Abstract
Phenol degradation under methanogenic conditions has long been studied, but the anaerobes responsible for the degradation reaction are still largely unknown. An anaerobe, designated strain UIT, was isolated in a pure syntrophic culture. This isolate is the first tangible, obligately anaerobic, syntrophic substrate-degrading organism capable of oxidizing phenol in association with an H2-scavenging methanogen partner. Besides phenol, it could metabolize p-cresol, 4-hydroxybenzoate, isophthalate, and benzoate. During the degradation of phenol, a small amount of 4-hydroxybenzoate (a maximum of 4 μM) and benzoate (a maximum of 11 μM) were formed as transient intermediates. When 4-hydroxybenzoate was used as the substrate, phenol (maximum, 20 μM) and benzoate (maximum, 92 μM) were detected as intermediates, which were then further degraded to acetate and methane by the coculture. No substrates were found to support the fermentative growth of strain UIT in pure culture, although 88 different substrates were tested for growth. 16S rRNA gene sequence analysis indicated that strain UIT belongs to an uncultured clone cluster (group TA) at the family (or order) level in the class Deltaproteobacteria. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., is proposed for strain UIT, and the novel family Syntrophorhabdaceae fam. nov. is described. Peripheral 16S rRNA gene sequences in the databases indicated that the proposed new family Syntrophorhabdaceae is largely represented by abundant bacteria within anaerobic ecosystems mainly decomposing aromatic compounds.
Phenols and phthalate isomers are among the most widely used chemicals and are often found in industrial wastewaters in abundance. These chemicals are known to be inhibitors for the growth of microorganisms in biological treatment processes and are regarded as priority pollutants on the U.S. Environmental Protection Agency list (19, 38). Phenol has long been known to be degraded anaerobically in the methanogenic environment (20, 43). Methanogenic degradation of phenol is significant considering the wastewater treatment processes of industrial waste chemicals and biogeochemistry of naturally occurring phenolic compounds in deep subsurface environments, such as an oil reservoir. However, despite the tremendous effort to search for phenol degraders in such environments, only a few examples are known. To date, only one species, Cryptanaerobacter phenolicus, has been isolated and characterized as an anaerobe able to metabolize phenol under methanogenic conditions (16). The organism is an anaerobic bacterium that can transform phenol into benzoate in the presence of as-yet-unidentified electron donors. However, no organisms that are genuinely capable of utilizing phenol as the sole energy source under methanogenic conditions have been isolated so far.
Recently, the enrichment and identification of mesophilic phthalate isomer-degrading bacteria from methanogenic sludges treating wastewater from the manufacture of terephthalic and isophthalic acids were reported (29). Through the enrichment, phthalate isomer-degrading microorganisms were found to be classified into two groups, the genus Pelotomaculum in the phylum Firmicutes and the clone cluster group TA in the class Deltaproteobacteria (29). Two strictly anaerobic, mesophilic, spore-forming, phthalate isomer-oxidizing strains belonging to the genus Pelotomaculum were proposed as new species in this genus (30). As part of continuing experiments, we report here the isolation and characterization of strain UIT obtained from one of the enrichment cultures that were previously made (29). To our knowledge, this is the first obligately anaerobic, syntrophic organism capable of growing on phenol under methanogenic conditions. Besides phenol, it could metabolize p-cresol, 4-hydroxybenzoate, isophthalate, and benzoate in association with an H2-scavenging methanogen partner. The bacterium is also the cultured representative of the clone cluster known as “group TA” at the family (or order) level in the class Deltaproteobacteria, and a new taxon is proposed for the isolate in this paper.
MATERIALS AND METHODS
Microorganisms, media, and cultivation.
Strain UIT was isolated from a methanogenic isophthalate-degrading enrichment culture (enrichment UI), as reported previously (29). Methanospirillum hungatei (DSM 864) and Methanothermobacter thermautotrophicus strain ΔH (DSM 1053) were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). The basal medium for cultivation was prepared as described previously (29, 34). All cultivations were performed anaerobically in 50-ml serum vials containing 20 ml medium at 37°C without shaking. Cultivations of M. hungatei and M. thermautotrophicus were conducted as previously reported (29). Desulfovibrio sp. strain UIS was cultured at 37°C with the medium supplemented with lactate (10 mM), sulfate (10 mM), and yeast extract (0.01%). The purity of strain UI in two coculture forms (DSM 17771T and JCM 13376T) was confirmed by the following examinations: (i) observation of cells by microscopy, (ii) incubation of the cultures with a variety of media that could support the growth of other fermentative anaerobes or sulfate reducers (30), (iii) fluorescence in situ hybridization (FISH) with 16S rRNA-targeted oligonucleotide probe UI178 specific for strain UIT (29), and (iv) 16S rRNA gene-based clone analysis (see below for more details).
Effects of pH, temperature, NaCl, and electron acceptor utilization.
The effects of pH, temperature, and NaCl concentration on the growth of strain UIT in coculture with M. hungatei (or triculture with M. hungatei and M. thermautotrophicus for the temperature test) was verified in the medium containing 2 mM isophthalate and 0.01% yeast extract (30, 34). All experiments were conducted with 50-ml serum vials (liquid volume, 20 ml) in duplicate, with exponential-phase cocultures of strain UIT grown on medium containing isophthalate as the inoculum (10% [vol/vol]). The growth was determined visually by observing turbidity and monitoring substrate depletion and production of methane and acetate. The utilization of electron acceptors of strain UIT was determined in the medium supplemented with 2 mM isophthalate (or 1 mM phenol) as an electron donor. In this test, 5 mM 2-bromoethane-sulfonate (2-BES) was added to inactivate methanogenesis in the coculture. The following electron acceptors were tested: sulfate (10 mM), thiosulfate (5 mM), sulfite (2 mM), elemental sulfur (5 mM), nitrate (10 mM), ferric iron [Fe(III)-nitrilotriacetic acid] (2 mM), fumarate (10 mM), anthraquinone-2,6-disulfonate (AQDS) (5 mM), and 4-hydroxybenzoate (5 mM).
