Abstract
Bacteria of the genus Methylobacterium are widespread in the environment, but their ecological role in ecosystems, such as the plant phyllosphere, is not very well understood. To gain better insight into the distribution of different Methylobacterium species in diverse ecosystems, a rapid and specific cultivation-independent method for detection of these organisms and analysis of their community structure is needed. Therefore, 16S rRNA gene-targeted primers specific for this genus were designed and evaluated. These primers were used in PCR in combination with a reverse primer that binds to the tRNAAla gene, which is located upstream of the 23S rRNA gene in the 16S-23S intergenic spacer (IGS). PCR products that were of different lengths were obtained due to the length heterogeneity of the IGS of different Methylobacterium species. This length variation allowed generation of fingerprints of Methylobacterium communities in environmental samples by automated ribosomal intergenic spacer analysis. The Methylobacterium communities on leaves of different plant species in a natural field were compared using this method. The new method allows rapid comparisons of Methylobacterium communities and is thus a useful tool to study Methylobacterium communities in different ecosystems.
Bacteria of the genus Methylobacterium are facultative methylotrophs capable of growth on one-carbon compounds, such as methanol, methylamine, formaldehyde, and formate (16). The members of this genus of Alphaproteobacteria are ubiquitous in nature. They have been detected in soil, dust, freshwater, lake sediments, and the air and on plants (16). They have also been found in association with humans (2), and some members of this genus have increasingly been reported to be a cause of opportunistic infections in immunocompromised patients (22). The genus Methylobacterium currently comprises 28 described species (28, 60). Several new species have been described during the last few years, including species from environments that have been analyzed for several decades, such as the plant phyllosphere (23, 27, 28, 38, 58). This suggests that the diversity of Methylobacterium is still not fully known. The high diversity of species in the genus Methylobacterium and their broad distribution in nature raises questions of whether different species are adapted to certain environments and whether they play similar ecological roles in different environments. To address these questions, a better understanding of the distribution of Methylobacterium strains between and within different ecosystems and analysis of Methylobacterium community compositions and dynamics in natural habitats are necessary.
One of the major habitats of Methylobacterium is represented by plants. Members of this genus have been detected by cultivation-dependent methods (8, 12, 39, 45, 59) and, to a lesser extent, by cultivation-independent methods (3, 24, 25, 47, 48) as epiphytic and endophytic colonizers of the plant phyllosphere, as intracellular colonizers of the buds of Scots pine (47), and as root-nodulating symbionts (26, 54). The focus of all the cultivation-independent studies was characterization of the whole bacterial community. So far, the Methylobacterium community on plants has not been characterized using cultivation-independent methods, despite the ubiquity of this bacterial genus on plants (21). However, a better understanding of plant colonization by Methylobacterium should help to elucidate the importance of this association for both partners, the bacteria and the plants. While it is evident that there is a symbiotic interaction between root-nodulating strains and their host plants, the importance of Methylobacterium in the plant phyllosphere is unknown. On the one hand, it has been shown that Methylobacterium species can benefit from methanol that is released by the plant and may also grow on other plant-derived carbon compounds (1, 15, 55). On the other hand, it has been shown that these bacteria produce phytohormones (31, 44, 56) and stimulate seed germination and growth of certain plants (1, 32, 36, 49).
To study Methylobacterium communities in their natural habitats, a rapid cultivation-independent method is needed. In this study we describe for the first time a genus-specific automated ribosomal intergenic spacer analysis (ARISA) method that allows generation of fingerprints from Methylobacterium communities. To evaluate the resolution of this method, we analyzed the potential of the 16S-23S rRNA intergenic spacer (IGS), also referred to as ribosomal intergenic transcribed spacer 1, to differentiate and identify Methylobacterium strains based on IGS sequence analysis of the strains. The established ARISA method was used to compare Methylobacterium communities on leaves of different plant species.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Methylobacterium strains were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) or were kindly provided by L. Chistoserdova, University of Washington, Seattle, S. Vuilleumier, Universite Louis Pasteur, Strasbourg, France, and Bernard Dreyfus, UMR LSTM, Montpellier, France. Bacterial strains were grown at 28°C on mineral salts medium (43) supplemented with 120 mM methanol or 30 mM succinate.
Plant-associated Methylobacterium strains were enriched and isolated from greenhouse-grown Arabidopsis thaliana Col-0 plants, agriculturally grown sunflower (Helianthus annuus), naturally grown cinquefoil (Potentilla reptans), and black locust (Robinia pseudoacacia). All of the plants were growing at the complex of the Institute Nationale de la Recherche Agronomique in Castanet-Tolosan, France. Leaf material was added to 1 ml of 20 mM potassium phosphate buffer, and bacteria were dislodged by sonication for 5 min. The plant material was then removed, and the bacteria were pelleted by centrifugation at 6,000 × g for 10 min. The pellet was resuspended in 150 μl of buffer, and a 10-fold dilution series was plated on the selective mineral salts agar medium supplemented with methanol and cycloheximide (50 mg liter−1). Pink colonies with different morphologies were restreaked from the highest positive dilutions and isolated. The purity of the isolates was checked on mineral salts medium agar plates supplemented with succinate and on nutrient agar plates.
Environmental samples.
