Abstract
hMSH2⋅hMSH6 heterodimer (hMutSα) and hMLH1⋅hPMS2 complex (hMutLα) have been implicated in the cytotoxic response of mammalian cells to a number of DNA-damaging compounds, including methylating agents that produce O6-methylguanine (O6MeG) adducts. This study demonstrates that O6MeG lesions, in which the damaged base is paired with either T or C, are subject to excision repair in a reaction that depends on a functional mismatch repair system. Furthermore, treatment of human cells with the SN1 DNA methylators N-methyl-N-nitrosourea or N-methyl-N′-nitro-N-nitrosoguanidine results in p53 phosphorylation on serine residues 15 and 392, and these phosphorylation events depend on the presence of functional hMutSα and hMutLα. Coupled with the previous demonstration that O6MeG⋅T and O6MeG⋅C pairs are recognized by hMutSα, these results implicate action of the mismatch repair system in the initial step of a damage-signaling cascade that can lead to cell-cycle checkpoint activation or cell death in response to DNA methylator damage.
The use of DNA-damaging agents remains one of the primary regimes in cancer chemotherapy. However, the utility of such agents is often compromised by the emergence of drug-resistant tumor cell populations. A number of previous studies have linked defects in the mammalian mismatch repair genes MSH2, MSH6, and MLH1 with resistance of cultured cells to the cytotoxic effects of SN1 DNA methylators, 6-thioguanine, cisplatin, and doxorubicin (1–6). These observations may have significant clinical ramifications, because early studies indicate that mismatch repair defects can also be associated with DNA methylator-resistant tumors in whole animals (7, 8).
In addition to their resistance to the cytotoxic effects of the agents mentioned above, cell lines deficient in the hMSH2⋅hMSH6 heterodimer (hMutSα) or the hMLH1⋅hPMS2 complex (hMutLα) also fail to elicit a G2 checkpoint response on treatment with sublethal levels of SN1 DNA methylators or 6-thioguanine (2, 9–12). SN1 DNA methylators are mutagenic and cytotoxic, with both activities attributable to production of O6-methylguanine (O6MeG) in DNA (13). The resistance of mismatch repair mutants to this class of drugs, which trigger an apoptotic response after G2-M arrest in normal cells (14), is because of the tolerance of DNA lesions that are otherwise lethal to mismatch repair proficient cells (9).
Such findings imply that functional hMutSα and hMutLα are required for certain DNA-damaging agents to elicit checkpoint activation and, at high lesion load, cell death. Although the molecular events involved in this damage response have not been established, the role of the mismatch repair system appears to be a direct one, because hMutSα has been found to specifically recognize O6MeG adducts and the 1,2-intrastrand purine–purine crosslink produced by SN1 DNA methylators and cisplatin, respectively (15–17). Because mismatch repair has also been implicated in the recognition and rectification of DNA biosynthetic and recombination errors (18–20), the repair system has been postulated to act as a sensor of genetic damage with a fairly broad specificity (2, 15, 16).
Two models have been proposed to explain the link between the mismatch repair system and the DNA-damage response. One attributes the cytotoxic effects to translesion DNA synthesis when the replication fork encounters a damaged nucleotide in the template strand (2, 9, 17). The resulting base pair anomaly triggers mismatch repair, but, because action of this system is restricted to the daughter DNA strand, the offending lesion can persist in the template. Excision repair triggered in this manner could lead to futile rounds of abortive repair, with persistent excision intermediates produced in this manner serving to activate the DNA-damage response. In the second model, assembly of a signaling complex of hMutSα, hMutLα, and other activities at the site of a lesion has a finite probability of directly triggering a damage response regardless of the DNA replication status (15, 16).
Experimental Procedures
Cell Lines.