Substrate utilization.
Utilization of phthalate isomers, benzoate, phenol, and 4-hydroxybenzoate by strain UIT in coculture with M. hungatei was tested as described previously (29). Strain UIT cocultured with M. hungatei in the medium containing isophthalate was used for inoculation (inoculum size, 20%). The influences of the presence of methanogens and excess amounts of hydrogen on the degradation of phenol and 4-hydroxybenzoate were determined by the method of Béchard et al. (1).
The utilization of substrates for growth was determined by monitoring the turbidity of cultures and substrate depletion and product formation (such as acetate and methane) in 50-ml serum vials (liquid volume, 20 ml) (10% inoculum). Autoclaved or filter-sterilized substrates were added to the basal medium to give final concentrations between 1 and 20 mM (30). Substrate utilization by a pure culture was determined in a parallel manner in the addition of 2-BES as an inhibitor for methanogenesis. All the experiments were performed at 37°C and pH 7.0 for over 4 months.
Analytical methods.
The concentrations of phthalate isomers, phenol, cresols (o-, m-, and p-isomers), benzoate, and 4-hydroxybenzoate were determined by high-performance liquid chromatography as described previously (28). Short-chain fatty acids, sulfate, sulfite, alcohols, methane, hydrogen, carbon dioxide, and other intermediate substances (such as succinate, malate, and lactate) were measured as described previously (15, 34).
Microscopy.
Cell morphology was examined by using a phase-contrast microscope (Olympus BX50F). The Gram staining reaction was performed by the method of Hucker (9). Transmission electron microscopy of strain UIT was performed using a Hitachi H-7000 transmission electron microscope as described previously (13).
FISH and cloning analysis.
FISH and cloning analysis were performed according to a previous report (29). For the construction of the 16S rRNA gene clone library, the universal bacterial primer set EUB8F/Uni1490R was used (41). For each of the two coculture forms (DSM 17771T and JCM 13376T) and a methanogenic phenol-degrading coculture, 20 clonal rRNA genes were randomly picked and then subjected to sequencing.
Sequencing of 16S rRNA gene and phylogenetic analysis.
DNAs from a pure culture of Desulfovibrio sp. strain UIS and cocultures of strain UIT with M. hungatei were obtained by the method of Hiraishi (14). PCR amplification of bacterial 16S rRNA genes was done as described previously (33). The PCR primers used in the amplification were the Bacteria-universal primers 8f and 1490r (41). PCR products were purified and sequenced as described previously (15). 16S rRNA gene sequences of strains UIT and UIS were determined by dye terminator cycle sequencing with a Quick Start kit (Beckman Coulter) and an automated sequence analyzer (CEQ-2000XL; Beckman Coulter). Sequence data were aligned with the ARB program package (26), and the aligned data were manually corrected on the basis of information about primary and secondary structures. The phylogenetic tree was constructed by the neighbor-joining method (31) implemented in the ARB program. Bootstrap resampling analysis (11) for 1,000 replicates was performed with the PAUP* 4.0 package (35).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA gene sequences of strains UIT and UIS are AB212873 and AB212874, respectively.
RESULTS
Isolation of strain UIT in two coculture forms.
The detailed enrichment procedure of strain UIT has been described in our previous study (29). Microscopic examination indicated that the methanogenic enrichment culture contained F420-autofluorescent rods resembling Methanospirillum and thin short rods that were later identified as strain UIT as the major morphotypes of cells. In addition, a small number of vibrio-shaped cells morphologically resembling the genus Desulfovibrio were always found in the primary enrichments (the vibrio-type cells seemed to account for less than 0.5% of the total cell population in the enrichment). When the growth medium was supplemented with 10 mM sulfate, large numbers of Desulfovibrio-like bacteria appeared and took over the role of the original H2-scavenger methanogenic partner. Syntrophic UIT culture with the Desulfovibrio-like bacterium was stable and grew faster than with methanogens. The vibrio-shaped bacterium could be isolated and identified as Desulfovibrio sp. strain UIS, and it was able to grow on lactate plus sulfate and also autotrophically on H2, CO2, and sulfate. Attempts were then made to isolate strain UIT in coculture with Desulfovibrio sp. A highly enriched methanogenic culture was inoculated into anaerobic serial roll tubes containing a medium with 2 mM isophthalate, 10 mM sulfate, and 0.01% yeast extract and a pure culture of Desulfovibrio sp. strain UIS. After 3 months of incubation, several very small colonies that were light brown, lens shaped, and 0.1 to 0.2 mm in diameter, were formed. This step was repeated several times until a defined coculture was obtained (DSM 17771T). The isolate was transferred to a medium containing isophthalate, sulfate, and yeast extract to check whether it was the targeted bacterium. Indeed, degradation of isophthalate occurred with the concomitant growth of strain UIT in coculture with Desulfovibrio sp. after 3 months of incubation.
For the isolation of strain UIT in coculture with a methanogenic archaeon or in pure culture, conventional isolation techniques, such as the roll tube method and the use of other external electron acceptors and other substrates that the known syntrophic bacteria typically used, were applied, but none of them were successful (29). Therefore, repeated serial dilutions of strain UIT enrichment culture in the medium containing isophthalate and 0.5 mM molybdate (as the inhibitor for Desulfovibrio sp.) and M. hungatei cells were successively conducted over years, and eventually, a pure syntrophic culture of strain UIT with M. hungatei (JCM 13376T) was obtained. 16S rRNA genes of purified organism (strain UIT) in both the cocultures were confirmed to be identical. The defined methanogenic coculture was then used for detailed morphological, physiological, and genetic studies.
Morphology.
