Abstract
The enzymatic permeabilization procedure described here allows the detection of intracellular bacteria in the thecate dinoflagellate Alexandrium minutum by using catalyzed reporter deposition-fluorescence in situ hybridization. The combined use of propidium iodide and calcofluor for confocal laser scanning microscopy, together with general and specific fluorescent bacterial probes, demonstrated the intracellular presence of bacteria, including members of the phylum Bacteroidetes.
Interactions between bacteria and algae are commonly observed in aquatic ecosystems. It has been suggested that dinoflagellate-associated bacteria (DABs) may have a role as algal growth and bloom dynamics regulators, that they can be involved in dinoflagellate toxin production, and that some have algicidal properties (4, 5, 8, 9, 10, 11, 14, 15, 17, 20, 21, 24, 28, 29). The relationship between dinoflagellates and DABs has generally been postulated to be symbiotic, as the two organisms coexist and, in some cases, the association is maintained for a long time (1, 5, 6, 10, 12, 16).
The presence of bacteria within dinoflagellate cells is still controversial since, even though the presence of bacteria-like particles inside cultured dinoflagellates, first described by Silva in 1962 (25), has been confirmed in several species (1, 7, 16, 18, 19, 23), it has not been possible to detect them by fluorescence in situ hybridization (FISH) in Alexandrium species (7). Therefore, further studies are needed to clarify whether intracellular bacteria are associated with these microalgae.
The localization, identification, and quantification of specific groups of DABs are important for evaluating the role of bacteria-dinoflagellate associations in marine ecosystems and for reaching a better understanding of toxic bloom dynamics. Microscopy used to assess the physical association between bacteria and phytoplankton or molecular biology techniques to identify DABs cannot provide a simultaneous identification and localization of DABs. For this purpose, in situ hybridization using oligonucleotide probes targeting 16S rRNA (1, 2, 7, 22, 26), combined with a precise detection method such as confocal laser scanning microscopy (CLSM), has been used with positive results (1, 7), though not in the case of thecate dinoflagellates such as Alexandrium spp. (7).
In this study, we have modified previously described catalyzed reporter deposition-FISH protocols and fluorochrome combinations (7, 16, 22, 26) to detect, localize, and identify intracellular bacteria within cells of the thecate dinoflagellate species Alexandrium minutum by using CLSM.
Alexandrium minutum AL10C cells from Estartit (Spain) (provided by S. Fraga) were grown in L1 medium (13) prepared with artificial marine water (Tropic Marin, Germany) under a 16:8 h light/dark cycle at an irradiance level of 60 μmol m−2 s−1 and a temperature of 20°C. Samples were harvested after 10 days of culture, fixed with 4% formaldehyde (Merck, Darmstadt, Germany) for 24 h at 4°C, and immobilized onto an 8-μm-pore-size, 25-mm-diameter Cyclopore polycarbonate membrane (Whatman International Ltd., Maidstone, England). Immobilized cells were rinsed with 1× phosphate-buffered saline (0.1 M NaCl, 2 mM KCl, 4 mM Na2HPO4, pH 7.4) and dehydrated for 5 min in each step of a cold 50%, 80%, 100% ethanol series (Merck, Darmstadt, Germany).
Some dinoflagellates, such as Alexandrium minutum, posses a strong theca made of cellulosic compound layers which constitute a thick wall surrounding the cell that may hinder the entrance of molecules used in FISH. The protocols used in previous studies seem to present no problem for the detection of bacteria inside nonthecate dinoflagellates or nuclear sequences in both nonthecate and thecate dinoflagellates. However, they were not able to detect intracellular bacteria in the thecate Alexandrium spp. (7). Since the nuclear probes used as positive controls gave positive hybridization signals, the lack of signal when using bacterial probes might be due to a lack of intracellular bacteria or to failure of the probes to enter bacterial cells. Thus, in our permeabilization protocol, we introduced changes to previous protocols to address both permeabilization of the theca and improvement of bacterial cell wall permeabilization, facilitating the entrance not only of the bacterial probe but also of lysozyme, which could then permeabilize the bacterial cells and allow hybridization to take place. In the works by Biegala et al. (1, 7), the lack of a theca of Gyrodinium instriatum cells may have facilitated the entrance of lysozyme and, thus, the positive hybridization with intracellular bacteria.
Several reagents and conditions of treatment for theca permeabilization were tested. The optimal condition for permeabilization that allowed an acceptable signal-to-noise ratio using the bacterial probe EUB338 (2) involved incubation at 20°C for 90 min with an enzymatic mix containing 1% hemicellulases (Sigma Chemical, St. Louis, MO), 1% cellulase (Sigma Chemical, St. Louis, MO), and 0.4 M mannitol (Merck, Darmstadt, Germany), adjusted to pH 5.8. Samples were then washed twice at room temperature with 1× phosphate-buffered saline for 20 min each and air dried.