Human lymphoblastoid B-cell lines TK6 and MT1 (9) were grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum (HyClone). The MLH1−/− human colorectal adenocarcinoma cell line HCT116 and a HCT116 hybrid clone containing a copy of a MLH1+ chromosome 3 (HCT116.ch3) were obtained from Thomas Kunkel (National Institute of Environmental Health Sciences) (3). HCT116 and HCT116.ch3 lines were maintained in McCoy’s 5A medium (HyClone) supplemented with 2 mM l-glutamine and 10% (vol/vol) fetal bovine serum, which also contained 400 μg/ml geneticin disulfate in the case of HCT116.ch3. The absence and presence of hMLH1 in HCT116 and HCT116.ch3 cells was verified by immunoblot analysis.
Drug Treatment and Immunoblot Analysis.
TK6 and MT1 cells were suspended in fresh medium at a density of 105 cells per ml, and 10-ml samples were distributed into 150-mm dishes, where they were subjected to 10 μM N-methyl-N-nitrosourea (MNU) or 10 μM etoposide for 40 min, or to 20 J/m2 of 254-nm UV light. HCT116 and HCT116.ch3 were treated in serum-free medium (105 cells/ml) with 5 μM N-methyl-N ′-nitro-N-nitrosoguanidine for 40 min according to Koi et al. (3). After exposure to DNA-damaging agents, cells were resuspended in fresh complete medium (105 cells/ml) and distributed into 150-mm dishes (106 cells per dish). Samples (4 × 106 cells) were harvested as indicated, collected by centrifugation, washed with phosphate-buffered saline, and the cell pellet resuspended in 100 μl of ice-cold buffer A (10 mM Hepes-KOH (pH 7.6)/10 mM KCl/0.5 mM MgCl2/10% glycerol/1 mM DTT/1 μg/ml leupeptin/5 μg/ml E64/1 μg/ml aprotinin/0.1% PMSF). After 10 min on ice, the suspension was passed through a 20-gauge needle and centrifuged at 12,000 × g for 5 min at 4°C. The supernatant (cytoplasmic extract) was saved on ice, and the pellet was extracted for 30 min at 0°C with 40 μl of buffer A supplemented with 250 mM KCl/50 mM NaF/50 mM β-glycerophosphate/1 mM wortmannin. Nuclei and cell debris were pelleted at 12,000 × g as above, and the supernatant was added to the cytoplasmic extract.
Cell extract (30 μg) was subjected to electrophoresis through a 10% polyacrylamide gel containing SDS and proteins transferred onto nitrocellulose membrane. The membrane was then immunoblotted with p53 antibody AB6 (Oncogene Research Products), anti-hMLH1 (PharMingen clone G168–15), or phospho-specific antibodies directed against p53 peptides containing phospho-Ser-15 or phospho-Ser-392 (21–23).
DNA Substrates.
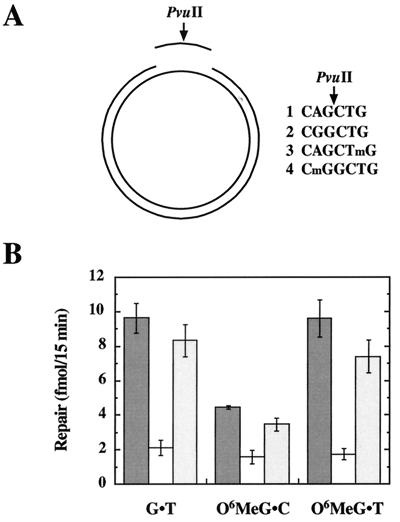
Bacteriophage f1MR56 was prepared by insertion of an oligonucleotide duplex containing an EcoRV site [d(CAAGCGGATATCAGCGT)⋅d(CTAGACGCTGATATCCGCTTGACG)] into f1MR1 (24) replicative form (RF) DNA that had been cleaved with AatII and XbaI. Phage f1MR57 was constructed by insertion of a synthetic duplex comprised of d(CCCCACTCCATAGGATGGATCCAGCTGGCGCCGGCTCGAGTATCCCACCCC) and d(GGGGTGGGATACTCGAGCCGGCGCCAGCTGGATCCATCCTATGGAGTGGGG) into EcoRV linearized f1MR56 RF DNA. The PvuII sequence within this insert was used as a site for placement of a G-T mismatch, and O6MeG-C or O6MeG-T lesions (Fig. 1), because presence of O6MeG