Abstract
We developed a method for concentrating pathogens from samples without enrichment. Immobilized gangliosides concentrated bacteria for detection with real-time PCR. A sensitivity of ∼4 CFU/ml (3 h) in samples without competing microflora was achieved. Samples with competing microflora had a sensitivity of 40,000 CFU/ml. The variance was less than one cycle.
The Centers for Disease Control and Prevention (CDC) estimates that food-borne diseases cause approximately 76 million illnesses, 325,000 hospitalizations, and 5,000 deaths in the United States each year, which in 2000 led to an estimated $6.9 billion burden to the U.S. economy for just the top five food-borne pathogens (http://www.ers.usda.gov/briefing/FoodSafety/). This avoidable burden can be reduced if the disease-causing organism is detected and remedial action taken quickly to control the spread of the disease. As such, many efforts to create rapid-detection tests (i.e., <24 h) have been undertaken in the last 10 years. The limitation in most methods is the need for a preenrichment step before detection to increase the population of interest with semiselective or selective media. Methods that eliminate the need for enrichment are needed to break this time barrier so that results are provided earlier in the disease process. This challenge can be addressed by creating assays that concentrate bacteria or by using detection methods that are very sensitive. More recently, use of genetic detection methods is widely available due to the large number of microbial genome sequences that are available.
The AOAC-approved rapid-detection technologies rely on either antibody-based tests or nucleic acid-based tests that claim detection limits as low as one organism, but these assays still require ≥24 to 48 h to provide a result. This time is somewhat shortened by affinity capture of the infectious agent by antibodies onto a solid matrix and then subsequent detection by PCR or real-time PCR (RT-PCR), but these tests still require enrichment, are still based on antibody reactions that can have false-positive results, and require extensive development time to ensure antibody quality. The approach presented in this work avoids the two most time-limiting steps in antibody-based assays, development and preenrichment.
An alternative approach to antibodies is the use of host receptors to capture the pathogen, which changes the paradigm of detection to finding those organisms that can bind host cells. Gangliosides are a large class of glycolipids that are common components of eukaryotic cells. Often, the specific classes are expressed in specific tissues that can be exploited to find organisms that will bind cells from specific tissues. In addition, the binding of bacteria and toxins to gangliosides has been documented in the literature (7, 13, 16, 22). To exploit this paradigm, we used host receptors (isolated mixed gangliosides) to mimic pathogen attachment onto the host cell to capture and concentrate pathogens from various samples onto a solid surface. With these molecules, a variety of pathogens can be captured onto a solid phase and subsequently be detected in a variety of different formats, depending on the amount of information needed during the testing regime. Here, we demonstrated that most common food-borne pathogens bind immobilized gangliosides onto 3-mm glass beads that are easily removed from the sample with physical separation. We determined the lower detection limit of the assay by using Escherichia coli O157:H7 as the model organism. After 5 min of binding, we detected up to 10 cells by RT-PCR within 3 h of sample collection. Spiked E. coli O157:H7 was detected in frozen spinach, pasteurized apple juice, and bottled water with a detection limit of 4 CFU/g of sample.
Gangliosides were purified from bovine buttermilk as described by Walsh and Nam (26). Briefly, the purification of gangliosides included ultrafiltration of fresh buttermilk (Gosners, Logan, UT) with a 1-kDa membrane to remove the lactose, followed by an organic extraction with chloroform-methanol-water (40:80:30). The purified gangliosides (6 μg) were used for immobilization with 100 g of 3-mm solid glass beads (catalog no. 11-312A; Fisher Scientific, St. Louis, MO), which produced bioactive beads (gangliobeads) with a mixture of gangliosides on the surface.
Organisms that bind gangliosides.
