suspensions using plate count agar medium and m-FC medium, respectively, according to standards ISO 6222 (19) and ISO 9308-1 (20).
Statistical analysis.
All statistical tests were performed using SigmaStat (version 2.03; SPSS, Inc.).
RESULTS
Attachment to wastewater and drinking water biofilms.
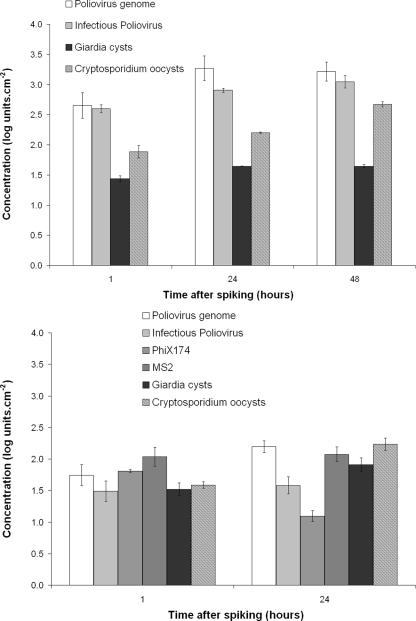
Viruses and parasites were detected in both drinking water and wastewater biofilms (Fig. 1) 1 h after spiking with the initial quantities given in Table 2. In the drinking water biofilm, parasites and viruses were detected at concentrations of 1.52 ± 0.10 (Giardia lamblia), 1.58 ± 0.05 (Cryptosporidium parvum), 1.49 ± 0.16 (infectious poliovirus), 1.74 ± 0.17 (poliovirus genome), 1.81 ± 0.02 (φX174), and 2.04 ± 0.15 (MS2) log units·cm−2 (n = 2). In the wastewater biofilm, parasites and viruses were detected at concentrations of 1.44 ± 0.05 (Giardia lamblia), 1.89 ± 0.10 (Cryptosporidium parvum), 2.60 ± 0.07 (infectious poliovirus), and 2.65 ± 0.21 (poliovirus genome) log units·cm−2 (n = 2). A difference in viral attachment was observed between the two contrasting biofilms. Indeed, the concentration of poliovirus was higher in the wastewater biofilm than in the drinking water biofilm (Mann-Whitney test, P < 0.05). On the contrary, concentrations of parasites were similar in the two biofilms (Mann-Whitney test, P > 0.05). In both the drinking water and the wastewater biofilms, most of the attachment occurred within 1 h (Fig. 1).
FIG. 1.
Virus and parasite concentrations in wastewater biofilm (upper panel) and drinking water biofilm (lower panel) in log units per cm2 plotted against the time after spiking into the reactor (n = 2).
Persistence in the drinking water biofilm.
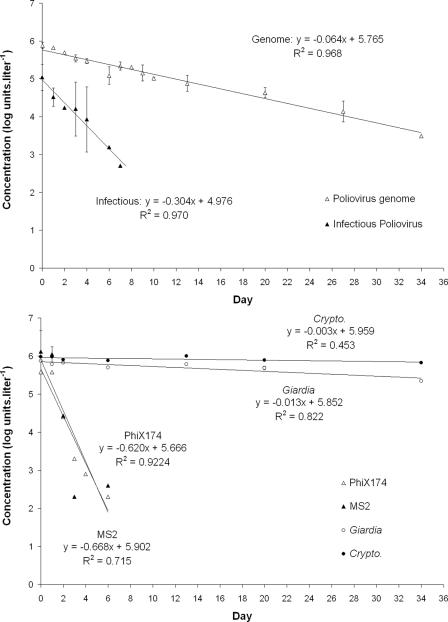
Persistence of parasites and viruses in the drinking water biofilm was assessed during a 34-day monitoring period. Results are presented in Fig. 2A. It is notable that draining operations were conducted once a week (at days 3, 7, 14, 21, 28, and 35) in order to remove unadsorbed particles. In the drinking water biofilm, infectious viruses (poliovirus, φX174, and MS2) were detected until day 6 while the poliovirus genome could be detected until day 34. Similarly, in the water control without biofilm (Fig. 3), no infectious viruses could be detected after day 6 (φX174 and MS2) or day 7 (infectious poliovirus) while the poliovirus genome was still detected at day 34. Parasites could be detected in the drinking water biofilm from day 1 (1 h after the parasites were spiked) to day 34 at concentrations that remained similar during the time of the experiment. Indeed, after day 1, the Giardia lamblia/Cryptosporidium parvum ratio remained constant (average on raw data = 0.51 ± 0.07; n = 5).