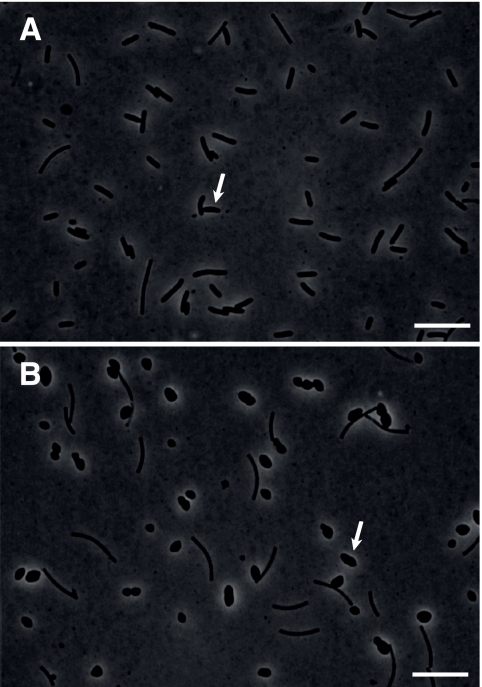
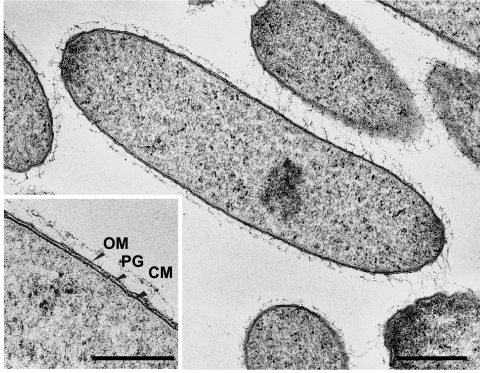
Cells of strain UIT were nonmotile and shaped like a thin rod, 0.4 to 0.8 μm wide, and 1.2 to 2.5 μm long (Fig. 1A). No spores were observed. The cells sometimes became longer in stationary-phase cultures or in exponential growth cultures after a very long lag phase. In these cultures, some of the cells elongated up to 5.0 μm. The morphology of strain UIT cells grown in the phenol-containing medium was largely different from that grown on the other substrates, such as isophthalate, benzoate, and 4-hydroxybenzoate. In cultures grown in the isophthalate-containing medium, the cells were relatively thin, short, and rod shaped (Fig. 1A). When phenol was used as the substrate, the cells appeared as oval rods. The average cell dimensions of the oval rods were as follows: width of 1.0 to 2.0 μm and a length of 1.2 to 3.0 μm (Fig. 1B). To confirm the identity of the cultures grown on isophthalate- or phenol-containing media, FISH with strain UIT-specific, 16S rRNA-targeted oligonucleotide probe (UI178) (29) was performed for the two cultures, confirming that the cells grown in both the cultures were identical at the strain level. Furthermore, 16S rRNA gene-based clone library analysis was performed for the two cultures. The sequences of 20 randomly selected clones were analyzed for each culture, and this resulted in the retrieval of only the sequence that was completely identical to that of strain UIT, supporting the identity of the strain found in the two cultures. The cells were Gram stain negative, and transmission electron microscopy indicated the presence of a gram-negative cell wall structure (Fig. 2).
FIG. 1.
Phase-contrast micrographs of strain UIT (indicated by white arrows) in coculture with M. hungatei, grown on 2 mM isophthalate (A) and grown on 2 mM phenol (B). Bars, 10 μm.
FIG. 2.
Thin-section electron micrographs of strain UIT in coculture with M. hungatei grown on 4-hydroxybenzoate (3 mM), illustrating typical gram-negative cell wall structure (OM, outer membrane; PG, peptidoglycan; CM, cytoplasmic membrane). The bar for the large panel represents 0.5 μm. The inset of the panel shows a magnified view of the cell wall structure (bar, 0.25 μm).
Growth properties.
The main physiological properties of strain UIT are listed in Table 1. Strain UIT was an obligately anaerobic microbe; no growth occurred in the presence of oxygen (air atmosphere). The addition of 0.01% yeast extract stimulated growth but was not necessarily required for growth. Sulfate, sulfite, thiosulfate, nitrate, fumarate, Fe(III)-nitrilotriacetic acid, and 4-hydroxybenzoate could not serve as external electron acceptors. However, strain UIT was capable of using AQDS (5 mM) as a terminal electron acceptor for oxidizing phenol (1 mM) or isophthalate (1 mM) as an electron donor with the concomitant growth of strain UIT within 6 weeks of incubation. The experiment was conducted in the presence of the methanogenic inhibitor, 2-BES, at a final concentration of 5 mM. Both substrates were completely converted into acetate by the 2-BES-added coculture in the presence of AQDS without the concomitant formation of methane. Attempts were made to isolate strain UIT in pure culture with phenol and AQDS, but these attempts were unsuccessful.
TABLE 1.