Plant material was obtained from grass (Poaceae), moss (Bryidae), and three different herbs, P. reptans, Bellis perennis, and Taraxacum officinale. All the plants grew on a lawn in Castanet-Tolosan, France (43°31′00.2"N, 1°30′12.4"E), and were obtained from a 2.25-m2 area at two different times, on 22 March and 12 April 2005. Each sample consisted of leaf material taken from a few different leaves of an individual plant. Between 12 and 184 mg of plant material was macerated and resuspended in 850 μl of phosphate buffer (120 mM, pH 8.0). One hundred microliters of this suspension was used to determine numbers of cells by serial dilution as described above. Numbers of cells were calculated based on the numbers of pink colonies counted for the highest dilutions after 8 days of incubation. The remaining suspension was frozen at −80°C for DNA extraction.
DNA extraction from bacterial cultures and environmental samples.
DNA was extracted from bacterial cultures using a method based on mechanical cell disruption (two 90-s treatments) with a Retsch Mixer Mill MM301 (Retsch GmbH, Haan, Germany), followed by ammonium acetate treatment and isopropanol precipitation of the DNA as described by Henckel et al. (19).
DNA was extracted from environmental samples using a FAST DNA spin kit (Bio 101, La Jolla, CA) according to the manufacturer's instructions. In brief, the samples were transferred to lysing matrix A tubes, 400 μl of buffer CLS-VF and 200 μl of PPS buffer were added, and cells were lysed with the Retsch Mixer Mill MM301 using two 90-s treatments. After centrifugation at 14,500 × g for 10 min at 4°C, the supernatant was removed and mixed with an equal volume of binding matrix. The matrix-bound DNA was purified by washing it twice with a 5.5 M guanidine isothiocyanate solution. The matrix was loaded onto a kit-supplied spin filter, and further purification of the DNA was performed according to the manufacturer's instructions. Final elution of the DNA from the filters was performed with 100 μl of DNase-free water.
Primer development and establishment of specific PCR assays.
Methylobacterium-specific primers were designed using the probe design tool of the ARB software package (37) and the 16S rRNA gene sequence database (release ssu_jan04_corr_opt), which was regularly updated with all publicly available Methylobacterium sequences. Primers 1319fGC20 (GCC CCC CGC CCC CGC CGC CCA CTC GRG TGC ATG AAG GCG G) and 444lof (CGG GAC GAT AAT GAC GGT ACC GGD DGA A) were developed and used to establish specific PCR assays. Primer 1319fGC20 contains a 20-bp GC clamp at the 5′ end. This clamp resulted in specificity of the primer that was greater than that of the primer without the clamp (data not shown). Several previously described primers that bind in the 5′ region of the 23S rRNA gene were evaluated in silico using the ARB software package (7, 40). Only two primers displayed no mismatches to the 23S rRNA gene sequences of Methylobacterium: ITSReub (7) and 23Sr (4). In addition, primers 21r (GCG CCA AGG CAT CCA CCG A) and 110r (GGG TTS CCC CAT TCG GAA ATC) were developed. These primers bind to the same regions as primers ITSReub and 23Sr but have higher melting temperatures that are comparable to the melting temperature of the specific forward primers. Moreover, primer 21r could be used for a nested cycle sequencing approach (described below). In addition, primer 45r (GAC GGG ATC GAA CCG ACG ACC) was developed, which binds to the tRNAAla gene.
To establish a specific PCR assay for the detection of Methylobacterium, genomic DNA of Methylobacterium extorquens AM1, Methylobacterium nodulans ORS2060T, and the type strain of the type species, Methylobacterium organophilum DSM 760T, which turned out to be a Methylobacterium radiotolerans strain after resequencing (29), were used. Among the nontarget organisms with the lowest number of mismatches, Methylosinus trichosporium OB3bT and Methylocystis sp. strain SC2 (grown as described previously [20]) were chosen as negative controls for PCR assays with primer 1319fGC20. Meiothermus ruber DSM 1279 (grown as described by the DSMZ) and Bradyrhizobium japonicum USDA110 (genomic DNA kindly provided by C. Bontemps, LIPM, Toulouse, France) were chosen as negative controls for PCR assays with primer 444lof.
The PCR assay mixture (total volume, 20 μl) contained 2 μl of Taq polymerase supplied 10× PCR buffer (Invitrogen), 1.25 mM of each deoxynucleoside triphosphate (Invitrogen), 0.5 μM of each primer (Invitrogen), 0.02 U μl−1 of Platinum Taq DNA polymerase (Invitrogen), and 0.5 μl of template DNA (DNA concentration, 5 ng μl−1). The MgCl2 concentration was adjusted to 7.5 mM for primer 1319fGC20 and to 4 mM for primer 444lof. The PCR was performed with an Eppendorf gradient Mastercycler. The PCR program consisted of initial denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at a specific temperature for 45 s, and elongation at 72°C for 2 min and then a final elongation at 72°C for 7 min. Specific amplification was achieved for primer 1319fGC20 in combination with primer 45r at an annealing temperature of 69.5°C after six cycles of touchdown from 72°C with a 0.5°C temperature decrease for each cycle. For primer 444lof the annealing temperature was set to 70°C after four cycles of touchdown from 72°C.
When the established PCR protocol was used to amplify DNA from environmental samples, 0.25 μg μl−1 of bovine serum albumin (Roche Diagnostics) was added to the assay mixtures. The reaction was performed using a 50-μl (total volume) mixture containing 2.5 μl of template DNA. To obtain sufficient PCR product from environmental samples, a two-step PCR was performed. The second PCR, consisting of 25 cycles, was performed using 1 μl of PCR product from the first PCR in a 25-μl (total volume) mixture.
The specificity of each primer set under the described PCR conditions was checked by constructing a clone library from an environmental sample (naturally grown moss) using a TOPO TA cloning kit (Invitrogen). PCR products were purified with a NucleoSpin Extract II purification kit (Machery-Nagel, Düren, Germany). Positive clones were picked and screened with M13 primers for the correct insert, and the 16S rRNA gene fragments were sequenced without further purification of the PCR products. The cycle sequencing reaction was performed with a nested primer (primer 1492r [20]) and an annealing temperature of 60°C instead of 55°C.