is known to prevent cleavage by this endonuclease (25). Circular heteroduplexes (6,400 bp) containing a G-T mismatch or an O6MeG lesion within a 51-residue oligonucleotide spanned by two single-strand breaks were prepared according to Yamaguchi et al. (26). Briefly, circular duplex DNA with a 51-nt gap was prepared by denaturing EcoRV-linearized f1MR56 and hybridizing to the circular viral DNA strand derived from f1MR57. These gapped circles were utilized for preparation of homoduplex, G-T heteroduplex, or O6MeG-C and O6MeG-T substrates, respectively, by hybridization of the 51-residue synthetic oligonucleotides: d(GGGGTGGGATACTCGAGCCGGCGCCAGCTGGATCCATCCTATGGAGTGGGG), d(GGGGTGGGATACTCGAGCCGGCGCCGGCTGGATCCATCCTATGGAGTGGGG), d(GGGGTGGGATACTCGAGCCGGCGCCAGCTmGGATCCATCCTATGGA- GTGGGG), or d(GGGGTGGGATACTCGAGCCGGCGCCmGGCTGGATCCATCCTATGGAGTGGGG), where mG indicates O6MeG. A 20-fold molar excess of oligomer was hybridized to gapped circular DNA in 20 mM Tris⋅HCl (pH 7.6)/100 mM NaCl/10 mM MgCl2 at 65°C for 30 min and the reaction allowed to slow cool to room temperature. Circular DNA products were purified by mixing with 0.1 packed volume of benzoylated naphthoylated DEAE-cellulose (BND) in 20 mM Tris⋅HCl (pH 7.6)/1 mM EDTA/1 M NaCl. The BND matrix was removed by centrifugation, and the DNA-containing supernatant was freed of small molecule contaminants by centrifugation through a Sephadex G50 spin column (Boehringer Mannheim). Purified substrates were concentrated by ethanol precipitation followed by resuspension in 20 mM Tris⋅HCl (pH 7.6)/1 mM EDTA at a concentration of approximately 100 μg/ml.
Figure 1.
O6MeG lesions are subject to processing by the human mismatch repair system. (A) Oligonucleotides (51 mer) containing PvuII sequence variants shown (mG indicates O6MeG) were hybridized to a circular 6,490-bp DNA containing a 51-nt gap in the complementary DNA strand (Experimental Procedures) to yield the four substrates used in this study. Hybridization of oligonucleotide 1 yields a homoduplex with a normal PvuII recognition sequence, whereas oligonucleotides 2, 3, or 4 generate substrates containing a G⋅T mismatch, an O6MeG⋅C bp, or an O6MeG⋅T lesion, respectively. Presence of the mismatch or O6MeG within the PvuII recognition site renders the DNA resistant to hydrolysis by this enzyme, with sensitivity restored on repair. (B) Mismatch repair assays (Experimental Procedures) scored repair on the incised strand that restored an intact PvuII recognition sequence. Dark grey bars, TK6 nuclear extract; white bars, MT1 nuclear extract; medium grey bars, hMutSα-supplemented MT1 nuclear extract. Values shown are the average of four determinations ± one standard deviation. Because TK6 and MT1 cells are deficient in the O6-alkylguanine-DNA alkyltransferase, complications caused by repair by this activity are obviated.
Nuclear Extracts and Repair Assays.
Nuclear extracts were prepared and mismatch repair reactions performed as described (10). Reactions contained 100 ng of heteroduplex DNA, 100 μg of nuclear extract, and, when indicated, 100 ng of near-homogeneous hMutSα (10). Incubation was for 15 min at 37°C.