FIG. 2.
Evolution of virus and parasite concentrations in the drinking water biofilm (A) or corresponding water phase (B); all samples were analyzed in duplicate except for parasites in the water phase, for which the total volume of the pilot was analyzed after concentration. Draining operations consisted in rinsing the reactor with 50 liters of tap water (100 ml·min−1 for 8 h under laminar flow). *, at day 27 (in biofilm) or day 6 (in drinking water), quantification of parasites was not determined.
FIG. 3.
Concentrations of poliovirus (infectious and genome), infectious bacteriophages (φX174 and MS2), and viable parasites (Giardia lamblia and Cryptosporidium parvum) in the drinking water control (without the presence of biofilm) plotted against time (n = 2, except for bacteriophages, where n = 1).
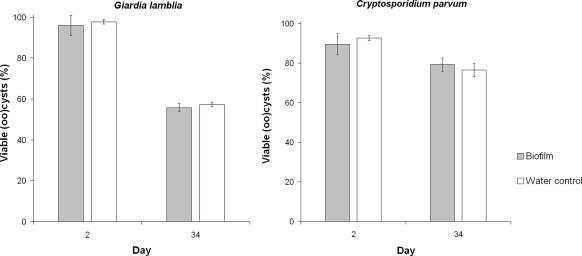
Between day 2 and day 34, viability of (oo)cysts attached to the biofilm decreased from 96% to 56% and from 90% to 79% for Giardia lamblia and Cryptosporidium parvum, respectively (Fig. 4). Similar results were observed in the water control without biofilm (Fig. 4), where viability of (oo)-cysts decreased from 98% to 57% and 93% to 76% between day 2 and day 34 for Giardia lamblia and Cryptosporidium parvum, respectively.
FIG. 4.
Percentages of viable Giardia lamblia and Cryptosporidium parvum (oo)cysts in the drinking water control compared to those in the drinking water biofilm at day 2 (24 h after parasite spiking day) and day 34 (n = 2).
Detachment from the drinking water biofilm under laminar regime.
In addition to the biofilm monitoring, concentrations of the parasites and viruses inoculated into the RAR were monitored in the water phase of the reactor under the laminar regime (Fig. 2B). Starting at day 3, draining operations were performed at days 3, 7, 14, 21, 28, and 35 in order to remove unadsorbed particles from the reactor and to assess the detachment of parasites and viruses occurring between two draining steps from the biofilm to the water phase under the laminar regime. All infectious viral particles could be detected in the water phase until day 2, where the concentrations were 4.77, 3.57, and 2.76 log units·liter−1 for poliovirus, φX174, and MS2, respectively. Immediately after each draining operation (starting at day 3), concentrations of all infectious viruses were below the detection limit in the water phase of the reactor (data not shown). Thus, it can be considered that draining operations successfully removed unadsorbed particles. Three days later (day 6), infectious poliovirus and φX174 were again detected at concentrations of 2.00 and 3.04 log units·liter−1, respectively. After the second draining operation (at day 7), no infectious viral particles could be detected in the water phase until the end of the experiment. In contrast, the poliovirus genome was detected until day 13 in the water phase and could not be detected afterward.
Giardia lamblia cysts and Cryptosporidium parvum oocysts could be detected in the 50 liters of water obtained after each draining operation. Detected parasites represent the number of parasites released from the biofilm between two draining operations. Both parasites were detected until day 34 in the water phase. Concentrations decreased regularly from day 2 (4.38 and 4.35 log units·liter−1 for Giardia lamblia and Cryptosporidium parvum, respectively) to day 34 (0.75 and 2.25 log units·liter−1 for Giardia lamblia and Cryptosporidium parvum, respectively).