In brief, all the organisms were grown to exponential phase in media and under conditions specific for the organism (Table 1). The organisms were washed twice in saline and diluted to an optical density at 600 nm of 0.2 in saline. This suspension (2 ml) was exposed to 10 gangliobeads for 10 min at 25°C with agitation and washed thrice with 50 mM Tris-Cl (pH 7.2) for 5 min each time. The presence or absence of the particular organism on the beads was confirmed either with specific antibody detection via a tagged tertiary antibody or by RT-PCR using universal bacterial 16S primers (IDT, Coralville, IA). Rabbit anti-Salmonella antibody (OEM Concepts, Toms River, NJ), goat anti-E. coli antibody (BioDesign International, Saco, ME), rabbit anti-Listeria monocytogenes antibody (BioDesign International), rabbit anti-Erwinia antibody (Abcam, Cambridge, MA), goat anti-Bacillus globigii spores (Dugway Proving Grounds, Dugway, UT), rabbit anti-Lactococcus antibody (Center for Integrated BioSystems, Utah State University, Logan, UT), and rabbit anti-Lactobacillus antibody (Biotechnology Center, Utah State University, Logan, UT) were all immunoglobulin G-type polyclonal antibodies. They were diluted as recommended by the manufacturer for an enzyme-linked immunosorbent assay or to a micromolar concentration if they were custom products. Tertiary anti-rabbit and anti-goat conjugated to alkaline phosphatase (Sigma, Saint Louis, MO) were diluted as recommended by the supplier. The substrate, p-nitrophenyl phosphate (Sigma, St. Louis, MO) in 0.1 M glycine buffer, 1 mM MgCl2, 1 mM ZnCl2, pH 10.4 (1 mg/ml), was added to each sample, and the plates were incubated for 20 min at 37°C. Color development was measured using a BioAssay 7000 plate reader (Perkin-Elmer, Norwalk, CT) at 405 nm. Each plate contained controls consisting of aliquots of each concentration of the specific antibody, with the appropriate tertiary antibody without bacteria (negative control), the tertiary antibodies alone, and the substrate alone. The signal-to-noise value was calculated by dividing the absorbance readings from each sample by the negative-control value. The entire experiment was done with three biological replicates.
TABLE 1.
Growth conditions and ganglioside binding specificities of selected bacteria
| Organism and strain | Mediuma | Temp (°C) | Growth condition | Ganglioside binding | Detection method(s)b |
|---|---|---|---|---|---|
| E. coli | |||||
| K-12 | NB | 37 | Shaking | + | 1, 2 |
| O157:H7 | NB | 37 | Shaking | + | 1, 2 |
| S. enterica serovar Enteritidis | |||||
| 8326 | NB | 37 | Shaking | + | 1 |
| 13076 | NB | 37 | Shaking | + | 1 |
| 13314 | NB | 37 | Shaking | + | 1 |
| 31194 | NB | 37 | Shaking | + | 1 |
| 49214 | NB | 37 | Shaking | + | 1 |
| 49215 | NB | 37 | Shaking | + | 1 |
| 49217 | NB | 37 | Shaking | + | 1 |
| 49218 | NB | 37 | Shaking | + | 1 |
| 49220 | NB | 37 | Shaking | + | 1 |
| S. enterica serovar Typhimurium 700720 | NB | 37 | Shaking | + | 1 |
| B. globigii sporesc | NA | NA | NA | + | 1 |
| L. lactis IL-1403 | MRS | 37 | Static | + | 1, 2 |
| L. acidophilus | |||||
| 4355 | MRS | 37 | Static | − | 1 |
| NCFM | MRS | 37 | Static | − | 2 |
| L. gasseri 33323 | MRS | 37 | Shaking | − | 2 |
| L. helveticus 10386 | MRS | 37 | Shaking | − | 1 |
| L. monocytogenes | |||||
| 43251 | BHI | 37 | Shaking | − | 1 |
| EGDe | BHI | 37 | Shaking | − | 2 |
| E. herbicola 33243 | NB | 37 | Shaking | − | 1 |
NB, nutrient broth; NA, not done; MRS, De Man, Rogosa, and Sharpe broth; BHI, brain heart infusion broth.
1, organism detected with a specific antibody; 2, organism detected with RT-PCR.