Results
Deficiency of hMutSα or hMutLα confers significant levels of resistance to the cytotoxic effects of a number of DNA-damaging agents, but the most dramatic effects have been observed with SN1 DNA methylators, where the 37% survival dose is increased more than 100-fold by mismatch repair defects (9). Because this implies that nearly all methylator-induced killing events depend on a functional mismatch repair system, we have examined the role of the repair system in the response to this type of damage. As noted above, killing by SN1 methylators is caused by production of O6MeG (13), and hMutSα has been shown to bind to O6MeG⋅C and O6MeG⋅T pairs (15). To determine whether such lesions are also subject to processing by the human mismatch repair system, nick-directed repair of heteroduplexes containing a single O6MeG⋅C or O6MeG⋅T base pair was tested in nuclear extracts derived from TK6 and MT1 lymphoblastoid cells (Fig. 1A). The methylation-tolerant MT1 cell line was derived from TK6 cells by single-step selection for high-level DNA methylator resistance and is deficient in hMutSα because of missense mutations in both alleles of MSH6 (9, 10, 27). As shown in Fig. 1B, O6MeG⋅T is repaired as well as a G⋅T mismatch by nuclear extract of TK6 cells, whereas the O6MeG⋅C lesion is rectified with about 50% the efficiency of the former pairing anomalies. As expected (2), repair of the G⋅T mismatch was greatly reduced in extracts of MSH6−/− MT1 cells, as was repair of both types of O6MeG lesion. However, repair in all cases was restored to near-normal levels on supplementation of MT1 nuclear extracts with near-homogeneous hMutSα. As previously observed for mismatch repair in nuclear extracts (28, 29), rectification of O6MeG adducts was inhibited by aphidicolin (not shown). The finding that these lesions are subject to excision repair by the mismatch repair system is consistent with the possibility that translesion synthesis and subsequent action of the repair system may be involved in eliciting the DNA-damage response.
Both the lesion recognition and translesion synthesis hypotheses described above attribute the drug-resistant phenotype of mismatch repair mutants to an inability to activate damage-signaling pathways because of the failure to recognize or process DNA adducts. This idea implies that the mismatch repair system acts upstream of one or more damage-signaling pathways. Because Ser-15 and Ser-392 phosphorylation of p53 has been implicated in cellular responses to several types of DNA damage (21–23, 30–36), we have used this tumor-suppressor protein as an in vivo substrate to determine whether damage-signaling kinases are activated in response to SN1 methylator damage in a hMutSα- and hMutLα-dependent manner.
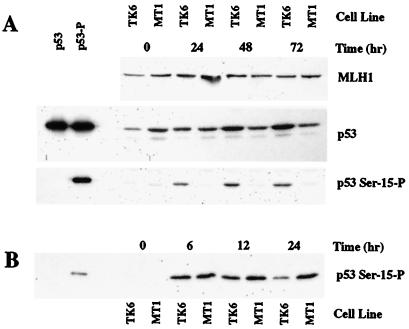
These experiments utilized repair-proficient TK6 lymphoblastoid cells that are wild type with respect to p53 function (37), and the repair-deficient MSH6−/− MT1 cell line derived from TK6 by selection for DNA methylator resistance (9). On treatment with 10 μM MNU, p53 levels increase in TK6 cells but not in its hMutSα-deficient derivative (Fig. 2A), confirming a previous observation to this effect (12). The p53 phosphorylation status at Ser-15 and Ser-392 was assessed in both cell lines by using phosphopeptide-specific polyclonal antibodies that recognize only phosphoserine-15 and phosphoserine-392 forms of the protein (21–23). As shown in Figs. 2A and 3A, the increase in p53 level that occurs on MNU treatment in TK6 cells is associated with phosphorylation of both Ser-15 and Ser-392. However, no detectable increase in phosphorylation of either residue was detected in mismatch repair-deficient MT1 cells.
Figure 2.