Detachment from biofilms as a result of regimen change from laminar to turbulent.
Detachment of parasites and viruses from the two contrasting biofilms was assessed after a turbulent regimen (380 rpm for 1 min) was applied. First, during the drinking water experiment, a turbulent regimen was applied at day 42 (1 week after the last draining operation, which occurred at day 35). After the change in flow velocity, the poliovirus genome, Giardia lamblia, and Cryptosporidium parvum (oo)cysts were detected in the water phase at concentrations of 3.62, 5.11, and 5.26 log units·liter−1, respectively. After the water phase (1 liter) has been centrifuged at 1,250 × g for 15 min, poliovirus genome was detected in the supernatant at a concentration of 1.79 log PCR units·liter−1 and in the pellet at a concentration of 3.62 log PCR units·liter−1. None of the spiked infectious viruses (poliovirus, φX174, and MS2) could be detected in the water phase either before or after the turbulent regimen was applied. Notably, Giardia lamblia and Cryptosporidium parvum (oo)cysts were still detected within the drinking water biofilm after the turbulent regimen at concentrations of 1.30 and 1.90 log units·cm−2, respectively.
As with the drinking water experiment, a turbulent regimen (380 rpm for 1 min) was applied during the wastewater experiment. Table 3 presents the concentrations of parasites and viruses in the water phase and in the biofilm before and after the flow velocity was increased. Detachment varied from 64% (poliovirus genome) to 88% (Giardia lamblia). It is interesting to note that the genome with 64% of release underestimated infectious poliovirus detachment (75%).
TABLE 3.
Particle concentrations in wastewater biofilms and corresponding water phase before and after applying a turbulent regime for 1 min (from 30 to 380 rpm)
| Microorganism | Size | Concn in wastewater biofilm (log units·cm−2)a:
|
% Detachmentb | Concn in water phase (log units·liter−1)c:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Before turbulent regimen
|
After turbulent regimen
|
Before turbulent regimen
|
After turbulent regimen
|
|||||||
| Avg | SD | Avg | SD | Avg | SD | Avg | SD | |||
| Giardia | 10 μm | 2.02 | 0.49 | 1.10 | 0.66 | 88 | 4.40 | 3.32 | 5.48 | 0.25 |
| Cryptosporidium | 5 μm | 2.67 | 0.51 | 1.78 | 0.67 | 87 | 4.40 | 0 | 5.59 | 0.16 |
| Total bacteria | 1 μm | 7.54 | 0.09 | 6.88 | 0.08 | 78 | ND | NA | ND | NA |
| Thermotolerant coliforms | 1 μm | 3.26 | 0.09 | 2.62 | 0.03 | 77 | 4.10 | NA | 5.93 | NA |
| Heterotrophic plate count bacteria | 1 μm | 5.80 | 0.04 | 5.22 | 0.03 | 74 | 7.85 | NA | 9.02 | NA |
| Infectious poliovirus | 30 nm | 3.05 | 0.10 | 2.44 | 0.01 | 75 | 5.58 | 0.37 | 5.91 | 0.09 |
| Poliovirus genome | 3.21 | 0.16 | 2.77 | 0.19 | 64 | 5.57 | 0.10 | 6.26 | 0.27 | |
For average concentrations, n = 3.
Percentages of detachment were calculated according to the following formula: % detachment = (1 − [10C after/10C before]) × 100, where “C after” and “C before” are the concentrations in wastewater biofilms before and after the turbulent regimen.
n = 2 for all parameters except for the heterotrophic plate count bacteria and thermotolerant coliforms, where n = 1. ND, not determined; NA, not applicable.