Main physiological characteristics of strain UIT
| Physiological characteristic | Result or value |
|---|---|
| Shape | Rod |
| Motility | Nonmotile |
| Spore formation | Not observed |
| Gram stain | Negative |
| pH range | 6.6-7.4 (optimum, 7.0) |
| Temp range (°C) | 25-37 (optimum, 35-37) |
| NaCl range (g liter−1) | 0-12.5 |
| Substrates utilized in coculture | |
| with M. hungatei (mM)a | Phenol (5), p-cresol (1), 4-hydroxybenzoate (5), isophthalate (2), and benzoate (5) |
The following substrates were also tested but were not used by the strain (concentrations are shown in parentheses and are millimolar concentrations, unless indicated otherwise): Casamino Acids (0.1%), tryptone (0.1%), yeast extract (0.5%), peptone (0.1%), H2-CO2 (80:20 [vol/vol]), glucose (20), ribose (20), xylose (20), arabinose (20), fructose (20), raffinose (20), sucrose (20), mannose (20), galactose (20), cellulose (20), starch (5 g liter−1), xylan (5 g liter−1), pectin (5 g liter−1), crotonate (10), formate (20), acetate (20), propionate (20), butyrate (20), isobutyrate (20), valerate (10), hexanoate (10), heptanoate (10), malate (20), lactate (20), pyruvate (20), succinate (20), oxalate (20), fumarate (5), adipate (5), pimelate (5), palmitate (5), stearate (5), citrate (5), acrylate (5), glutarate (5), glycine (10), ethylene glycol (10), glycerol (5), ethanol (20), methanol (20), 1-propanol (20), 2-propanol (20), 1-butanol (20), 1-pentanol (20), 1,2-butanediol (20), 2,3-butanediol (20), 1,3-propanediol (20), pyrogallol (1), 2-hydroxybenzoate (5), 3-hydroxybenzoate (5), 2,5-dihydroxybenzoate (5), 2-methylbenzoate (1), 4-methylbenzoate (1), 2-aminobenzoate (1), 3-aminobenzoate (1), 4-aminobenzoate (1), hydroquinone (1), resorcinol (1), catechol (1), o-cresol (1), m-cresol (1), cyclohexane carboxylate (1), 1-cyclohexene carboxylate (1), 3-phenylpropionate (1), aniline (1), 2-chlorobenzoate (1), 3-chlorobenzoate (1), 4-chlorobenzoate (1), protocatechuate (1), syringate (1), ferulate (1), malonate (1), 4-pentenoate (1), gallate (1), cinnamate (1), vanillate (1), cyclohexanone (1), and cyclohexene (1).
Substrate utilization.
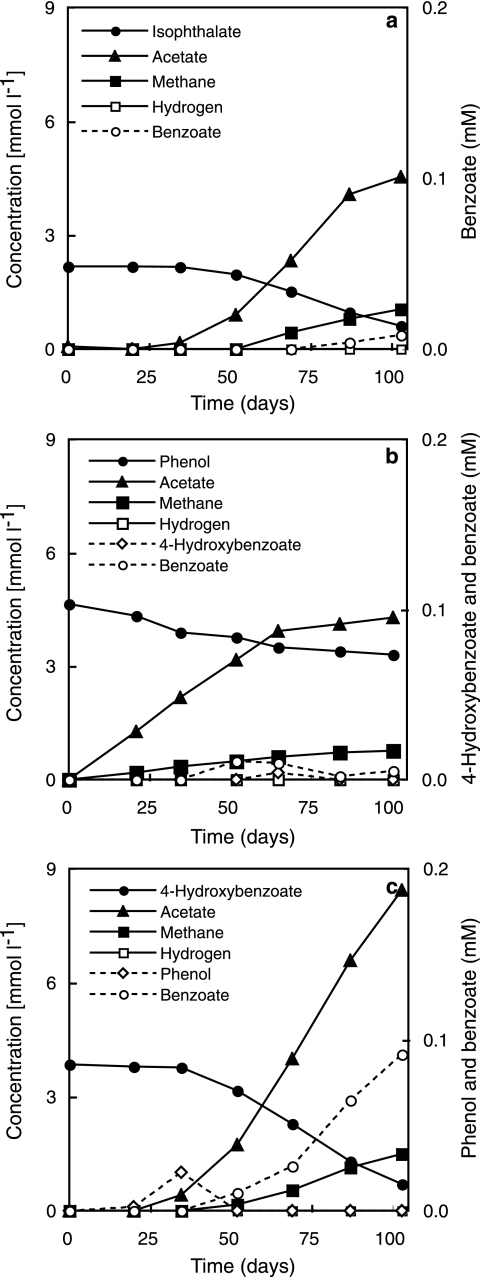
In syntrophic coculture with M. hungatei, strain UIT could utilize isophthalate, benzoate, phenol, p-cresol, and 4-hydroxybenzoate for growth and produce acetate and methane as end products. No substrates that support the fermentative growth of strain UIT in pure culture (in the presence of 2-BES) were found (Table 1). The time course of isophthalate, phenol, and 4-hydroxybenzoate degradations is illustrated in Fig. 3. The stoichiometry of substrate degradation is shown in Table 2. The addition of 5 mM 2-BES to a coculture with M. hungatei completely inhibited the degradation of isophthalate, phenol, and 4-hydroxybenzoate. Additionally, the degradation of phenol and 4-hydroxybenzoate was completely stopped when the cultures were exposed to a gas phase containing a high partial pressure of hydrogen (H2-CO2-N2 ratio of 10:10:80, respectively).
FIG. 3.
Isophthalate (a), phenol (b), and 4-hydroxybenzoate (c) degradation by strain UIT in coculture with M. hungatei. The concentrations are shown in millimoles liter-culture−1 (left) and in millimolar (right).
TABLE 2.
Stoichiometry of substrate degradation by strain UIT cocultured with M. hungatei (incubated for 100 days)
| Substrate | Initial substrate concn (mM) | Maximum concn of the following transient intermediate detected
|
Final substrate or product concn
|
Degradability in 100 days (%) | Electron recovery (%)b | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hydrogen (Pa) | Benzoate (mM) | Phenol (mM) | 4-Hydroxybenzoate (mM) | Substrate (mM) | Acetate (mM) | Methanea | ||||
| Isophthalate | 2.17 | 20 | 8 | 0.61 | 4.53 | 1.04 | 72 | 95 | ||
| Phenol | 4.64 | 20 | 11 | 4 | 3.31 | 4.11 | 0.71 | 31 | 104 | |
| 4-Hydroxybenzoate | 3.85 | 20 | 92 | 20 | 0.70 | 8.43 | 1.49 | 82 | 90 | |
The methane values are expressed as millimoles of methane formed in 1 liter of culture.