The Bacillus spores were provided as a powder, and no growth was needed.
Detection with RT-PCR used washed gangliobeads that were added to 100 μl distilled water and boiled for 10 min to release the DNA. An aliquot of the boiled sample (12.5 μl) was used for RT-PCR with universal bacterial 16S primers (IDT, Coralville, IA), using a DyNAmo HS Sybr green quantitative PCR kit (MJ Research, Waltham, MA) per the manufacturer's recommendations and a DNA Engine Opticon 2 thermal cycler (MJ Research). The thermal cycler was run for 50 cycles of 15 s at 95°C (melting), 30 s at 60°C (annealing), and 45 s at 72°C (extension and plate reading). The threshold cycle (CT) number was reported using Opticon Monitor analysis software (version 2.02; MJ Research). Appropriate negative controls for probing for nonspecific binding (using 3-mm glass beads instead of gangliobeads) and sterility in the beads and buffers (using gangliobeads with saline instead of bacterial suspension) were also used along with the test samples. The experiment was done with three biological replicates, using duplicate RT-PCRs.
Capture and detection of E. coli 0157:H7 from buffers and food.
To check the lower detection limit of this approach, we exposed 2-ml 10-fold serial dilutions of log-phase E. coli O157:H7 cells (108 to 100 CFU/ml; cell population was determined using plate counts with nutrient agar) to 10 gangliobeads. The beads were either unwashed or washed before testing with RT-PCR. The organisms on the beads were detected using the commercially available Genevision rapid pathogen detection system for E. coli O157:H7 (Warnex Diagnostics, Quebec, Canada) per the manufacturer's instructions. Pasteurized apple juice, commercial bottled water, sterile ultrahigh-temperature milk, salami, hamburger patties, and frozen spinach were purchased from the local grocery store, plated on nutrient agar (as described in the Bacteriological Analytical Manual [17]), and spiked with log-phase E. coli O157:H7 cells at the levels of 106, 104, 102, 101, 100, and 0 CFU/25-g sample after the pure-culture population was determined by plating on nutrient agar. Spinach, hamburger patties, and salami were diluted 1:1 (weight to volume) with saline in a filtered stomacher bag and stomached at high speed using the Seward stomacher laboratory system (Lab3; Bristol, England) for 15 min, and the sample rinse was spiked with the above-mentioned E. coli O157:H7 population. Food samples (25 g) were exposed to 30 gangliobeads on a rocking platform for 30 min. After 30 min, the beads were removed by decanting the liquid from the sample and added to 400 μl of lysis buffer supplied with the Genevision rapid pathogen detection kit, and PCR was performed per the manufacturer's instructions. The aerobic plate counts of all the foods were estimated prior to spiking with E. coli O157:H7 by using nutrient agar at 32°C as described in the Bacteriological Analytical Manual (17). The average of three replicate plate counts was reported. The experiment was done with three biological replicates, using RT-PCR in duplicate.
We screened 20 different bacterial strains for their abilities to specifically bind immobilized gangliosides (Table 1). Ten strains of Salmonella, two strains of Escherichia coli, Lactococcus lactis, and Bacillus globigii spores bound to the beads. However, Lactobacillus acidophilus, Lactobacillus gasseri, Lactobacillus helveticus, Listeria monocytogenes, and Erwinia herbicola did not bind to the immobilized ganglioside mixture. All of the tested enterobacteriaceae bound the gangliobeads in this study. While variability among Salmonella strains with respect to binding gangliosides is known, we focused on Salmonella enterica serovar Enteritidis, where all the strains tested bound. Of the firmicutes tested, only L. lactis IL-1403 bound the beads, indicating that organisms used routinely in fermented foods will not interfere with this assay. None of the Listeria strains tested bound the beads.