ARISA of pure cultures and environmental samples.
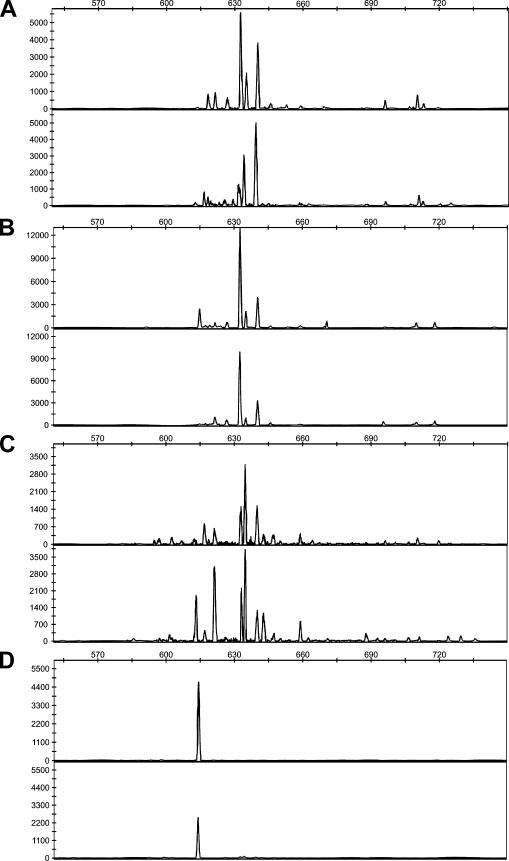
DNA was amplified as described above with primer 1319fGC20 and primer 45r labeled at the 5′ end with 6-carboxyfluorescein. For analysis of the lengths of the PCR products, 2 μl of fivefold-diluted PCR product (a mixture of two independently prepared PCR assay mixtures for environmental samples) was mixed with 7.9 μl of Hi-Di formamide (Applied Biosystems) and 0.1 μl of GeneScan-1000 ROX size standard (Applied Biosystems). The samples were denatured by heating them at 95°C for 8 min and were analyzed with a 3730 ABI capillary sequencer filled with polymer POP7. Data analysis was done using the Genemapper Software, version 4.0 (Applied Biosystems). Peaks with a difference of at least 2 bp were clearly baseline separated in the chromatograms (see Fig. 3). Double peaks were considered to represent two PCR products with a 1-bp difference, and each half of the peak was integrated separately. For comparison of ARISA patterns, fragments that were the same size were grouped together. All peaks with a height of ≥50 fluorescent units were included in further analyses. Samples were standardized using the method of Sait et al. (50) by eliminating all peaks whose relative abundance was less than 1% in each sample.
FIG. 3.
ARISA chromatograms representing the Methylobacterium communities on leaf samples taken from individual plants of B. perennis (A), T. officinale (B), moss (C), and grass (D) at the second sampling event. Representative samples of each plant species were taken and are indicated in Fig. 4.
Sequencing and phylogenetic analysis.
The Methylobacterium isolates obtained from the plant phyllosphere were identified by sequencing of a 16S rRNA gene fragment amplified with primers 9f and 1492r using a 3730 ABI capillary sequencer as described previously (20). Only the central part of the 16S rRNA gene was sequenced for all isolates. Therefore, the cycle sequencing reaction was performed with nonpurified PCR products using the nested primer 533f (19) at an annealing temperature of 60°C instead of 55°C. Nearly complete 16S rRNA gene sequences were generated only for strains with different partial 16S rRNA gene sequences.
Sequencing of the IGS of Methylobacterium strains was performed after amplification of the IGS region using primers 907f (reverse complement of 907b [35]) and 110r. Nonpurified PCR products were sequenced with nested primers 1492f (reverse complement of 1492r) and 21r. In order to analyze IGS sequence variability in strains with multiple different rrn operons, the different IGS copies of a few type strains (Methylobacterium aquaticum DSM 16371T, Methylobacterium fujisawaense 0-31T, and Methylobacterium mesophilicum A47T) for which PCR products of different sizes had been detected were analyzed by cloning PCR products as described above and sequencing eight randomly selected clones.