The electron balance was calculated based only on the substrate utilized and acetate and methane formed (biomass was excluded).
16S rRNA sequence analysis.
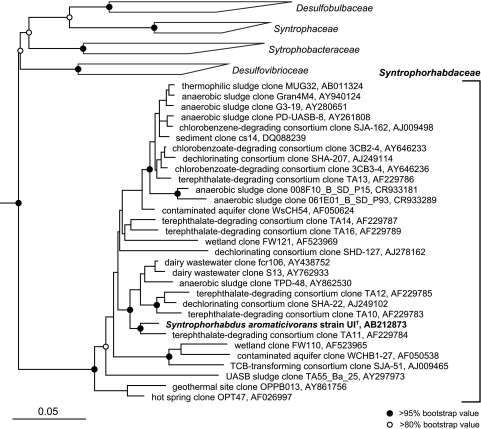
A total of 1,482 base pairs of the 16S rRNA gene from strain UIT were sequenced and compared with other relative sequences in the databases. Phylogenetic analysis indicated that strain UIT was affiliated with the clone cluster called group TA in the class Deltaproteobacteria, with less than 83% similarity to those of previously isolated bacteria within the class (Fig. 4). The 16S rRNA gene of strain UIT was most closely related to the clonal sequence TA11 (accession number AF229784; similarity value of 97%), which was obtained from a terephthalate-degrading anaerobic granular sludge. Strain UIS was 100% (1,465 bp compared) matched with strain Desulfovibrio sp. strain PA35E4 and 99.5% matched with Desulfovibrio alcoholovorans (DSM 5433).
FIG. 4.
Phylogenetic position of strain UIT among members of the class Deltaproteobacteria. Group TA (Syntrophorhabdaceae) previously consisted only of environmental 16S rRNA gene clone sequences that were mainly retrieved from anaerobic sludge treating terephthalate-containing wastewater. The tree was calculated on the basis of a distance matrix analysis of 16S rRNA gene sequences (neighbor-joining tree). The scale bar represents the number of changes in nucleotide per sequence position. The numbers at the nodes show the bootstrap values (as a percentage) obtained with 1,000 resampling analysis. The GenBank accession numbers are shown after the clone. TCB, trichlorobenzene; UASB, upflow anaerobic sludge blanket.
DISCUSSION
Isolation of uncultured clone cluster group TA.
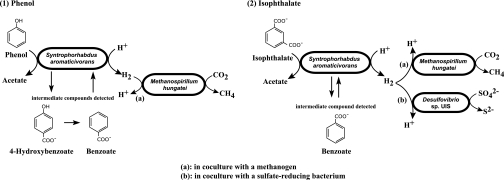
In a previous study, “group TA” was recognized as a subclass-level monophyletic group in the class Deltaproteobacteria with no cultivated representatives (42). The group currently consists solely of over 30 16S rRNA gene clones that were retrieved mainly from anoxic ecosystems, such as anaerobic upflow anaerobic sludge blanket samples (33, 42), a thermophilic anaerobic hybrid reactor degrading terephthalate (7), a contaminated aquifer site (10), an anaerobic trichlorobenzene-transforming consortium (39), and an anaerobic dichloropropane-dechlorinating consortium (32). Importantly, some of these environmental clones were abundantly found in anaerobic sludge treating terephthalate-containing wastewater (42); by cultivation-independent molecular approaches, it was found that the most predominant bacterial populations (67% of the total bacterial clones) in the anaerobic terephthalate-treating sludge community were affiliated with the group TA (42). Group TA was also found in a phenol-degrading anaerobic granular sludge accounting for 7% of the total bacterial clones analyzed for an anaerobic sludge community (44). These results had suggested the functional importance of group TA organisms as aromatic compound degraders. In fact, strain UIT is the cultured representative of group TA, degrading benzoate, isophthalate, phenol, p-cresol, and 4-hydroxybenzoate in a methanogenic syntrophic culture.
The following physiological traits of strain UIT would suggest why group TA organisms are difficult to culture: (i) low growth rate (the methanogenic coculture of strain UIT grew at a very low growth rate of μmax of 0.025 day−1 with isophthalate as the substrate [doubling time = 20 days, Fig. 3a, calculated on the basis of methane production]), (ii) limited substrate range (strain UIT could utilize only a limited range of aromatic compounds in syntrophic coculture with M. hungatei or in the presence of AQDS axenically), and (iii) requirement of almost obligately syntrophic cooperation with methanogens.
Physiological novelty.
The class Deltaproteobacteria contains the major lineages of gram-negative sulfate reducers and also contains well-known species of syntrophic substrate-degrading anaerobes such as those of the genera Syntrophobacter, Syntrophus, and Smithella. Species of the genus Syntrophobacter show the ability to utilize sulfate as an external electron acceptor, but their growth by sulfate reduction is known to be very slow (40). The inability to reduce sulfate is a common trait of syntrophic species of the genera Syntrophus and Smithella (24, 27); however, they could grow in a pure culture with limited substrates such as crotonate. Strain UIT exhibited syntrophic growth with a methanogen, but it did not show the ability to grow by itself with sulfate, showing a lack of sulfate-respiring apparatus. However, one of the most interesting features of strain UIT was that it could use AQDS, a humic quinone moiety model compound, as a terminal electron acceptor for the anaerobic oxidation of phenolic compounds. When the growth of the partner methanogen was inhibited by 2-BES, AQDS was able to replace methanogen as an electron acceptor and only acetate (and CO2) was produced. This has been predicted by Cervantes et al. (5, 6), using methanogenic phenol-degrading enrichment cultures. These findings strongly indicated that AQDS could serve as an alternate electron acceptor in the group TA members. However, one of the significant differences between strain UIT and previously isolated organisms that can use AQDS is that all of the former isolates are members of Geobacteracea that could also use iron (III) as a terminal electron acceptor (8), whereas strain UIT cannot.