MNU-induced Ser-15 phosphorylation of p53 depends on MSH6 function, but that induced by etoposide does not. TK6 or MT1 cells were treated with 10 μM MNU (A) or 8 μM etoposide (B) for the indicated times, and extract (30 μg) subjected to SDS gel electrophoresis and Western blotting by using antibodies against hMLH1, p53, or affinity-purified antibody specific for the Ser-15 phosphorylated form of p53 (Experimental Procedures). The left two lanes in each p53 blot correspond to 2–5 ng of purified recombinant human p53 protein produced in Escherichia coli that was either untreated or phosphorylated in vitro by using near-homogeneous DNA protein kinase and Ku protein. This kinase is known to phosphorylate p53 on Ser residues 15 and 37 (58).
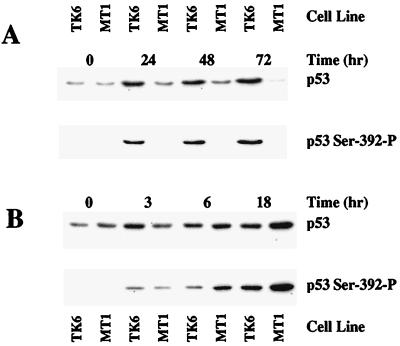
Figure 3.
Phosphorylation of p53 at Ser-392 is induced by methylnitrosourea in a MSH6-dependent manner. TK6 or MT1 cells were treated with 10 μM MNU for the indicated time (A) or exposed to 20 J/m2 of 254 nm UV light and then incubated in complete medium for the times indicated (B). Samples of extract (30 μg) were analyzed by SDS gel electrophoresis and immunological blot as described in Fig. 2, except that phosphopeptide-specific antibody was directed against the phosphorylated form of Ser-392.
The differential phosphorylation of p53 observed in these two cell lines depends on the nature of the DNA damage. In contrast to results obtained after DNA methylator exposure, Ser-15 and Ser-392 were phosphorylated to a similar degree in both cell lines when DNA damage was produced by etoposide treatment or UV radiation (Figs. 2B and 3B). The finding that Ser-392 phosphorylation in response to UV irradiation is independent of the mismatch repair system is of interest, because hMutSα has been shown to recognize some UV photoproducts (38, 39). This observation was not unexpected because, in contrast to DNA methylator, cisplatin, and 6-thioguanine resistance conferred by hMutSα or hMutLα deficiency, mismatch repair defects are associated with enhanced sensitivity to killing by UV irradiation (40).
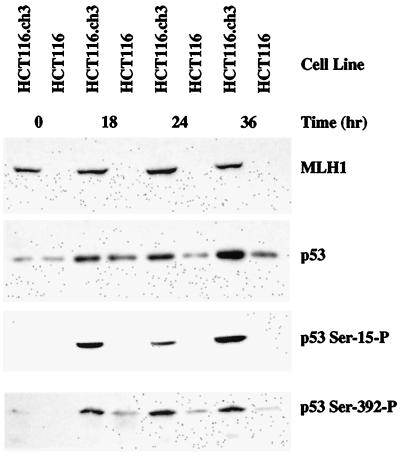
Although the MSH6−/− MT1 cell line was isolated by single-step selection from TK6 cells (9), its mutator phenotype raises the possibility that the p53 phosphorylation defect might be caused by a secondary mutation. To evaluate this possibility and to assess potential involvement of hMutLα in p53 phosphorylation, two additional cell lines were tested. HCT116 colorectal tumor cells, which harbor wild-type p53 loci (41), are defective in mismatch repair because of inactivation of both alleles of MLH1 and hMutLα deficiency (42, 43). This defect is associated with high-level resistance to SN1 DNA methylators and 6-thioguanine, but introduction of a single copy of human chromosome 3 with a wild-type MLH1 gene restores mismatch repair proficiency, genetic stability, and drug sensitivity (3, 11). As shown in Fig. 4, phosphorylation of p53 serines 15 and 392 was not observed on treatment of MLH1−/− HCT116 cells with 5 μM N-methyl-N′-nitro-N-nitrosoguanidine, an SN1 DNA methylator. However, phosphorylation of both residues did occur in the HCT116 hybrid clone that contains a copy of human chromosome 3 with a wild-type MLH1 allele. These findings confirm upstream involvement of the mismatch repair system in the phosphorylation of p53 serine residues 15 and 392 in response to DNA methylator damage and imply that both hMutSα and hMutLα are required for this effect. Inasmuch as hMutSα specifically recognizes O6MeG lesions (15), and because such lesions are subject to processing by the mismatch repair system (Fig. 1), it is clear that hMutSα and hMutLα function during the earliest steps in the cellular response to DNA methylator damage, with their presence required for activation of the kinase(s) that phosphorylate p53 in response to such lesions.