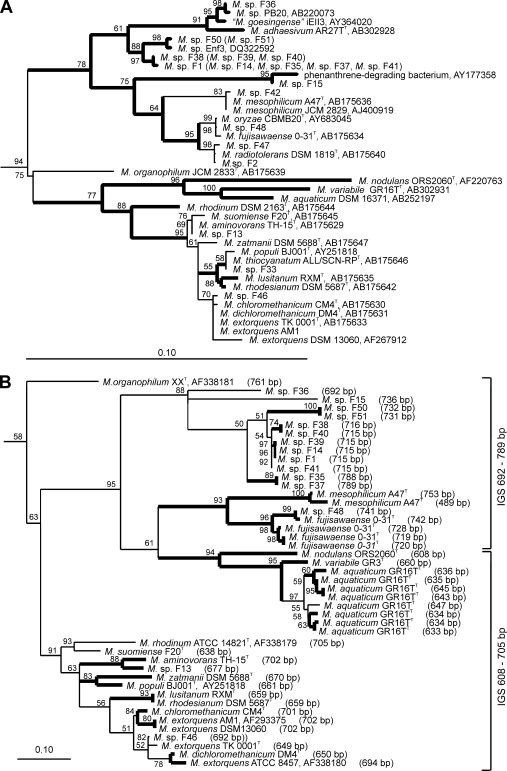
Sequence assembly was performed with Vector NTI (Invitrogen). Published 16S rRNA gene sequences closely related to the sequences of the isolates were identified by BLAST searches (October 2007). Results were confirmed by comparative sequence analysis with the ARB database. IGS sequences were aligned using ClustalW (implemented in the ARB software package), and the alignment was manually corrected. Treepuzzle trees (51) were calculated using the ARB software package. The evolutionary model of Schöniger and von Haeseler (52) was used for the 16S rRNA gene-based tree, and the HKY evolutionary model of Hasegawa et al. (18) was used for the IGS sequence-based tree. The topology of the Treepuzzle trees was compared to the topology of bootstrapped maximum likelihood trees, neighbor joining trees with Jukes-Cantor correction, and parsimony trees calculated with the Phylip 3.65 software package (http://evolution.genetics.washington.edu/phylip.html). Sequences of other Alphaproteobacteria were included in tree reconstruction analyses and used to define an outgroup (not shown in Fig. 1); sequences of Rhodopseudomonas palustris, Bradyrhizobium japonicum, and Afipia felis were used (accession numbers X87279, AP005940, and AF338177 for the 16S rRNA gene-based tree and accession numbers AF338178, BA000040, and AF338177 for the IGS sequence-based tree).
FIG. 1.
(A) 16S rRNA gene sequence-based tree showing the phylogenetic placement of the phyllosphere isolates obtained in this study. The tree was calculated based on 1,356 nucleotide positions. Isolates in parentheses were not included in the tree calculation process but were added to facilitate comparison with the 16S-23S rRNA IGS sequence-based tree. (B) Phylogenetic tree calculated based on aligned IGS sequences (1,244 nucleotide positions). The full length of the IGS, deduced from the nucleotide sequence, is shown in parentheses. Both trees were constructed using the Treepuzzle algorithm. Branch points that were recovered in <50% of 10,000 reconstructed trees are shown as multifurcations; values for branches with ≥50% recovery are indicated. Branch points in bold type are supported by at least two other algorithms (maximum parsimony, maximum likelihood, or neighbor joining). Bars = 0.10 change per nucleotide position. The 16S rRNA gene sequence of M. extorquens AM1 was obtained from the draft genome sequence (www.integratedgenomics.com/genomereleases.html#6).
Statistical data analysis.
Significant differences in numbers of Methylobacterium cells on different plants were analyzed by an analysis of variance of log-transformed data using SPSS 14.0 (SPSS Inc., Chicago, IL). Significant differences between numbers of cells of different plant species were reported based on Dunnett's T3 post hoc tests, a test that can be used when there are unequal variances.
To visualize similarities between ARISA patterns of different samples, a cluster analysis was performed using SYSTAT 12.0 (SYSTAT Software Inc., Chicago, IL). The tree presented was calculated based on pairwise Pearson similarity coefficients and was constructed based on a hierarchical clustering procedure using flexible beta linkage with a β value of −0.25.
Nucleotide sequence accession numbers.
Representative nearly complete 16S rRNA gene sequences obtained in this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AM910530 to AM910541. IGS sequences have been deposited under accession numbers AM910542 to AM910581.
RESULTS AND DISCUSSION
In order to study Methylobacterium communities in environmental samples by cultivation-independent methods, (i) a suitable target in the Methylobacterium genome that allows specific amplification of Methylobacterium DNA in complex microbial communities is needed and (ii) the nucleotide sequence of the target has to be sufficiently different in diverse Methylobacterium species or even strains to obtain good resolution of the diversity of a community. We observed that 16S rRNA gene-based fingerprinting methods, such as terminal restriction fragment length polymorphism analysis or denaturing gradient gel electrophoresis, do not allow adequate resolution of the Methylobacterium diversity (data not shown), a finding that is consistent with the results of studies that focused on other bacterial groups (11, 30, 40). Therefore, we evaluated the potential of ARISA, a fast and high-resolution fingerprinting method, for differentiation of Methylobacterium strains (13). The 16S-23S rRNA intergenic spacer, which is targeted by this method, shows more variation in sequence and length than the 16S rRNA gene, allowing better differentiation of isolates of a particular bacterial species or genus (17). The IGS has frequently been targeted in studies of phylogeny, molecular evolution, or population genetics to differentiate between very closely related bacteria (5, 40, 41). The length variability of the IGS has been used as a basis for more complex community profiling methods with environmental samples (13, 14, 40, 41).
Development of Methylobacterium-specific ARISA.