DISCUSSION
Our results support other studies showing that Cryptosporidium oocysts (24, 41, 52) and viruses (42) are able to attach to biofilms. In addition, this study demonstrates that Giardia lamblia cysts are able to attach and persist in a drinking water biofilm. Concerning viruses, our results are in agreement with those of Hock and Botzenhart (C. Hock and K. Botzenhart, presented at the 3rd IWA World Water Congress, Melbourne, Australia, 2002) who showed that infectious MS2, φX174, and poliovirus type 1 were able to attach to drinking water biofilms within 1 h. In their experiment, however, concentrations decreased in the biofilm after 24 h, but no information on chlorine, which can enhance viral inactivation, was given. It can be underlined that under our experimental conditions, the rate of attachment of MS2 to the drinking water biofilm was higher than the attachment rate of φX174 or poliovirus type 1, which attached similarly. This observation could be explained by the difference in surface properties for each virus. MS2 phage exhibits an isoelectric point of 3.9, whereas φX174 and poliovirus present higher isoelectric points of 6.8 and 6.6, respectively (14, 16, 51). Giardia and Cryptosporidium exhibit low isoelectric points of 2.2 to 3.6 and 2.2 to 3.9, respectively (11, 18, 34). MS2 phage and parasites both express a negative charge at neutral pH. Nevertheless, parasites adsorbed significantly less than MS2 1 h after inoculation (difference = 0.5 log unit·cm−2) but similarly after 24 h. Attachment of particles depends not only on surface properties but also on their size. The smallest particles may find adsorption sites more rapidly on complex and irregular surfaces than larger ones.
The highest attachment rate for viruses and parasites occurred within the first hour after inoculation regardless of the nature of the biofilm. For instance, in the drinking water biofilm, the lowest attachment of 1.49 log units·cm−2 was observed after 1 h for infectious poliovirus. By comparison, the maximum increase between 1 and 24 h was observed for Cryptosporidium parvum, with an increase of 0.65 log unit·cm−2. It can be noticed that for both viruses and parasites, the increase in the concentration in the biofilms that occurred during the first hour of the experiment was associated with a decrease in the concentration in the water phase of the reactor, corroborating the transfer of viruses and parasites from the water to the biofilm.
A slight difference in attachment between the two contrasting biofilms was observed for poliovirus but not for parasites. Indeed, poliovirus adsorption was higher with the wastewater biofilm than with the drinking water biofilm, suggesting that viral adsorption depends on the structure/composition of the biofilm. Our wastewater biofilm may exhibit more voids and cavities than our drinking water biofilm, offering larger adsorption sites for smaller particles like viruses (47). Conversely, Giardia lamblia and Cryptosporidium parvum (oo)cysts attach similarly to both biofilms. Their attachment may be less influenced by the presence of voids and cavities due to their size being larger than that of viruses. The concentration of infectious viruses, including poliovirus, φX174, and MS2, decreased in the drinking water biofilm, and no infectious particles could be detected after day 6. Similarly, infectious viral particles could not be detected in the water control after day 6 (φX174 and MS2) or day 7 (infectious poliovirus). It is worth underlining that under our dynamic conditions, the decrease in the infectious virus concentration in the biofilm corresponds to the cumulative effect of inactivation and detachment. Also, the water control was analyzed daily, whereas the biofilm was analyzed weekly due to the limitation in the number of coupons available. The comparison of viral inactivation in the water and in the biofilm is therefore problematic. Results published earlier (41) showed that wastewater biofilms protect viruses from inactivation. The level of viral protection of a biofilm may depend on parameters such as the composition/thickness/structure of the biofilm. Moreover, it is important to note that a protective effect of biofilms has commonly been demonstrated in the presence of antimicrobial agents (28, 45). Since during our experiment, the chlorine concentration was below the detection threshold (<0.02 mg·liter−1), it is not possible to state whether the drinking water biofilm would have played a protective role in the presence of disinfectants.