Figure 4.
Phosphorylation of p53 at Ser-15 and Ser-392 is induced by N-methyl-N′-nitro-N-nitrosoguanidine in a MLH1-dependent manner. HCT116 or HCT116.ch3 were treated with 5 μM N-methyl-N ′-nitro-N-nitrosoguanidine (Experimental Procedures) and then incubated in complete medium for the indicated time. Samples of extract (30 μg) were analyzed by SDS gel electrophoresis and immunological blot by using the antisera indicated.
Discussion
The resistance of mismatch repair mutants to SN1 DNA methylating agents has been known for some time (1–4), but the mechanism underlying this effect is poorly understood. Exposure of mismatch repair-proficient cells to SN1 DNA methylators or 6-thioguanine evokes G2-M arrest, which occurs in the second cycle after drug exposure (9, 11). In the case of DNA methylation damage, killing at high lesion load has been attributed to an apoptotic response that occurs subsequent to arrest at the second G2 (14). MSH6−/− MT1 cells and MLH1−/− HCT116 cells used in the work described here fail to elicit either of these responses (9, 11), implicating hMutSα and hMutLα in G2 arrest and apoptosis triggered by these agents. Occurrence of the damage response in the second G2 after exposure to these agents suggests that these effects depend on translesion synthesis by a replication fork (2, 9, 17). As discussed above, this model attributes G2 arrest and the death response to abortive turnover of a newly synthesized strand by the mismatch repair system because of the presence of lesions in template DNA. Although hMutSα has been previously shown to recognize O6MeG lesions (15), the work described here shows that such lesions are also subject to excision repair by the mismatch repair system, as required by this model.
We have also observed hMutSα- and hMutLα-dependent activation of one or more protein kinases that phosphorylate the p53 tumor-suppressor protein in response to DNA methylation damage. It is pertinent to note that the p53 phosphorylation events that we have observed are independent of MutSβ (MSH2⋅MSH3 heterodimer), because this activity is present in the MSH6−/− MT1 cells used in this study (10, 44). Although use of p53 as an in vivo reporter substrate has clearly linked the mismatch repair system to kinase activation in response to DNA methylation damage, the identity of the kinase(s) responsible for the Ser-15 and Ser-392 phosphorylation events described here is not known. However, it is noteworthy that members of the phosphoinositide-3 kinase-related kinase superfamily have been implicated in the phosphorylation of Ser-15. Thus, ATM is believed to phosphorylate Ser-15 in response to γ-irradiation. (21, 30, 31, 33), whereas ATR apparently functions in Ser-15 phosphorylation after γ-irradiation or UV exposure (36). p53 is also phosphorylated in vitro at Ser-15 by the DNA-dependent protein kinase, but involvement of this activity in the response to DNA damage is controversial (34, 35, 45). Phosphorylation of Ser-392 also occurs in response to UV but not ionizing irradiation (23). Although the kinase involved in the Ser-392 damage response has not been identified, p53 is phosphorylated on this residue in vitro by casein kinase II (46) and the cyclin-dependent kinase-activating kinase (32). These activities are obvious candidates for the hMutSα- and hMutLα-dependent p53 phosphorylation events described here, and tests of their potential involvement are in progress.