In order to PCR amplify the IGS specifically from Methylobacterium strains, at least one genus-specific primer is needed. We used the 16S rRNA gene as a target for the specific detection of Methylobacterium, because a much larger database for the development of such a genus-specific primer is available for this gene than for the IGS region. Several specific primers and probes that target the 16S rRNA gene of Methylobacterium have been described previously (6, 33, 42, 57), but all of them have shortcomings. Either they no longer cover the entire diversity known for the genus, they detect only a specific subset of species, or they are not highly specific, especially if they are not used in combination in PCR assays. Thus, we evaluated the potential of the 16S rRNA gene for the development of novel genus-specific primers. In silico analysis using the ARB probe design tool revealed two suitable regions. Different primers that bind to these regions were designed. Two of these primers, primers 1319fGC20 and 444lof, showed the greatest specificity in PCR assays. The percentage of Methylobacterium sequences showing mismatches with the developed primers was less than 4% (see Table S1 in the supplemental material). Both primers had no mismatches with some nontarget organisms (i.e., bacteria belonging to other genera) (see Table S2 in the supplemental material). Most nontarget organisms that contained sequences matching primer 1319fGC20 belonged to the genera Acidicaldus and Sphingomonas, and most nontarget organisms that contained sequences matching primer 444lof belonged to the Acidobacteriaceae. All other nontarget genera were excluded by primer 444lof with at least two mismatches, while primer 1319fGC20 had just one mismatch with some nontarget genera. Based on in silico analyses, both primers that were developed should detect the different Methylobacterium species more specifically than the previously described primers detect these species, especially in PCR assays in which only one specific primer is used. To generate PCR products for ARISA, the specific forward primers were tested in combination with different reverse primers that targeted either the 23S rRNA gene or the tRNAAla gene, which is located upstream of the 23S rRNA gene within the IGS. Amplification of DNA from nontarget organisms could be excluded most efficiently when reverse primer 45r was used, which binds to the tRNAAla gene. In combination with this reverse primer, primer 1319fGC20 was more suitable for ARISA than primer 444lof, since PCR products less than 1,000 bp long could be obtained. Smaller PCR products are preferable in ARISA because their lengths can be determined more precisely.
In order to evaluate primer specificity, both primers were used to amplify DNA from a plant phyllosphere sample. The plant sample showing the greatest peak diversity in ARISA (a moss sample) was selected for this analysis. Twenty-three clones, constructed using amplicons of primers 1319fGC20 and 45r, were sequenced. Sequence analysis of the 16S rRNA gene fragment showed that 3 of the 23 sequences were not related to Methylobacterium but rather were related to Sphingomonas strains. The in silico analysis had already shown that some Sphingomonas strains have no mismatches with primer 1319fGC20 (see Table S2 in the supplemental material). Sequence analysis of the cloned 16S rRNA gene fragments generated with primers 444lof and 45r showed that primer 444lof was less specific (data not shown). Therefore, primer 1319fGC20 was used in combination with primer 45r in further studies.
Potential of the 16S-23S rRNA IGS for differentiation and identification of Methylobacterium strains.
The 16S-23S rRNA IGS has been used previously for differentiation of Methylobacterium isolates (24, 59). However, the suitability of the IGS for differentiation of Methylobacterium strains based on length heterogeneity and its potential as a phylogenetic marker for identification of strains in environmental samples have not been evaluated so far. In order to address these questions and to determine whether it is possible to differentiate closely related strains of the same species, it was first necessary to obtain different strains and species of Methylobacterium. Hence, Methylobacterium strains were isolated from the phyllosphere of different plants (Table 1). Phylogenetic analysis of partial 16S rRNA gene sequences (Escherichia coli positions 560 to 1451) revealed that all isolates belonged to the genus Methylobacterium and that 12 different sequence types were obtained (Fig. 1 and Table 1). The nearly complete 16S rRNA gene sequence of most isolates had 99 to 100% identity to the sequences of described type strains, such as the type strains of Methylobacterium aminovorans, M. extorquens, Methylobacterium thiocyanatum, M. mesophilicum, and M. radiotolerans. Isolates F1, F38, and F50 had 97.6 to 97.9% identity to “Methylobacterium goesingense” iEII3 and 97.6 to 97.8% identity to Methylobacterium adhaesivum AR27T. Isolate F15 was most closely related (99.6%) to a phenanthrene-degrading Methylobacterium strain (accession number AY177358); the most closely related type strains were the type strains of M. mesophilicum (96.8%), M. radiotolerans (96.5%), and M. fujisawaense (96.4%), suggesting that this strain may represent a new species.
TABLE 1.
Methylobacterium isolates obtained from the plant phyllospherea
| Species (based on 16S rRNA gene sequence)a | Strain | Sequence to which the 16S rRNA gene sequence is identical | Isolation source | ARISA pattern (length[s] of detected fragments [bp])b |
|---|---|---|---|---|
| M. radiotolerans | F2 | F2 | A. thaliana | 621, 695 |
| M. radiotolerans | F9 | F2 | A. thaliana | 629 |
| M. radiotolerans | F12 | F2 | A. thaliana | 603, 617, 695 |
| M. radiotolerans | F23 | F2 | A. thaliana | 603, 683 |
| M. radiotolerans | F47 | F47 | A. thaliana | 603, 617, 639, 652, 696 |
| M. thiocyanatum | F53 | F33 | R. pseudoacacia | 593, 608 |
| M. thiocyanatum | F18 | F33 | A. thaliana | 605 |
| M. thiocyanatum | F33 | F33 | A. thaliana | 592, 606 |
| M. thiocyanatum | F19 | F33 | A. thaliana | 589, 611 |
| M. thiocyanatum | F34 | F33 | A. thaliana | 592, 606 |
| “M. goesingense”c | F36 | F36 | P. reptans | 616 |
| M. aminovorans | F13 | F13 | A. thaliana | 588 |
| M. extorquens | F46 | F46 | A. thaliana | 602 |
| M. mesophilicum | F42 | F42 | H. annuus | 641 |
| M. oryzaed | F48 | F48 | A. thaliana | 638 |
| Methylobacteriumsp. | F15 | F15 | A. thaliana | 639 |
| Methylobacteriumsp. | F1 | F1 | A. thaliana | 627 |
| Methylobacterium sp. | F14 | F1 | A. thaliana | 628 |
| Methylobacterium sp. | F35 | F1 | P. reptans | 700 |
| Methylobacterium sp. | F37 | F1 | P. reptans | 698 |
| Methylobacterium sp. | F41 | F1 | P. reptans | 629 |
| Methylobacteriumsp. | F38 | F38 | P. reptans | 628 |
| Methylobacterium sp. | F39 | F38 | P. reptans | 630 |
| Methylobacterium sp. | F40 | F38 | P. reptans | 629 |
| Methylobacteriumsp. | F50 | F50 | A. thaliana | 632 |
| Methylobacterium sp. | F51 | F50 | A. thaliana | 633 |
Bold type indicates strains that are representative of all strains with identical partial 16S rRNA gene sequences.