Infectious poliovirus could not be detected in the biofilm after day 6, whereas the poliovirus genome was still detected at day 34, corresponding to the last day of the monitoring period. Our results are in agreement with those of Lehtola et al. (27), who showed a high persistence of the canine calicivirus genome, used as a surrogate for noroviruses in a drinking water biofilm. The authors detected the norovirus genome for 3 weeks, corresponding to the length of their experiment. However, our results show that only a part of the total virus abundance (estimated on the basis of the genome quantification) corresponds to infectious particles (15, 40). Since the genome was detected much longer than infectious particles, especially in biofilm, our results raise the question of the significance of detecting viral genomes in drinking water biofilms for risk assessment. According to our results, in biofilms, viral inactivation seems to occur faster than the corresponding genome degradation, suggesting that molecular techniques overestimate the risk associated with the presence of infective particles. However, it cannot be excluded that deeply entrapped infectious particles could not be eluted and therefore remained undetectable by the culturing method. In order to test this hypothesis, further investigations should be performed (e.g., testing different elution protocols in order to improve viral recovery). As with the poliovirus genome, parasites were detected within the drinking water biofilm during the whole experiment. The evaluation of parasite persistence was based on a viability assessment using the propidium iodide exclusion technique. Viable (oo)cysts are considered to be potentially infectious, though it is known that the dye exclusion technique can overestimate the infectivity of Cryptosporidium oocysts assessed by using cell culture (43). The propidium iodide technique thus gives only a rough estimate of infectivity. After 34 days, 56% and 79% of (oo)cysts are still viable in biofilm for Giardia lamblia and Cryptosporidium parvum, respectively. At day 34, the percentages of viable (oo)cysts are similar in the biofilm and in the water control, suggesting that a similar viability loss occurred. Thus, as with poliovirus, biofilm cannot be considered a shelter for these parasites. Moreover, our results confirm that Cryptosporidium oocysts survive longer than Giardia cysts (23, 33) in both water and biofilm. In the drinking water experiment, viruses and parasites that were detected in the water phase 1 week after each draining step corresponded to those that were released from the biofilm during this period. No infectious poliovirus or bacteriophages could be detected just after the first draining step, while they were detectable 1 week later, so that it is clear that a release of infectious viruses from the biofilm occurred during this period. Similarly, both parasites were detected in the water phase after each draining step, corroborating the release of (oo)cysts from the biofilm during the whole length of the experiment. The concentration of released Cryptosporidium parvum oocysts measured in the water phase was always higher than the concentration of Giardia lamblia cysts. This may be explained by the higher concentration of Cryptosporidium parvum than of Giardia lamblia in the biofilm. The viability of detached parasites was not assessed, since they underwent an additional treatment (a concentration step using an Envirocheck cartridge followed by an elution and a concentration by centrifugation) compared to the (oo)cysts obtained from the biofilm or from the water control (for which only one concentration by centrifugation was needed), which could have distorted the results, making comparisons problematic. Nevertheless, considering that more than half of the (oo)cysts present in the biofilm after 34 days were still viable, it is likely that part of the released (oo)cysts were viable as well.
Our results confirmed that an increase in the flow rate from the laminar to the turbulent regimen leads to a massive transfer of viruses and parasites from the biofilm to the water phase. For instance, during the drinking water experiment, no poliovirus genome was detected in the water phase just before the application of a turbulent regimen, while it was detected after. In the wastewater experiment, the simultaneous increase in bacterial concentrations (heterotrophic plate count bacteria and thermotolerant coliforms) in the water phase combined with the decrease in these bacterial concentrations in the biofilm confirms the occurrence of sloughing events and suggests that biofilm-associated viruses and parasites are simultaneously released.
This is in agreement with previous results showing a massive detachment of biofilm after a turbulent regimen is applied (46, 49). Furthermore, the poliovirus genome concentration was 68 times higher in the pellet of the low-speed-centrifuged water than in the supernatant, indicating that most of the poliovirus genome was associated with pieces of biofilm able to settle at a low centrifugation speed. Thus, viral particles may be released as single particles, but our results suggest that most of the released viruses are associated with pieces of biofilm. No poliovirus genome was detected in the drinking water biofilm after the turbulent regimen, while conversely, Giardia lamblia cysts and Cryptosporidium parvum oocysts were still detected.