A treatment with lysozyme at a high concentration, during a longer incubation time, and at a higher temperature than previously reported was used to facilitate the entrance of probes to the bacterial cells. Thus, before hybridizations were performed, 10 ml of 10 mg/ml lysozyme (Fluka; Sigma-Aldrich) in 1 M Tris-HCl, pH 7.7, 0.5 M EDTA (Merck, Darmstadt, Germany) was placed over the membranes in a petri dish and kept at 37°C for 90 min in order to partially digest the prokaryotic cell walls. The enzyme reaction was stopped by rinsing the filters in 5 ml sterile water for 5 min. Then, samples were serially dehydrated for 5 min in each step of a 50%, 80%, 100% ethanol series (Merck, Darmstadt, Germany) and kept at 4°C in the dark until FISH experiments were performed.
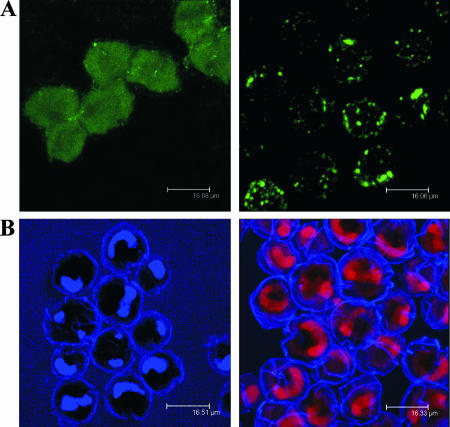
Dinoflagellates show strong autofluorescence, complicating the identification of DABs by nonamplified FISH by masking the fluorescent signal to levels inadequate for this purpose (Fig. 1A, left). Usually the autofluorescence can be diminished by ethanol washes, as described previously (7, 26). In some cases, the serial ethanol washes used were not enough to completely remove autofluorescence and a further treatment was needed. Incubation with 70% ethanol at −20°C for 24 to 48 h diminished the levels of autofluorescence, making the samples workable.
FIG. 1.
(A) CLSM images of hybridization signals from FISH experiments. Assay of A. minutum cells using probe EUB338 without (left) or with (right) the autofluorescence removal and permeabilization pretreatments and tyramide signal amplification. (B) A. minutum cells stained with calcofluor and either DAPI (left) or propidium iodide (right).
Hybridizations were carried out with the horseradish peroxidase-conjugated oligonucleotide probes EUB338 (2) and CFB319a (19) (Biomers.net, Germany), with the signal being amplified by the use of a tyramide signal amplification (TSA) protocol (3, 22, 27). Whole-cell in situ hybridization by using TSA-FISH was adapted from the methods of Amann et al. (3) and Biegala et al. (7). The probes were used at a final concentration of 5 μM. The TSA reaction was carried out by using a TSA kit (Invitrogen, Eugene, OR) in which tyramide was labeled with Alexa Fluor 488, following the specifications of the manufacturer. After the TSA reaction was performed, the filters were dried and kept in the dark at 4°C until the counterstain step.
To be able to assess the three-dimensional localization of the bacterial cells, we tried calcofluor as a theca-staining agent and two alternative DNA staining compounds, 4′,6′-diamidino-2-phenylindole (DAPI) and propidium iodide. The initial experiments were performed by combining calcofluor and DAPI (Fig. 1B, left) as previously described (1, 7), but since the two fluorochromes have very close excitation wavelengths and produce the same blue fluorescence emission (470 nm), it was difficult to clearly distinguish the inside from the outside of the dinoflagellate cells. The use of propidium iodide, which is excited at 537 nm, producing red fluorescence, allowed the outside (blue) and the inside (red) of the dinoflagellate cells to be distinguished (Fig. 1B, right). The following protocol was used for staining: dried samples on filters were incubated with 15 μl of a mix containing 100 μg/ml of calcofluor (Sigma Chemical, St. Louis, MO) and 1.5 μM propidium iodide (Invitrogen, Eugene, OR) for 10 min at room temperature in the dark. The filters were rinsed in 5 ml sterile water for 5 min, dried, mounted with ProLong gold antifade reagent (Invitrogen, Eugene, OR), and covered with coverslips. The whole-cell in situ preparations were kept at −20°C in the dark until use. Fluorescence images were acquired with a Leica TCS SP2 confocal spectral microscope. The scanning speed was 400 Hz, and the slices were 1 μm thick. Oil immersion objectives of 40× (numerical aperture, 1.25 to 0.75; Leica) and 63× (numerical aperture, 1.4 to 0.6; Leica) were used. The micrographs were taken with a Leica DFC 350 FX (3.3 megapixels and 12 bits).