Due to methodological variation, slight differences (±2 bp) in the fragment length are possible. Thus, isolates may have identical ARISA patterns even if the lengths of the fragments in the patterns are slightly different.
The most closely related validly described type strain is M. adhaesivum AR27T.
The phylogenetic placement of the strain is not absolutely clear. The levels of 16S rRNA gene sequence identity are 100% with M. oryzae and 99.93% with M. fujisawaense.
The potential of the IGS for differentiation of Methylobacterium strains on the basis of IGS sequence length differences was evaluated in silico using IGS sequence analysis of several type strains and phyllosphere isolates and by ARISA using the established PCR assays for DNA amplification of all phyllosphere isolates and type strains. IGS sequence analysis showed that all strains harbored rrn operons, in which two tRNA genes, the tRNAIle and tRNAAla genes, were located in the IGS, a configuration that is known for other bacterial taxa (5). M. mesophilicum A47T has in addition an rrn operon(s), in which both tRNA genes are missing. The total lengths of the intergenic spacers containing two tRNA genes were in the range from 608 bp (M. nodulans ORS2060T) to 789 bp (isolate F37) (Fig. 1), which is rather long compared to the lengths of spacers in several other bacterial taxa (5). A longer IGS increases its suitability for differentiation of strains, based on differences in length and sequence. Indeed, most species and even strains of the same species showed differences in IGS sequence length (Fig. 1); for example, this was true for different M. extorquens strains plus Methylobacterium chloromethanicum CM4 and Methylobacterium dichloromethanicum DM4, which were recently reclassified as M. extorquens strains (29).
ARISA patterns were obtained for all type strains and phyllosphere isolates (Tables 1 and 2). Fragments of more than one size were observed for several of the strains, due to the presence of multiple, different rrn operons in these Methylobacterium strains. Separation of strains with shorter (561- to 618-bp) or longer (603- to 726-bp) fragments in correlation to their phylogenetic placement was observed. The full length of the IGS varied in the same way among the different species, as shown in Fig. 1. The ARISA patterns were different for all analyzed species and often also for different strains (e.g., for several of the M. extorquens strains or for M. radiotolerans strains) (Tables 1 and 2). Thus, differentiation and classification of isolates based on IGS sequence length differences by the established ARISA method are possible. Moreover, ARISA should generate different patterns for different Methylobacterium communities and should therefore allow rapid comparison of Methylobacterium communities in different samples. When ARISA is used to characterize Methylobacterium communities in environmental samples, the possibility that peak diversity does not perfectly reflect the diversity of the community must be considered. On the one hand, the presence of different rrn operons in some strains can lead to overestimation of diversity. On the other hand, a peak of a particular size can represent different strains. The latter observation reveals that in most cases it is difficult to link a particular peak size to a particular bacterial strain or species. Similar to other fingerprinting methods used to characterize microbial communities in environmental samples, the construction and analysis of clone libraries are indispensable for linking bacterial strains to peaks in ARISA patterns.
TABLE 2.
ARISA patterns of Methylobacterium type strains
| Strain | No. of analyses | Exptl size of detected peaks (bp) (relative peak ht)a
|
||||
|---|---|---|---|---|---|---|
| Peak 1 | Peak 2 | Peak 3 | Peak 4 | Peak 5 | ||
| M. hispanicum GP34T | 4 | 621 ± 0.18 (0.21 ± 0.017) | 623 ± 0.18 (0.56 ± 0.061) | 625 ± 0.18 (0.18 ± 0.018) | 703 ± 0.15 (0.11 ± 0.081)b | |
| M. mesophilicum A47T | 4 | 639 ± 0.11 | ||||
| M. fujisawaense 0-31T | 4 | 622 ± 0.13 (0.59 ± 0.027) | 623 ± 0.17 (0.11 ± 0.041) | 637 ± 0.12 (0.30 ± 0.015) | ||
| M. radiotolerans DSM 1819T | 3 | 605 ± 0.26 (0.49 ± 0.033) | 696 ± 0.14 (0.34 ± 0.020) | 726 ± 0.69 (0.17 ± 0.013) | ||
| M. aquaticum DSM 16371T | 4 | 586 ± 0.08 (0.37 ± 0.021) | 589 ± 0.11 (0.10 ± 0.006) | 595 ± 0.15 (0.16 ± 0.004) | 598 ± 0.11 (0.12 ± 0.004) | 600 ± 0.11 (0.25 ± 0.020) |
| M. variabile GR16TT | 3 | 598 ± 0.26 | ||||
| M. nodulans ORS2060T | 4 | 570 ± 0.03 | ||||
| M. rhodinum DSM 2163T | 4 | 613 ± 0.32 (0.31 ± 0.003) | 615 ± 0.33 (0.69 ± 0.003) | |||
| M. podarium FM4T | 4 | 570 ± 0.05 (0.28 ± 0.007) | 606 ± 0.15 (0.16 ± 0.003) | 613 ± 0.18 (0.56 ± 0.007) | ||
| M. thiocyanatum ALL/SCN-RPT | 3 | 592 ± 0.06 (0.26 ± 0.018) | 607 ± 0.03 (0.56 ± 0.016) | 613 ± 0.05 (0.18 ± 0.003) | ||
| M. zatmanii DSM 5688T | 3 | 615 ± 0.37 (0.79 ± 0.004) | 618 ± 0.37 (0.21 ± 0.004) | |||
| M. populi BJ001T | 4 | 605 ± 0.11 (0.45 ± 0.009) | 611 ± 0.09 (0.55 ± 0.009) | |||
| M. lusitanum RXMT | 3 | 572 ± 0.14 | ||||
| M. rhodesianum DSM 5687T | 3 | 571 ± 0.12 | ||||
| M. suomiense F20T | 4 | 588 ± 0.10 | ||||
| M. aminovorans TH-15T | 4 | 615 ± 0.19 | ||||
| M. chloromethanicum CM4T | 4 | 612 ± 0.21 | ||||
| M. dichloromethanicum DM4T | 4 | 561 ± 0.02 | ||||
| M. extorquens TK 0001T | 4 | 561 ± 0.04 | ||||
| M. extorquens AM1 | 4 | 613 ± 0.07 | ||||
| M. extorquens DSM 13060 | 4 | 612 ± 0.14 | ||||
The values are means ± standard errors of the means for three or four independent analyses (different PCR assays and subsequent length determination).