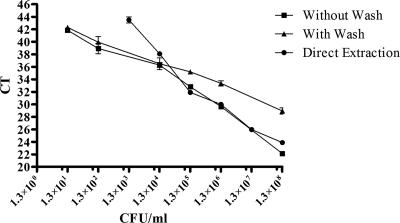
Subsequently, we focused on testing the detection limit by using E. coli O157:H7 with RT-PCR (Genevision) as a proof of concept for this system (Fig. 1). The lower detection limit of the Genevision assay (i.e., without addition of gangliobeads for concentration) using direct extraction was 1,000 CFU/ml. This detection limit was improved with addition of the gangliobead capture step to 10 CFU/ml. Washing beads prior to detection via RT-PCR caused a loss in detection for E. coli populations with ≥104 CFU/ml, but at lower populations, no significant difference (P > 0.05) was observed. Washing the beads led to loss of organisms that were not tightly bound to the bead surface, just as described by Blake and Weimer (3), who used a solid-phase enzyme-linked immunosorbent assay for Bacillus spores from water. To further increase the binding capacity, more beads may be added to increase the number bacteria that will be bound (3). Hence, gangliobeads efficiently concentrated E. coli O157:H7 from solution, even when the organisms were present at 10 CFU/ml.
FIG. 1.
CT numbers observed when gangliobeads were exposed to different cell populations of E. coli O157:H7 with or without washing of the beads. The error bars represent the SEMs of the CT values from six observations. A CT of >45 was considered no detection. A lower CT is equal to a faster detection time, which indicates a higher population.
The inability of the Genevision system alone (without the use of a solid-phase concentration) to detect organisms at <1,000 CFU/ml is due to the need for at least 1 CFU/10 μl (the volume of cell lysate used in each PCR), which equates to 1,000 CFU/ml. This limitation was overcome by the addition of gangliobeads to the sample to concentrate low cell numbers from a large volume. This resulted in a volume reduction that is more compatible with sample processing requirements for PCR (i.e., microliter volumes rather than milliliter volumes).
After ascertaining the ability of gangliobeads to concentrate E. coli O157:H7 from solution, we tested the ability of this approach to detect spiked organisms directly from food (Table 2). We detected spiked E. coli O157:H7 in bottled water, apple juice, and frozen spinach with a lower detection limit of 4 CFU/g. The lower detection limit for hamburger was 400 CFU/g, while for salami and milk the limit was 40,000 CFU/g. Except with milk, a significant correlation (r2 = 1; P < 0.0001) between the level of background microflora and the minimum detection limit for each food was observed (Table 2). Hence, it is likely that the background microorganisms out-competed the spiked E. coli O157:H7 for binding to gangliosides, which explains the higher detection limits for hamburger and salami. The higher detection limit for milk with no competing flora is likely due to the presence of milk fat globule membranes that contain gangliosides and lactoferrin, which binds gangliosides. In either case, this would lead to competitive binding with E. coli O157:H7 for the gangliosides (26). Weimer et al. (27) did not observe competition for solid-phase capture when using antibodies immobilized onto glass beads. In light of these data, the efficiency of binding with gangliobeads is affected by the levels of background microflora and other molecules in the foods that bind gangliosides. Hence, ganglioside-immobilized beads would be suitable for food systems containing low initial microbial populations and foods of nonanimal origin (e.g., bread, fruit juices, ready-to-eat salad, water, fruits, and vegetables).
TABLE 2.
Minimum detection limits of spiked E. coli O157:H7 in the food types used in this study and levels of food-associated background microflora
| Food type | Aerobic plate count before E. coli O157:H7 spike (CFU/g) | Lower detection limit (no. of CFU/g) | SEMa at detection limit | % CV at detection limit |
|---|---|---|---|---|
| Water | 0 | 4 | 0.69 | 3.3 |
| Apple juice | 8 | 4 | 0.34 | 1.6 |
| Spinach | 577 | 4 | 1.32 | 6.8 |
| Hamburger | 24,856 | 400 | 0.70 | 3.4 |
| Salami | 9,350,000 | 40,000 | 0.27 | 1.6 |
| Milk | 0 | 40,000 | 0.29 | 2.0 |
| Mean | 0.60 | 3.1 |
SEM of the CT value for the RT-PCR detection step.