In conclusion, this work supports other studies showing that pathogenic viruses and parasites can attach to drinking water biofilms. Though under our experimental conditions we have not observed a protective effect of the drinking water biofilm toward viruses and parasites, we confirm that attached viable parasites and infectious viruses can be released from biofilms either after a change in the flow velocity from laminar to turbulent or under a laminar regimen for a long period of time. Our work highlights that the detection of viral genome in biofilm is not sufficient to assess a risk associated with the presence of infectious particles. Our study also brings information on Giardia lamblia cyst interactions with a drinking water biofilm. Regarding the similar behaviors of parasites and viruses toward two contrasting, complex matrices, our results might represent a wide range of environmental biofilms. Mechanisms involved in pathogen-biofilm interactions should be further investigated. Also, lower concentrations of pathogenic viruses and parasites have to be tested in order to approach the conditions that might occur in drinking water networks.
Acknowledgments
This work was funded by the FNR (National Research Fund) from Luxembourg (SECAL Program, KAWA Project; FNR/03/07/07).
We thank Nicolas Bonjean, Delphine Collard, and Julie Mathu for their excellent technical assistance, Christophe Bouillon for scanning electron microscopy observations, Thomas Udelhoven for statistical advice, and Romain Muno for kind collaboration at the wastewater treatment plant of Pétange.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Alleman, B. C., B. E. Logan, and R. L. Gilbertson. 1995. Degradation of pentachlorophenol by fixed films of white rot fungi in rotating tube bioreactors. Water Res. 29:61-67. [Google Scholar]
- 2.Angles, M. L., J. P. Chandy, P. T. Cox, I. H. Fisher, and M. R. Warnecke. 2007. Implications of biofilm-associated waterborne Cryptosporidium oocysts for the water industry. Trends Parasitol. 23:352-356. [DOI] [PubMed] [Google Scholar]
- 3.Braganra, S. M., N. F. Azevedo, L. C. Simoes, C. W. Keevil, and M. J. Vieira. 2007. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 55:387-393. [DOI] [PubMed] [Google Scholar]
- 4.Brennen, C. E. 1994. Radial and rotordynamic forces. In C. E. Brennen (ed.), Hydrodynamics of pumps. Oxford University Press and Concepts NREC, White River Junction, VT.
- 5.Camper, A. K., W. L. Jones, and J. T. Hayes. 1996. Effect of growth conditions and substratum composition on the persistence of coliforms in mixed-population biofilms. Appl. Environ. Microbiol. 62:4014-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauret, C., C. Volk, R. Creason, J. Jarosh, J. Robinson, and C. Warnes. 2001. Detection of Aeromonas hydrophila in a drinking-water distribution system: a field and pilot study. Can. J. Microbiol. 47:782-786. [PubMed] [Google Scholar]
- 7.Choi, Y. C., and E. Morgenroth. 2003. Monitoring biofilm detachment under dynamic changes in shear stress using laser-based particle size analysis and mass fractionation. Water Sci. Technol. 47:69-76. [PubMed] [Google Scholar]
- 8.Cloete, T. E., D. Westaard, and S. J. van Vuuren. 2003. Dynamic response of biofilm to pipe surface and fluid velocity. Water Sci. Technol. 47:57-59. [PubMed] [Google Scholar]
- 9.Dailloux, M., M. Albert, C. Laurain, S. Andolfatto, A. Lozniewski, P. Hartemann, and L. Mathieu. 2003. Mycobacterium xenopi and drinking water biofilms. Appl. Environ. Microbiol. 69:6946-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly, B., W. B. Betts, A. P. Brown, and J. G. O'Neill. 1998. Bacterial loss from biofilms exposed to free chlorine. Microbios 96:7-21. [PubMed] [Google Scholar]
- 11.Drozd, C., and J. Schwartzbrod. 1996. Hydrophobic and electrostatic cell surface properties of Cryptosporidium parvum. Appl. Environ. Microbiol. 62:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emtiazi, F., T. Schwartz, S. M. Marten, P. Krolla-Sidenstein, and U. Obst. 2004. Investigation of natural biofilms formed during the production of drinking water from surface water embankment filtration. Water Res. 38:1197-1206. [DOI] [PubMed] [Google Scholar]
- 13.Ford, T. E. 1999. Microbiological safety of drinking water: United States and global perspectives. Environ. Health Perspect. 107:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujito, B. T., and C. D. Lytle. 1996. Elution of viruses by ionic and nonionic surfactants. Appl. Environ. Microbiol. 62:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gassilloud, B., L. Schwartzbrod, and C. Gantzer. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl. Environ. Microbiol. 69:3965-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerba, C. P. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133-168. [DOI] [PubMed] [Google Scholar]
- 17.Howe, A. D., S. Forster, S. Morton, R. Marshall, K. S. Osborn, P. Wright, and P. R. Hunter. 2002. Cryptosporidium oocysts in a water supply associated with a cryptosporidiosis outbreak. Emerg. Infect. Dis. 8:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, B. M., and C. Huang. 2004. Influence of ionic strength and pH on hydrophobicity and zeta potential of Giardia and Cryptosporidium. Colloid Surf. A: Physicochem. Eng. Aspect 201:201-206. [Google Scholar]
- 19.International Organization for Standardization. 1999. Water quality—enumeration of culturable micro-organisms—colony count by inoculation in a nutrient agar culture medium. ISO 6222. International Organization for Standardization, Geneva, Switzerland.