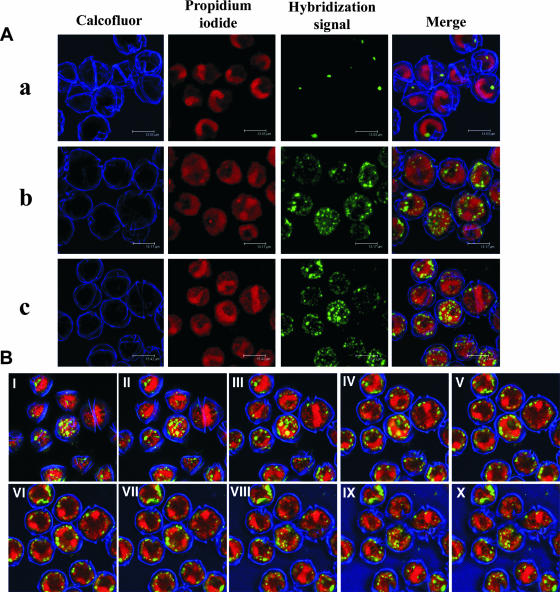
The combination of various small but significant modifications of previous methods resulted in a substantial increase in FISH sensitivity for assaying thecate dinoflagellates compared with the sensitivities observed in previous studies (1, 7, 22). The protocol used has allowed us to show that horseradish peroxidase-labeled oligonucleotide probes targeting 16S rRNA can successfully access the inside of thecate dinoflagellate cells and enter bacterial cells (Fig. 1A, right), as well as to demonstrate unequivocally and for the first time that bacteria from the Cytophaga/Flavobacterium/Bacteroides group, among others, are present inside the dinoflagellate cells (Fig. 2).
FIG. 2.
CLSM images of TSA-FISH of A. minutum cells. (A) Assay without probe (negative control) (a) and with probe CFB319a (b) or EUB338 (c). (B) Ten consecutive 1-μm-thick optical sections (I to X) of the sample shown in row c of panel A.
Acknowledgments
We thank S. Fraga for providing the A. minutum strains used in this work; A. Fernández-Villamarín and D. Morales for technical assistance; and R. Martinez, J. Field, and J. P. Abad for critical readings of the manuscript.
L. Palacios was funded through a fellowship from the Comunidad de Madrid. This work was supported by grant CTM2004-04078.C03-03/MAR from the MEC awarded to I.M. and an institutional grant from Fundación Ramón Areces to the Centro de Biologia Molecular Severo Ochoa.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Alverca, E., I. C. Biegala, G. M. Kennaway, J. Lewis, and S. Franca. 2002. In situ identification and localization of bacteria associated with Gyrodinium instriatum (Gymnodiales, Dinophyceae) by electron and confocal microscopy. Eur. J. Phycol. 37:523-530. [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. Zarda, D. A. Stahl, and K.-H. Schleifer. 1992. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 58:3007-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaro, A. M., M. S. Fuentes, S. R. Ogalde, J. A. Venegas, and B. J. Suárez-Isla. 2005. Identification and characterization of potentially algal-lytic marine bacteria strongly associated with the toxic dinoflagellate Alexandrium catenella. J. Eukaryot. Microbiol. 52:191-200. [DOI] [PubMed] [Google Scholar]
- 5.Baker, T. R., G. J. Doucette, C. L. Powell, G. L. Boyer, and F. G. Plumley. 2003. GTX4 imposters: characterization of fluorescence compounds synthesized by Pseudoalteromonas stutzeri SF/P and Pseudomonas/Alteromonas PTB-1, symbionts of saxitoxin-producing Alexandrium spp. Toxicon 41:339-347. [DOI] [PubMed] [Google Scholar]
- 6.Bell, W. H., and R. Mitchell. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143:265-277. [Google Scholar]
- 7.Biegala, I., G. Kennaway, E. Alverca, J.-F. Lennon, D. Vaulot, and N. Simon. 2002. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescence in situ hybridization and confocal microscopy. J. Phycol. 38:404-411. [Google Scholar]
- 8.Doucette, G. J., and C. L. Powell. 1998. Algal-bacterial interactions: can they determine the PSP-related toxicity of dinoflagellates?, p. 406-409. In B. Reguera, J. Blanco, M. L. Fernández, and T. Wyatt (ed.), Harmful algae. Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Santiago de Compostela, Spain.