The variability of the assay was measured for each approach with buffer and foods. The standard errors of the means (SEMs) for the decreasing concentrations of E. coli ranged from 0.03 to 0.81 cycles in unwashed samples, while washed samples had a range of 0.36 to 0.92 cycles (Fig. 1, error bars). This translated into coefficients of variation (CVs) of between 1.9 and 3.9% over the concentrations in the wash treatments. For the food samples, the variance and percents CV differed by sample, but all SEMs were less than 1.32 and all CVs less than 6.8%. The average CV for concentrations in all foods was 3.1%.
The specificity and sensitivity of PCR-based rapid-detection systems have driven their increased popularity in the last 2 decades (6). There are several PCR-based pathogen detection systems available commercially, such as BAX (DuPont Qualicon, Wilmington, DE), Probelia (Sanofi Diagnostic Pasteur, Marnes La Coquette, France), and Genevision (Warnex Diagnostics, Quebec, Canada), but all are hindered by a detection limit ranging from 103 to 106 CFU/ml tied to the presence of PCR inhibitors in the sample, sample dilution prior to cell lysis, or large sample volumes that do not allow a representative sample for PCR. To overcome these limitations, the sample is preenriched in laboratory media for 6 to 48 h prior to detection in at least 2 liters of media. The enrichment procedures are labor-intensive and do not allow quantitative or high-throughput automated screening procedures. In addition, all the commercially available PCR-based systems require at least 24 to 48 h to report a result. To lower the detection times, various researchers have used antibody-coated immunomagnetic beads (4, 8-10, 15, 20, 25, 28) to capture a variety of pathogens, such as Salmonella, E. coli, Campylobacter, Listeria, Yersinia, and Clostridia, for subsequent detection of them with PCR or RT-PCR. These studies achieved sensitivities of 5 to 1,000 CFU/ml within 3 to 10 h in food systems. The advantage of using gangliosides over using antibody-coated beads is that one can capture a broad repertoire of pathogens by using a single reagent. The specificity of detection can be adjusted by using PCR for each pathogen of interest with various primer designs and combinations. Simultaneous detection of various pathogens in one PCR is also possible by employing multiplex PCR (19). Additionally, coupling gangliobead capture with RT-PCR without preenrichment for detection makes the assay quantitative even at a low population of 4 CFU/g. Furthermore, the specificities of ganglioside binding to polyomavirus, simian virus 40 (12), Newcastle disease virus (11), human immunodeficiency virus (21), adenovirus (2), and human influenza virus (23) suggest that this technology has potential use for viral detection when coupled with real-time reverse transcriptase PCR (14). Ganglioside-immobilized beads could also be used for capturing specific toxins, such as botulinum toxin (1, 24), cholera toxin (5), and E. coli heat-labile enterotoxin (18), with subsequent detection via a solid-phase immunoassay, such as that reported by Weimer et al. (27). Mason et al. (16) successfully used liposomes with encapsulated DNA reporters and ganglioside receptors embedded in the bilayer to detect botulinum toxin with a lower detection limit of 0.02 fg/ml. Coupling of ganglioside-immobilized beads with the ImmunoFlow system (27), which uses a fluidized bead bed for capture, may increase the sensitivity to as low as 1 CFU independent of the sample size, as demonstrated for Escherichia coli O157:H7 and Bacillus globigii spores by using antibody-coated beads.
In conclusion, we demonstrated the ability of gangliosides immobilized on 3-mm glass beads to concentrate pathogenic organisms and Bacillus spores directly from samples without preenrichment. Use of gangliobeads prior to a commercial RT-PCR kit for food pathogen detection improved the sensitivity 100-fold in samples that did not contain competing bacteria or molecules that bind gangliosides. However, with competing microflora and biomolecules the detection limit was 40,000 CFU/g. This approach concentrated E. coli O157:H7 directly from foods containing low initial bacterial counts and foods of nonanimal origin with a detection limit of 4 CFU/g in ≤3 h.