Modulation of p53 as a transcriptional activator occurs in response to both DNA damage and environmental stress (47–50). The nature of the p53 response, which can depend on the identity of the activating signal, is believed to be caused in part by posttranslational modification of the protein at a number of distinct sites. Thus, different types of DNA lesions may elicit an overlapping set of p53 modification events by distinct signaling pathways. As noted above, this appears to be the case with respect to differential ATR and ATM involvement in Ser-15 phosphorylation in response to UV or γ-irradiation. In a similar vein, we have found that, whereas phosphorylation of serines Ser-15 and Ser-392 in response to DNA methylation damage requires hMutSα and hMutLα (Figs. 2 and 3), modification of these residues in response to etoposide damage or UV irradiation is independent of the mismatch repair system.
Although we have used p53 simply as an in vivo substrate to monitor activation of damage-dependent signaling activities, the Ser-15 and Ser-392 phosphorylation events that we have scored may bear on the G2 arrest and apoptosis that occur in response to DNA methylation damage. p53 activation has been implicated in the apoptotic response (47–49), and recent work has implicated the protein in maintenance of G2-M arrest after DNA damage (41, 51, 52). If p53 activation does play a role in the response to methylation damage, then alternate signaling pathways must also exist, because p53-deficient HeLa cells are subject to killing by SN1 DNA methylators, and methylation-tolerant cell lines defective in mismatch repair have been isolated in this background (53, 54).
The cytotoxic response of mammalian cells to cisplatin also depends on a functional mismatch repair system (5, 6), and hMutSα has been shown to recognize cisplatin lesions (15–17). The degree of resistance conferred by mismatch repair defects is considerably less dramatic for cisplatin than for DNA methylators, suggesting that multiple lesion sensing/processing pathways are involved in cisplatin killing. Nevertheless, cisplatin treatment elicits activation of the c-Abl kinase and c-jun NH2-terminal kinase, and this response is absent or reduced in mismatch repair-deficient cells (55). Recently, Gong et al. (56) demonstrated accumulation of p73 after cisplatin exposure, an effect that depends on functional MLH1 and c-Abl activation. Although p53 was also found to accumulate after cisplatin treatment, this increase is independent of MLH1. Inasmuch as p53 has also been implicated in the apoptotic response to cisplatin (57), this drug apparently induces at least two proapoptotic pathways, one involving p53, the other involving p73, with only the latter depending on a functional mismatch repair system. It remains to be determined whether p73 is also activated in response to SN1 DNA methylation damage.
Acknowledgments
P.M. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- hMutSα
hMSH2⋅hMSH6 heterodimer
- hMutLα
MLH1⋅PMS2 complex
- O6MeG
O6-methylguanine
- MNU
N-methyl-N-nitrosourea
References
- 1.Branch P, Aquilina G, Bignami M, Karran P. Nature (London) 1993;362:652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- 2.Kat A, Thilly W G, Fang W H, Longley M J, Li G M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 4.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 5.Drummond J T, Anthoney A, Brown R, Modrich P. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 6.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen R D, Boland C R, Koi M, Fishel R, et al. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 7.Friedman H S, Johnson S P, Dong Q, Schold S C, Rasheed B K, Bigner S H, Ali-Osman F, Dolan E, Colvin O M, Houghton P, et al. Cancer Res. 1997;57:2933–2936. [PubMed] [Google Scholar]
- 8.Friedman H S, McLendon R E, Kerby T, Dugan M, Bigner S H, Henry A J, Ashley D M, Krischer J, Lovell S, Rasheed K, et al. J Clin Oncol. 1998;16:3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 9.Goldmacher V S, Cuzick R A, Thilly W G. J Biol Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 10.Drummond J T, Li G-M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 11.Hawn M T, Umar A, Carethers J M, Marra G, Kunkel T A, Boland C R, Koi M. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 12.D’Atri S, Tentori L, Lacal P M, Graziani G, Pagani E, Benincasa E, Zambruno G, Bonmassar E, Jiricny J. Mol Pharmacol. 1998;54:334–341. doi: 10.1124/mol.54.2.334. [DOI] [PubMed] [Google Scholar]