- 20.International Organization for Standardization. 1990. Water quality—detection and enumeration of coliform organisms, thermotolerant coliform organisms and presumptive Escherichia coli—part 1: membrane filtration method. ISO 9308-1. International Organization for Standardization, Geneva, Switzerland.
- 21.International Organization for Standardization. 2001. Water quality—detection and enumeration of bacteriophages—part 1: enumeration of F-specific RNA bacteriophages. ISO/FDIS 10705-1. International Organization for Standardization, Geneva, Switzerland.
- 22.International Organization for Standardization. 2001. Water quality—detection and enumeration of bacteriophages—part 2: enumeration of somatic coliphages. ISO/FDIS 10705-2. International Organization for Standardization, Geneva, Switzerland.
- 23.Johnson, D. C., C. E. Enriquez, I. L. Pepper, T. L. Davis, C. P. Gerba, and J. B. Rose. 1997. Survival of Giardia, Cryptosporidium, Poliovirus and Salmonella in marine waters. Water Sci. Technol. 35:261-268. [Google Scholar]
- 24.Keevil, C. W. 2003. Rapid detection of biofilms and adherent pathogens using scanning confocal laser microscopy and episcopic differential interference contrast microscopy. Water Sci. Technol. 47:105-116. [PubMed] [Google Scholar]
- 25.Kukkula, M., L. Maunula, E. Silvennoinen, and C. H. von Bonsdorff. 1999. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 180:1771-1776. [DOI] [PubMed] [Google Scholar]
- 26.Larsen, T. A., and P. Harremoes. 1994. Combined reactor and microelectrode measurements in laboratory grown biofilms. Water Res. 28:1435-1441. [Google Scholar]
- 27.Lehtola, M. J., E. Torvinen, J. Kusnetsov, T. Pitkanen, L. Maunula, C. H. von Bonsdorff, P. J. Martikainen, S. A. Wilks, C. W. Keevil, and I. T. Miettinen. 2007. Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl. Environ. Microbiol. 73:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 29.Maul, A. 1991. Aspects statistiques des méthodes de quantification en virologie, p. 143-171. In L. Schwartzbrod (ed.), Virologie des milieux hydriques. Lavoisier, Paris, France.