Footnotes
Published ahead of print on 8 February 2008.
Approved as journal paper no. 7743 by the Utah Agricultural Experiment Station, Utah State University, Logan, UT 84322-4810.
REFERENCES
- 1.Ahn-Yoon, S., T. R. DeCory, and R. A. Durst. 2004. Ganglioside-liposome immunoassay for the detection of botulinum toxin. Anal. Bioanal. Chem. 378:68-75. [DOI] [PubMed] [Google Scholar]
- 2.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Blake, M. R., and B. C. Weimer. 1997. Immunomagnetic detection of Bacillus stearothermophilus spores in food and environmental samples. Appl. Environ. Microbiol. 63:1643-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, J. E., M. Blanco, A. Mora, C. Prado, M. Rio, L. Fernandez, M. J. Fernandez, V. Sainz, and J. Blanco. 1996. Detection of enterohaemorrhagic Escherichia coli O157:H7 in minced beef using immunomagnetic separation. Microbiologia 12:385-394. [PubMed] [Google Scholar]
- 5.Blanco, L. P., and V. J. DiRita. 2006. Bacterial-associated cholera toxin and GM1 binding are required for transcytosis of classical biotype Vibrio cholerae through an in vitro M cell model system. Cell. Microbiol. 8:982-998. [DOI] [PubMed] [Google Scholar]
- 6.Bohaychuk, V. M., G. E. Gensler, R. K. King, J. T. Wu, and L. M. McMullen. 2005. Evaluation of detection methods for screening meat and poultry products for the presence of foodborne pathogens. J. Food Prot. 68:2637-2647. [DOI] [PubMed] [Google Scholar]
- 7.Charych, D., Q. Cheng, A. Reichert, G. Kuziemko, M. Stroh, J. O. Nagy, W. Spevak, and R. C. Stevens. 1996. A ‘litmus test’ for molecular recognition using artificial membranes. Chem. Biol. 3:113-120. [DOI] [PubMed] [Google Scholar]
- 8.Cudjoe, K. S., and R. Krona. 1997. Detection of Salmonella from raw food samples using Dynabeads anti-Salmonella and a conventional reference method. Int. J. Food Microbiol. 37:55-62. [DOI] [PubMed] [Google Scholar]
- 9.Cudjoe, K. S., R. Krona, and E. Olsen. 1994. IMS: a new selective enrichment technique for detection of Salmonella in foods. Int. J. Food Microbiol. 23:159-165. [DOI] [PubMed] [Google Scholar]
- 10.de Leon, L., F. Siverio, and A. Rodriguez. 2006. Detection of Clavibacter michiganensis subsp. michiganensis in tomato seeds using immunomagnetic separation. J. Microbiol. Methods 67:141-149. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira, L., E. Villar, and I. Munoz-Barroso. 2004. Gangliosides and N-glycoproteins function as Newcastle disease virus receptors. Int. J. Biochem. Cell Biol. 36:2344-2356. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, J., J. Dahl, C. Riney, J. You, C. Cui, R. Holmes, W. Lencer, and T. Benjamin. 2005. Ganglioside GD1a restores infectibility to mouse cells lacking functional receptors for polyomavirus. J. Virol. 79:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansson, G. C., K. A. Karlsson, G. Larson, N. Stromberg, and J. Thurin. 1985. Carbohydrate-specific adhesion of bacteria to thin-layer chromatograms: a rationalized approach to the study of host cell glycolipid receptors. Anal. Biochem. 146:158-163. [DOI] [PubMed] [Google Scholar]
- 14.Houde, A., E. Guevremont, E. Poitras, D. Leblanc, P. Ward, C. Simard, and Y. L. Trottier. 2007. Comparative evaluation of new TaqMan real-time assays for the detection of hepatitis A virus. J. Virol. Methods 140:80-89. [DOI] [PubMed] [Google Scholar]
- 15.Lynch, M. J., C. G. Leon-Velarde, S. McEwen, and J. A. Odumeru. 2004. Evaluation of an automated immunomagnetic separation method for the rapid detection of Salmonella species in poultry environmental samples. J. Microbiol. Methods 58:285-288. [DOI] [PubMed] [Google Scholar]
- 16.Mason, J. T., L. Xu, Z. Sheng, and T. J. O'Leary. 2006. A liposome-PCR assay for the ultrasensitive detection of biological toxins. Nat. Biotechnol. 24:555-557. [DOI] [PubMed] [Google Scholar]
- 17.Maturin, L., and J. T. Peeler. 1998. Aerobic plate count. In G. J. Jackson, R. Merker, and R. Bandler (ed.), FDA bacteriological analytical manual, 8th ed. U.S. Food and Drug Administration, Rockville, MD. http://www.cfsan.fda.gov/∼ebam/bam-3.html.