- 13.Karran P, Bignami M. Nucleic Acids Res. 1992;20:2933–2940. doi: 10.1093/nar/20.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 15.Duckett D R, Drummond J T, Murchie A I H, Reardon J T, Sancar A, Lilley D M J, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mello J A, Acharya S, Fishel R, Essigmann J M. Chem Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 17.Yamada M, O’Regan E, Brown R, Karran P. Nucleic Acids Res. 1997;25:491–496. doi: 10.1093/nar/25.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 19.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 20.Radman M. Genome. 1989;31:68–73. doi: 10.1139/g89-014. [DOI] [PubMed] [Google Scholar]
- 21.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 23.Lu H, Taya Y, Ikeda M, Levine A J. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su S-S, Lahue R S, Au K G, Modrich P. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 25.Wu R S, Hurst-Calderone S, Kohn K W. Cancer Res. 1987;47:6229–6235. [PubMed] [Google Scholar]
- 26.Yamaguchi M, Dao V, Modrich P. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 27.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K, Kinzler K W, et al. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 28.Holmes J, Clark S, Modrich P. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas D C, Roberts J D, Kunkel T A. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 30.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 31.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Fisher R P, Bailey P, Levine A J. Mol Cell Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, et al. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 34.Woo R A, McLure K G, Lees-Miller S P, Rancourt D E, Lee P W. Nature (London) 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez G S, Bryntesson F, Torres-Arzayus M I, Priestley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo P A, Taccioli G E, et al. Nature (London) 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- 36.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips E N, Xia F, Kelsey K T, Liber H L. Radiat Res. 1995;143:255–262. [PubMed] [Google Scholar]
- 38.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Lawrence C W, Li G M, Hays J B. J Biol Chem. 1999;274:16894–16900. doi: 10.1074/jbc.274.24.16894. [DOI] [PubMed] [Google Scholar]
- 40.Mellon I, Rajpal D K, Koi M, Boland C R, Champe G N. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 41.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 42.Papadopoulos N, Nicolaides N C, Wei Y-F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 43.Li G-M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genschel J, Littman S J, Drummond J T, Modrich P. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 45.Araki R, Fukumura R, Fujimori A, Taya Y, Shiloh Y, Kurimasa A, Burma S, Li G C, Chen D J, Sato K, et al. Cancer Res. 1999;59:3543–3546. [PubMed] [Google Scholar]
- 46.Hall S R, Campbell L E, Meek D W. Nucleic Acids Res. 1996;24:1119–1126. doi: 10.1093/nar/24.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 48.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 49.Morgan S E, Kastan M B. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 50.Giaccia A J, Kastan M B. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 51.Agarwal M L, Agarwal A, Taylor W R, Stark G R. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winters Z E, Ongkeko W M, Harris A L, Norbury C J. Oncogene. 1998;17:673–684. doi: 10.1038/sj.onc.1201991. [DOI] [PubMed] [Google Scholar]
- 53.Aquilina G, Hess P, Fiumicino S, Ceccotti S, Bignami M. Cancer Res. 1995;55:2569–2575. [PubMed] [Google Scholar]
- 54.Aquilina G, Fiumicino S, Zijno A, Martinelli S, Overkamp W J, Zdzienicka M Z, Oshimura M, Wild C P, Bignami M. Mutat Res. 1997;385:115–126. doi: 10.1016/s0921-8777(97)00037-2. [DOI] [PubMed] [Google Scholar]
- 55.Nehme A, Baskaran R, Aebi S, Fink D, Nebel S, Cenni B, Wang J Y, Howell S B, Christen R D. Cancer Res. 1997;57:3253–3257. [PubMed] [Google Scholar]
- 56.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. Nature (London) 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 57.Gallagher W M, Cairney M, Schott B, Roninson I B, Brown R. Oncogene. 1997;14:185–193. doi: 10.1038/sj.onc.1200813. [DOI] [PubMed] [Google Scholar]
- 58.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]