Another unique phenotypic characteristic of strain UIT is its ability to degrade phenol via 4-hydroxybenzoate and benzoate and finally to acetate and methane. In this study, phenol was found to serve as the carbon and energy source for the growth of strain UIT. The inhibition of methanogenesis influenced the degradation of phenol and 4-hydroxybenzoate, and the presence of hydrogen prevented the degradation of phenol and 4-hydroxybenzoate. These results undoubtedly suggest that these reactions need tight coupling with syntrophic hydrogen (and/or formate) scavenging reaction by hydrogenotrophic organisms, which is in agreement with previous reports (36, 37). The equations of the theoretical reactions are shown as follows (17). The actual degradation and product formation by strain UIT coculture were nearly equivalent to the theoretical stoichiometry.
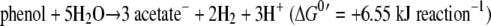 |
 |
For hydrogenotrophic methanogenesis,
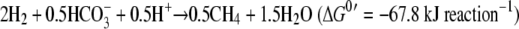 |
The overall reactions were as follows:
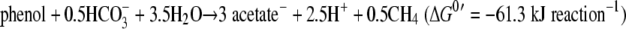 |
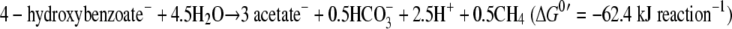 |
Thermodynamically, the oxidation of phenol to acetate, carbon dioxide, and hydrogen is unfavorable, and phenol is thought to be metabolized only at a very low hydrogen partial pressure under methanogenic conditions. However, the formation of benzoate from phenol may be a thermodynamically favorable reaction: i.e., C6H5OH + CO2 + H2 → C6H5COO− + H+ + H2O (ΔG0′ = −40.8 kJ reaction−1) (22). Hence, a fermenting bacterium could theoretically use phenol in pure culture when external electron donors (such as hydrogen) are provided. In fact, the transformation of phenol to benzoate by methanogenic consortia has been reported, and several investigators enriched anaerobic consortia containing such organisms (16, 20, 22, 23, 25). Karlsson et al. (18) have obtained a nonmethanogenic, pasteurized enrichment culture from a methanogenic consortium, transforming phenol to benzoate, acetate, and butyrate, although the organism was not isolated and characterized. The influence of H2 and CO2 on the degradation of phenol by the culture was investigated in their study, indicating that the addition of H2 does not hamper transformation. Recently, a mesophilic, nonsyntrophic bacterium, Cryptanaerobacter phenolicus, was isolated in pure culture (16). C. phenolicus can transform phenol and 4-hydroxybenzoate into benzoate under anoxic conditions; this was the only organism isolated that catalyzed phenol transformation under anoxic (methanogenic) conditions. However, C. phenolicus needs complex supplements, such as yeast extract and proteose peptone plus supernatant from the culture of Clostridium sporogenes M55 as essential factors for growth on phenol and for the transformation of phenol to benzoate (16, 22). Therefore, it is assumed that phenol is transformed into benzoate via an electron-accepting reaction, in which the intermediate product, i.e., 4-hydroxybenzoate, could serve as an external electron acceptor, and that the transformation requires uncertain electron donors present in the complex supplements (16). The reversible conversion of phenol and 4-hydroxybenzoate also occurs in Sedimentibacter hydroxybenzoicus (previously known as Clostridium hydroxybenzoicum) with resting cell suspensions and cell extracts (4, 46). S. hydroxybenzoicus is a spore-forming bacterium that decarboxylates 4-hydroxybenzoate to phenol for growth but does not further metabolize phenol in pure culture (45). Similar to C. phenolicus, proteose peptone or yeast extract is essential for the transformation of phenol by S. hydroxybenzoicus cells. Therefore, the uniqueness of strain UIT among these known phenol-transforming bacteria is its ability to syntrophically degrade phenol to form acetate and methane with no other external energy sources.
Under methanogenic conditions, the degradation pathways of phenol and its intermediates, such as benzoate, are well documented (1, 16, 21). It was presumed that phenol is first carboxylated to produce 4-hydroxybenzoic acid, which is then dehydroxylated to form benzoic acid (2, 3, 12, 16). The actual mechanisms employed by strain UIT cells for the metabolism of phenol remain unclear, but the phenol degradation pathway may include steps similar to those of known phenol transformers, since small amounts of 4-hydroxybenzoate (4 μM) and benzoate (11 μM) were detected during the transformation of phenol by strain UIT cells, and these were then utilized by the cells (Fig. 5).
FIG. 5.
Models of syntrophic phenol degradation and isophthalate degradation by strain UIT with different partners and intermediate compounds formed in the processes. The electron shuttle for interspecies electron transfer was assumed to be a proton in the models, since an increase in hydrogen partial pressures (ca. 20 Pa) was observed in all degradation processes. However, it may be possible that other external electron carriers can also intermediate in the syntrophic reactions. Intermediate compounds detected experimentally and possible degradation processes of these compounds were also shown in the models.
Since these characteristics suggest the phylogenetic and physiological novelty of strain UIT, the name Syntrophorhabdus aromaticivorans gen. nov., sp. nov., is proposed for strain UIT, and the family Syntrophorhabdaceae is also proposed for the phylogenetic group TA in Deltaproteobacteria.
Description of Syntrophorhabdaceae fam. nov.
Syntrophorhabdaceae (Syn.tro′pho.rhab.da.ce′ae. N.L. fem. n. Syntrophorhabdus, the type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Syntrophorhabdaceae, the Syntrophorhabdus family). The type genus (and only genus) is Syntrophorhabdus.
Description of Syntrophorhabdus gen. nov.