- 18.Minke, W. E., C. Roach, W. G. Hol, and C. L. Verlinde. 1999. Structure-based exploration of the ganglioside GM1 binding sites of Escherichia coli heat-labile enterotoxin and cholera toxin for the discovery of receptor antagonists. Biochemistry 38:5684-5692. [DOI] [PubMed] [Google Scholar]
- 19.Park, Y. S., S. R. Lee, and Y. G. Kim. 2006. Detection of Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus and Listeria monocytogenes in kimchi by multiplex polymerase chain reaction (mPCR). J. Microbiol. 44:92-97. [PubMed] [Google Scholar]
- 20.Pyle, B. H., S. C. Broadaway, and G. A. McFeters. 1999. Sensitive detection of Escherichia coli O157:H7 in food and water by immunomagnetic separation and solid-phase laser cytometry. Appl. Environ. Microbiol. 65:1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat, S. S., S. A. Gallo, J. Eaton, T. D. Martin, S. Ablan, V. N. KewalRamani, J. M. Wang, R. Blumenthal, and A. Puri. 2004. Elevated expression of GM3 in receptor-bearing targets confers resistance to human immunodeficiency virus type 1 fusion. J. Virol. 78:7360-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stromberg, N., M. Ryd, A. A. Lindberg, and K. A. Karlsson. 1988. Studies on the binding of bacteria to glycolipids. Two species of Propionibacterium apparently recognize separate epitopes on lactose of lactosylceramide. FEBS Lett. 232:193-198. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, Y., M. Matsunaga, Y. Nagao, T. Taki, Y. Hirabayashi, and M. Matsumoto. 1985. Ganglioside GM1b as an influenza virus receptor. Vaccine 3:201-203. [DOI] [PubMed] [Google Scholar]
- 24.Tsukamoto, K., T. Kohda, M. Mukamoto, K. Takeuchi, H. Ihara, M. Saito, and S. Kozaki. 2005. Binding of Clostridium botulinum type C and D neurotoxins to ganglioside and phospholipid. Novel insights into the receptor for clostridial neurotoxins. J. Biol. Chem. 280:35164-35171. [DOI] [PubMed] [Google Scholar]
- 25.Varshney, M., L. Yang, X. L. Su, and Y. Li. 2005. Magnetic nanoparticle-antibody conjugates for the separation of Escherichia coli O157:H7 in ground beef. J. Food Prot. 68:1804-1811. [DOI] [PubMed] [Google Scholar]
- 26.Walsh, M. K., and S. H. Nam. 2001. Affinity enrichment of bovine lactoferrin in whey. Prep. Biochem. Biotechnol. 31:229-240. [DOI] [PubMed] [Google Scholar]
- 27.Weimer, B. C., M. K. Walsh, C. Beer, R. Koka, and X. Wang. 2001. Solid-phase capture of proteins, spores, and bacteria. Appl. Environ. Microbiol. 67:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, L. S., J. Uknalis, and S. I. Tu. 2001. Immunomagnetic separation methods for the isolation of Campylobacter jejuni from ground poultry meats. J. Immunol. Methods 256:11-18. [DOI] [PubMed] [Google Scholar]