The peak was detected in only two replicate analyses.
To evaluate the suitability of the IGS as a phylogenetic marker, all available IGS sequences from Methylobacterium strains were used to construct phylogenetic trees based on different algorithms. The resulting consensus tree reflected quite well the topology of the 16S rRNA gene-based tree (Fig. 1). The different species were separated from each other, and the sequences of different strains of a species clustered together. Likewise, the different IGS sequences of strains with multiple different rrn operons clustered together. However, the differences between the IGS sequences of a single strain can be greater than the differences between the sequences of a strain and the sequences of another strain of the same species. This was observed for one of the IGS sequences of M. fujisawaense 0-31T, which was most similar to the sequence of Methylobacterium sp. isolate F48, the most closely related strain analyzed. Strains with identical 16S rRNA gene sequences could be clearly differentiated by their IGS sequences, but two pairs of strains with different 16S rRNA gene sequences had identical IGS sequences. These strains were Methylobacterium lusitanum RXMT and Methylobacterium rhodesianum DSM 5687T, which were recently reclassified as different strains of the same species (29), and M. extorquens AM1 and M. extorquens DSM 13060. This finding suggests that strains may still be different even if their IGS sequences are identical. In conclusion, the 16S-23S rRNA IGS sequence allows better differentiation of similar strains than the 16S rRNA gene sequence, and the IGS can be used for phylogenetic analyses. However, the presence of multiple different IGS copies in some strains does not allow IGS sequencing without cloning of the PCR-amplified fragments. In cultivation-independent studies, it should in most cases be possible to identify the number of different species present in a sample, but the number of different strains cannot be determined if several highly similar sequence types are detected, due to the presence of multiple, different IGS copies in some strains.
Analysis of the Methylobacterium community in the plant phyllosphere.
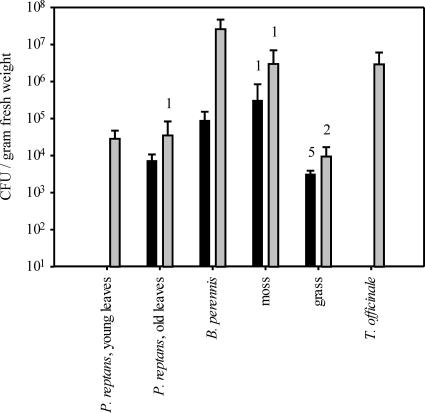
The established ARISA method was used to compare the Methylobacterium communities in the phyllospheres of different plant species. In addition to the cultivation-independent characterization of the Methylobacterium community, the size of the population was estimated by a cultivation-dependent method. Methylobacterium was detected on 80% of all analyzed plants, and the largest population sizes were up to 5.1 × 107 cells per g (fresh weight) on B. perennis (Fig. 2). The numbers of cells varied significantly among different plant species (P < 0.001). The grass and P. reptans plants were colonized by smaller populations than B. perennis and T. officinale (P < 0.05). Use of the Methylobacterium-specific primer system allowed successful amplification of DNA from all samples, and the mixed PCR products could be separated by ARISA (Fig. 3). Peaks in the range from 480 to 733 bp were detected in the analyzed samples. The lowest peak diversity was observed for the grass samples (1 to 8 peaks) and the highest peak diversity was seen for moss samples (11 to 22 peaks), suggesting that different plant species may harbor Methylobacterium communities with different complexities (Fig. 3 and 4).
FIG. 2.
Abundance of Methylobacterium on different plant species collected at two different time points (black bars, 22 March; gray bars, 12 April). The bars indicate the means for five samples from individual plants, and the error bars indicate standard deviations. The numbers of samples with numbers of CFU below the detection limit are indicated above the columns. The detection limit was dependent on the amount of plant material analyzed and ranged from 8.7 × 102 to 1.3 × 104 cells per g (fresh weight). For the samples with numbers of cells below the detection limit a number of cells just below the detection limit was assumed.