- 9.Doucette, G. J., E. R. McGovern, and J. A. Babinchak. 1999. Alguicidal bacteria active against Gymnodinium breve (Dinophyceae). I. Bacterial isolation and characterisation of killing activity. J. Phycol. 35:1447-1454. [Google Scholar]
- 10.Furuki, M., and M. Kobayashi. 1991. Interactions between Chattonella and bacteria and prevention of this red tide—EMECS '90. Mar. Pollut. Bull. 23:189-193. [Google Scholar]
- 11.Gallacher, S., and E. A. Smith. 1999. Bacteria and paralytic shellfish toxins. Protist 150:245-255. [DOI] [PubMed] [Google Scholar]
- 12.Gold, K., and U. Polingher. 1971. Occurrence of endosymbiotic bacteria in marine dinoflagellates. J. Phycol. 7:264-265. [Google Scholar]
- 13.Guillard, R. R. L., and P. E. Hargraves. 1993. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32:234-236. [Google Scholar]
- 14.Hold, G. L., E. A. Smith, T. H. Birkbeck, and S. Gallacher. 2001. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 36:223-234. [DOI] [PubMed] [Google Scholar]
- 15.Kim, M. C., I. Yoshinaga, I. Imai, K. Nagasaki, S. Itakura, and Y. Ishida. 1998. A close relationship between alguicidal bacteria and termination of Heterosigma akashiwo (Raphidophyceae) bloom in Hiroshima Bay, Japan. Mar. Ecol. Prog. Ser. 170:25-32. [Google Scholar]
- 16.Lewis, J., G. Kennaway, S. Franca, and E. Alverca. 2001. Bacteria-dinoflagellate interactions: investigative microscopy of Alexandrium spp. (Gonyaulacales. Dinophyceae). Phycologia 40:280-285. [Google Scholar]
- 17.Lovejoy, C., J. P. Bowman, and G. M. Hallegraeff. 1998. Alguicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, Gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl. Environ. Microbiol. 64:2806-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas, I. A. N. 1982. Observations on Noctiluca scintillians Macartney (Ehrenb.) (Dinophyceae) with notes on an intracellular bacterium. J. Plankton Res. 4:401-409. [Google Scholar]
- 19.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleider. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacterium-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 20.Mayali, X., and F. Azam. 2004. Alguicidal bacteria in the sea and their impact on algal bloom. J. Eukaryot. Microbiol. 51:139-144. [DOI] [PubMed] [Google Scholar]
- 21.Palacios, L., D. Arahal, B. Reguera, and I. Marin. 2006. Hoeflea alexandrii sp. nov., isolated from the toxic dinoflagellate Alexandrium minutum AL1V. J. Syst. Evol. Microbiol. 56:1991-1995. [DOI] [PubMed] [Google Scholar]
- 22.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausch de Traunbenberg, C., M.-L. Geraud, M. O. Soyer-Gobillard, and D. Emdadi. 1995. The toxic dinoflagellate Prorocentrum lima and its associated bacteria. Eur. J. Protistol. 31:318-326. [Google Scholar]
- 24.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva, E. S. 1962. Some observations on marine dinoflagellate cultures. I. Prorocentrum micans Ehr. and Gymnodinium spinifera (Clap.and Lach.) dies., Gonyaulax tamarensis Leb., and Peridinium trochoideum (Stein) Lemm. Notas Estudos Inst. Biol. Mar. 26:1-21. [Google Scholar]
- 26.Tujula, N. A., C. Holmstöm, M. Mußmann, R. Amann, S. Kjelleberg, and G. R. Crocetti. 2006. A CARD-FISH protocol for the identification and enumeration of epiphytic bacteria on marine algae. J. Microbiol. Methods 65:604-607. [DOI] [PubMed] [Google Scholar]
- 27.Urdea, M. S., B. D. Warner, J. A. Running, M. Stempien, J. Clyne, and T. Horn. 1998. A comparison of nonradioisotopic hybridization assay methods using fluorescent, chemiluminiscent and enzyme labelled synthetic oligodeoxyribonucleotide probes. Nucleic Acids Res. 16:4937-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uribe, P., and R. T. Espejo. 2003. Effect of associated bacteria on the growth and toxicity of Alexandrium catenella. Appl. Environ. Microbiol. 69:659-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, C. H., Y. Y. Wang, Y. Y. Sun, and X. T. Xie. 2006. Effect of antibiotic treatment on toxin production by Alexandrium tamarense. Biomed. Environ. Sci. 16:340-347. [PubMed] [Google Scholar]