Syntrophorhabdus (Syn.tro′pho.rhab′dus. Gr. adj. syn, together with; Gr. n. trophos, one who feeds; Gr. fem. n. rhabdus, rod; N.L. fem. n. Syntrophorhabdus, rod which feeds together with (another species). Obligately anaerobic bacterium. Nonmotile, rod-shaped cells. Sulfate, sulfite, thiosulfate, nitrate, nitrite, elemental sulfur, or ferric iron cannot serve as an electron acceptor, while anthraquinone-2,6-disulfonate can. Syntrophic oxidation of aromatic compounds, such as benzoate. Syntrophorhabdus belongs to the class Deltaproteobacteria. The type species is Syntrophorhabdus aromaticivorans.
Description of Syntrophorhabdus aromaticivorans sp. nov.
Syntrophorhabdus aromaticivorans (ar.o.ma′ti.ca. N.L. adj. aromaticivorans, aromatic, referring to the property of being able to utilize aromatic compounds). An obligately anaerobic mesophilic organism. Cells are normally thin and rod shaped (0.4 to 0.8 μm in width and 1.2 to 2.5 μm in length), while they appear as fatter rods when phenol is used as a substrate (1.0 to 2.0 μm in width and 1.2 to 3.0 μm in length). Gram negative, nonmotile. Spore formation was never observed. The temperature range for growth is 25 to 37°C (optimum, 35 to 37°C). The pH range is 6.6 to 7.4 (optimum, 7.0). Growth occurs in the presence of 0 to 1.25% NaCl but does not occur in the presence of more than 1.5% NaCl. In syntrophic association with a hydrogenotrophic methanogen, the strain can utilize phenol, p-cresol, isophthalate, benzoate, and 4-hydroxybenzoate. No substrates tested support the growth in pure culture. Sulfate, sulfite, thiosulfate, nitrate, elemental sulfur, fumarate, ferric iron, or 4-hydroxybenzoate cannot be used as an electron acceptor, while AQDS can. Habitat is granular sludge from an upflow anaerobic sludge bed reactor treating wastewater from manufacturing terephthalic acid. The type strain is UIT (= JCM 13376T [in coculture with M. hungatei DSM 864] = DSM 17771T [in coculture with Desulfovibrio sp. strain UIS]).
Acknowledgments
We thank Xian-Ying Meng at the National Institute of Advanced Industrial Science and Technology (AIST) for the transmission electron microscopy studies. We also thank Jean Euzéby for expert advice on bacterial nomenclature and etymology.
This study was financially supported by a research grant for the support of young researchers subsidized by the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants from the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Foreign Researchers.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Béchard, G., J.-G. Bisaillon, R. Beaudet, and M. Sylvestre. 1990. Degradation of phenol by a bacterial consortium under methanogenic conditions. Can. J. Microbiol. 36:573-578. [Google Scholar]
- 2.Bisaillon, J. G., F. Lépine, and R. Beaudet. 1991. Study of the methanogenic degradation of phenol via carboxylation to benzoate. Can. J. Microbiol. 37:573-576. [Google Scholar]
- 3.Bisaillon, J. G., F. Lépine, R. Beaudet, and M. Sylvestre. 1993. Potential for carboxylation-dehydroxylation of phenolic compounds by a methanogenic consortium. Can. J. Microbiol. 39:642-648. [DOI] [PubMed] [Google Scholar]
- 4.Breitenstein, A., J. Wiegel, C. Haertig, N. Weiss, J. R. Andreesen, and U. Lechner. 2002. Reclassification of Clostridium hydroxybenzoicum as Sedimentibacter hydroxybenzoicus gen. nov., comb. nov., and description of Sedimentibacter saalensis sp. nov. Int. J. Syst. Evol. Microbiol. 52:801-807. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes, F. J., S. van der Velde, G. Lettinga, and J. A. Field. 2000. Quinones as terminal electron acceptors for anaerobic microbial oxidation of phenolic compounds. Biodegradation 11:313-321. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes, F. J., W. Dijksma, T. Duong-Dac, A. Ivanova, G. Lettinga, and J. A. Field. 2001. Anaerobic mineralization of toluene by enriched sediments with quinones and humus as terminal electron acceptors. Appl. Environ. Microbiol. 67:4471-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. L., H. Macarie, I. Ramirez, A. Olmos, S. L. Ong, O. Monroy, and W. T. Liu. 2004. Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 150:3429-3440. [DOI] [PubMed] [Google Scholar]
- 8.Coates, J. D., D. J. Ellis, E. L. Blunt-Harris, C. V. Gaw, E. R. Roden, and D. R. Lovley. 1998. Recovery of humic-reducing bacteria from a diversity of environments. Appl. Environ. Microbiol. 64:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doetsch, R. N. 1981. Determinative methods of light microscopy, p. 21-33. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC.