- 30.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin, P., A. Camper, W. Jones, D. Gatel, and J. C. Goldman. 1996. Colonization and disinfection of biofilms hosting coliform-colonized carbon fines. Appl. Environ. Microbiol. 62:4428-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu, T. R., and J. R. Lawrence. 1997. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microbiol. Ecol. 24:11-25. [Google Scholar]
- 33.Olson, M. E., J. Goh, M. Phillips, N. Guselle, and T. A. McAllister. 1999. Giardia cyst and Cryptosporidium oocyst survival in water, soil, and cattle feces. J. Environ. Qual. 28:1991-1996. [Google Scholar]
- 34.Ongerth, J. E., and J. P. Pecoraro. 1996. Electrophoretic mobility of Cryptosporidium oocysts and Giardia cysts. J. Environ. Eng. 122:228-231. [Google Scholar]
- 35.Quignon, F., M. Sardin, L. Kiene, and L. Schwartzbrod. 1997. Poliovirus-1 inactivation and interaction with biofilm: a pilot-scale study. Appl. Environ. Microbiol. 63:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, L. J., T. Forberg, L. Hermansen, B. K. Gjerde, J. O. Alvsvag, and N. Langeland. 2006. Cryptosporidium parvum infections in Bergen, Norway, during an extensive outbreak of waterborne giardiasis in autumn and winter 2004. Appl. Environ. Microbiol. 72:2218-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schvoerer, E., F. Bonnet, V. Dubois, A. M. Rogues, J. P. Gachie, M. E. Lafon, and H. J. Fleury. 1999. A hospital outbreak of gastroenteritis possibly related to the contamination of tap water by a small round structured virus. J. Hosp. Infect. 43:149-154. [DOI] [PubMed] [Google Scholar]
- 39.Searcy, K. E., A. I. Packman, E. R. Atwill, and T. Harter. 2006. Capture and retention of Cryptosporidium parvum oocysts by Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 72:6242-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skraber, S., B. Gassilloud, L. Schwartzbrod, and C. Gantzer. 2004. Survival of infectious poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and poliovirus-1 genome. Water Res. 38:2927-2933. [DOI] [PubMed] [Google Scholar]
- 41.Skraber, S., K. Helmi, R. Willame, M. Ferreol, C. Gantzer, L. Hoffmann, and H. M. Cauchie. 2007. Occurrence and persistence of bacterial and viral faecal indicators in wastewater biofilms. Water Sci. Technol. 55:377-385. [DOI] [PubMed] [Google Scholar]
- 42.Skraber, S., J. F. Schijven, C. Gantzer, and A. M. Husman. 2005. Pathogenic viruses in drinking-water biofilms: a public health risk? Biofilms 2:1-13. [Google Scholar]
- 43.Slifko, T. R., D. E. Huffman, and J. B. Rose. 1999. A most-probable-number assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 65:3936-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, P. S., T. Griebe, R. Srinivasan, C. I. Chen, F. P. Yu, D. deBeer, and G. A. McFeters. 1994. Comparison of respiratory activity and culturability during monochloramine disinfection of binary population biofilms. Appl. Environ. Microbiol. 60:1690-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart, P. S., J. Rayner, F. Roe, and W. M. Rees. 2001. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J. Appl. Microbiol. 91:525-532. [DOI] [PubMed] [Google Scholar]
- 46.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey, M. V., and N. J. Ashbolt. 2003. Enteric virions and microbial biofilms—a secondary source of public health concern? Water Sci. Technol. 48:97-104. [PubMed] [Google Scholar]
- 48.Szewzyk, U., R. Szewzyk, W. Manz, and K. H. Schleifer. 2000. Microbiological safety of drinking water. Annu. Rev. Microbiol. 54:81-127. [DOI] [PubMed] [Google Scholar]
- 49.Telgmann, U., H. Horn, and E. Morgenroth. 2004. Influence of growth history on sloughing and erosion from biofilms. Water Res. 38:3671-3684. [DOI] [PubMed] [Google Scholar]
- 50.U.S. Environmental Protection Agency. 2001. Cryptosporidium and Giardia in water filtration/IMS/FA. EPA no. 821-R-01-025. Method 1623. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 51.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. Estes, and S. M. I. Lemon. 2000. Virus taxonomy: the classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 52.Warnecke, M. 2006. Cryptosporidium oocyst interactions with drinking water pipe biofilms. Research report 5, p. 1-65. Cooperative Research Center for Water Quality and Treatment, Adelaide, Australia. http://www.waterquality.crc.org.au/.
- 53.Watson, C. L., R. J. Owen, B. Said, S. Lai, J. V. Lee, S. Surman-Lee, and G. Nichols. 2004. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J. Appl. Microbiol. 97:690-698. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, T., and P. L. Bishop. 1994. Density, porosity, and pore structure of biofilms. Water Res. 28:2267-2277. [Google Scholar]