- 10.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits of phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Genthner, B. R., G. T. Townsend, and P. J. Chapman. 1991. para-Hydroxybenzoate as an intermediate in the anaerobic transformation of phenol to benzoate. FEMS Microbiol. Lett. 62:265-269. [DOI] [PubMed] [Google Scholar]
- 13.Hattori, S., Y. Kamagata, S. Hanada, and H. Shoun. 2000. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 50:1601-1609. [DOI] [PubMed] [Google Scholar]
- 14.Hiraishi, A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:210-213. [DOI] [PubMed] [Google Scholar]
- 15.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada. 2000. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl. Environ. Microbiol. 66:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juteau, P., V. Côte, M. F. Duckett, R. Beaudet, F. Lépine, R. Villemur, and J. G. Bisaillon. 2005. Cryptanaerobacter phenolicus gen. nov., sp. nov., an anaerobe that transforms phenol into benzoate via 4-hydroxybenzoate. Int. J. Syst. Evol. Microbiol. 55:245-250. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser, J. P., and K. W. Hanselmann. 1982. Fermentative metabolism of substituted monoaromatic compounds by a bacterial community from anaerobic sediments. Arch. Microbiol. 133:185-194. [Google Scholar]
- 18.Karlsson, A., J. Ejlertsson, and B. H. Svensson. 2000. CO2-dependent fermentation of phenol to acetate, butyrate and benzoate by an anaerobic, pasteurised culture. Arch. Microbiol. 173:398-402. [DOI] [PubMed] [Google Scholar]
- 19.Kleerebezem, R., L. W. Hulshoff Pol, and G. Lettinga. 1999. Anaerobic degradation of phthalate isomers by methanogenic consortia. Appl. Environ. Microbiol. 65:1152-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoll, G., and J. Winter. 1989. Degradation of phenol via carboxylation to benzoate by a defined, obligate syntrophic consortium of anaerobic bacteria. Appl. Microbiol. Biotechnol. 30:318-324. [Google Scholar]
- 21.Lépine, F., J. G. Bisaillon, S. Milot, H. K. Tawfiki, R. Beaudet, and R. Villemur. 1996. Transformation of phenol into phenylalanine by a methanogenic consortium. Appl. Environ. Microbiol. 62:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letowski, J., P. Juteau, R. Villemur, M. F. Duckett, R. Beaudet, F. Lépine, and J. G. Bisaillon. 2001. Separation of a phenol carboxylating organism from a two-member, strict anaerobic co-culture. Can. J. Microbiol. 47:373-381. [PubMed] [Google Scholar]
- 23.Li, T., J.-G. Bisaillon, R. Villemur, L. Létourneau, K. Bernard, F. Lépine, and R. Beaudet. 1996. Isolation and characterization of a new bacterium carboxylating phenol to benzoic acid under anaerobic conditions. J. Bacteriol. 178:2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y., D. L. Balkwill, H. C. Aldrich, G. R. Drake, and D. R. Boone. 1999. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 49:545-556. [DOI] [PubMed] [Google Scholar]
- 25.Londry, K. L., and P. M. Fedorak. 1992. Benzoic acid intermediates in the anaerobic biodegradation of phenols. Can. J. Microbiol. 38:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mountfort, D. O., W. J. Brulla, L. R. Krumholz, and M. P. Bryant. 1984. Syntrophus buswellii gen. nov., sp. nov.: a benzoate catabolizer from methanogenic ecosystems. Int. J. Syst. Bacteriol. 34:216-217. [Google Scholar]
- 28.Qiu, Y. L., Y. Sekiguchi, H. Imachi, Y. Kamagata, I. C. Tseng, S. S. Cheng, A. Ohashi, and H. Harada. 2003. Sporotomaculum syntrophicum sp. nov., a novel anaerobic, syntrophic benzoate-degrading bacterium isolated from methanogenic sludge treating wastewater from terephthalate manufacturing. Arch. Microbiol. 179:242-249. [DOI] [PubMed] [Google Scholar]
- 29.Qiu, Y.-L., Y. Sekiguchi, H. Imachi, Y. Kamagata, I.-C. Tseng, S.-S. Cheng, A. Ohashi, and H. Harada. 2004. Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing. Appl. Environ. Microbiol. 70:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu, Y. L., Y. Sekiguchi, S. Hanada, H. Imachi, I. C. Tseng, S. S. Cheng, A. Ohashi, H. Harada, and Y. Kamagata. 2006. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185:172-182. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Schlötelburg, C., F. von Wintzingerode, R. Hauck, W. Hegemann, and U. B. Göbel. 2000. Bacteria of an anaerobic 1,2-dichloropropane-dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int. J. Syst. Evol. Microbiol. 50:1505-1511. [DOI] [PubMed] [Google Scholar]
- 33.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 34.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 2000. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int. J. Syst. Evol. Microbiol. 50:771-779. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 36.Szewzyk, U., R. Szewzyk, and B. Schink. 1985. Methanogenic degradation of hydroquinone and catechol via reductive dehydroxylation to phenol. FEMS Microbiol. Ecol. 31:79-87. [Google Scholar]
- 37.Tschech, A., and B. Schink. 1986. Fermentative degradation of monohydroxybenzoates by defined syntrophic cocultures. Arch. Microbiol. 145:396-402. [Google Scholar]
- 38.Veeresh, G. S., P. Kumar, and I. Mehrotra. 2005. Treatment of phenol and cresols in upflow anaerobic sludge blanket (UASB) process: a review. Water Res. 39:154-170. [DOI] [PubMed] [Google Scholar]
- 39.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallrabenstein, C., E. Hauschild, and B. Schink. 1995. Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Arch. Microbiol. 164:346-352. [Google Scholar]
- 41.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, J. H., W. T. Liu, I. C. Tseng, and S. S. Cheng. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373-382. [DOI] [PubMed] [Google Scholar]
- 43.Young, L. Y., and M. D. Rivera. 1985. Methanogenic degradation of four phenolic compounds. Water Res. 19:1325-1332. [Google Scholar]
- 44.Zhang, T., and H. H. P. Fang. 2004. Microbial characteristics of a methanogenic phenol-degrading sludge, p. 161-165. In S. R. Guiot, S. G. Pavlostathis, and J. B. van Lier (ed.), Anaerobic digestion X. Selected Proceedings of the 10th IWA Congress on Anaerobic Digestion, held in Montréal, Canada, 28 September-1 October 2004. International Water Association, London, United Kingdom. [PubMed]
- 45.Zhang, X., L. Mandelco, and J. Wiegel. 1994. Clostridium hydroxybenzoicum sp. nov., an amino acid-utilizing, hydroxybenzoate-decarboxylating bacterium isolated from methanogenic freshwater pond sediment. Int. J. Syst. Bacteriol. 44:214-222. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X., and J. Wiegel. 1994. Reversible conversion of 4-hydroxybenzoate and phenol by Clostridium hydroxybenzoicum. Appl. Environ. Microbiol. 60:4182-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]