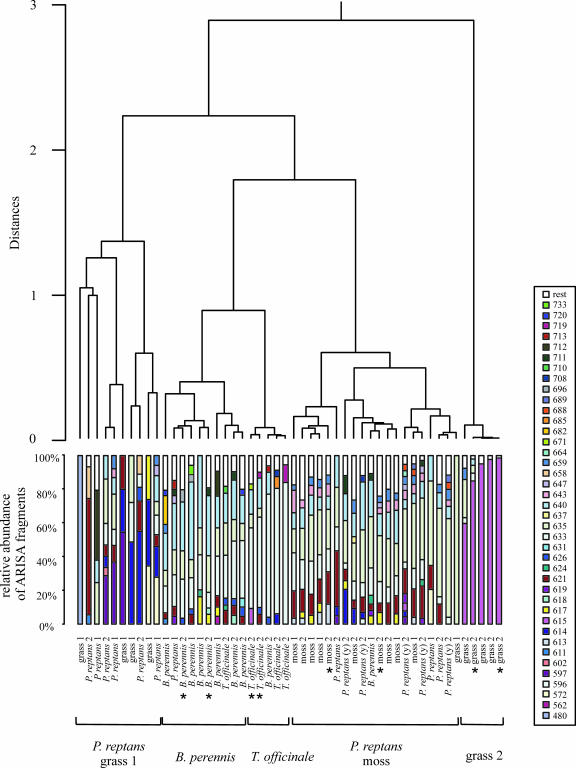
FIG. 4.
Cluster analysis based on ARISA patterns obtained from individual plants of different plant species, taken at two different time points (indicated by 1 and 2). Young leaves of P. reptans, which were taken during the second sampling, are indicated by (y). The ARISA pattern of each sample shows the relative abundance of ARISA fragments. Peaks that contributed less than 3% to a pattern are represented by the fraction called “rest.” Fragment sizes are expressed in experimentally determined numbers of base pairs. The different clusters are grouped together and labeled according to the dominant plant species. Samples whose ARISA chromatograms are shown in Fig. 3 are indicated by an asterisk.
A cluster analysis was performed to group the different samples based on the similarity of the fingerprints (Fig. 4). With the exception of the grass samples, samples of each plant species taken at the two different time points clustered together, suggesting that the community compositions on the plant species remained stable between the two sampling events, although the population size increased significantly (P < 0.005) between the first and second sampling times, especially on B. perennis (Fig. 2). The grass samples taken at the second sampling time were clearly separated from all other samples in the cluster analysis. The 615-bp peak was detected as the major peak (60 to 99%) in all these grass samples, but it contributed <10% to the patterns of a few other plant samples (Fig. 4). No peak was detected for all representatives of a single plant species but for no other plant species, suggesting that plant colonization by particular Methylobacterium strains is not highly exclusive.
In approximately two-thirds of all analyzed samples, particularly samples taken from B. perennis, T. officinale, and moss and some of the P. reptans samples, the major detected peaks were at 621, 633, 635, and/or 640 bp, suggesting that similar Methylobacterium communities were present in these samples. Despite these similarities, almost all samples of B. perennis and T. officinale were separated from the moss and P. reptans samples and also from each other in the cluster analysis tree. This finding suggests that the Methylobacterium communities were more similar in samples from the same plant species than in samples from different plant species. Such a finding is in agreement with the findings of other studies of general bacterial diversity in which the total bacterial community of the plant phyllosphere was analyzed by cultivation-independent methods, and specific microbial communities were observed on different plant species in all these studies (34, 46, 61). In one of the studies the dominant subpopulations (the genera Burkholderia and Serratia) were analyzed in detail. Again, plant species-specific colonization patterns were observed; they were even similar for plants taken from different sampling sites (46). Furthermore, plant species-dependent differences in the total microbial community and in subpopulations have been observed in the rhizosphere (9, 10, 53).
By contrast, very different Methylobacterium communities were detected on some of the P. reptans samples and the grass samples. It is well known from cultivation-dependent studies that the structures of microbial communities can differ substantially among leaves within a plant canopy and that types of bacteria and their relative abundances vary not only from one leaf to another but also within individual leaves (21). This observation is probably valid not only for the whole microbial community but also for subgroups of this community. The differences on individual leaves of the same plant species may have been detected in this study because rather small amounts of leaf material were analyzed.
In conclusion, the ARISA method developed in this study allowed generation of fingerprints of the Methylobacterium communities from phyllosphere samples and comparison of these communities on leaves of different plant species. This first analysis showed that some of the plant species harbored very similar Methylobacterium communities, which differed from those of other plant species grown at the same sampling site, while leaves of some other plant species harbored rather different communities. The complexities of the communities were also different on different plant species. To draw general conclusions concerning Methylobacterium plant colonization patterns, the phyllosphere community needs to be studied more carefully, and the parameters which affect the Methylobacterium community composition on different plants need to be determined. The results of this study show that ARISA is a tool that is well suited for characterization of Methylobacterium communities in environmental samples. In combination with other specific primers, the method may also be used for characterization of other phylogenetic groups at the subdomain level.
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique and ETH Zurich.
We acknowledge Laurent Sauviac, who shared with us the idea and gave advice about how to sequence nonpurified PCR products.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abanda-Nkpwatt, D., M. Müsch, J. Tschiersch, M. Boettner, and W. Schwab. 2006. Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J. Exp. Bot. 57:4025-4032. [DOI] [PubMed] [Google Scholar]
- 2.Anesti, V., J. Vohra, S. Goonetilleka, I. R. McDonald, B. Straubler, E. Stackebrandt, D. P. Kelly, and A. P. Wood. 2004. Molecular detection and isolation of facultatively methylotrophic bacteria, including Methylobacterium podarium sp. nov., from the human foot microflora. Environ. Microbiol. 6:820-830. [DOI] [PubMed] [Google Scholar]
- 3.Araujo, W. L., J. Marcon, W. Maccheroni, Jr., J. D. Van Elsas, J. W. Van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, S. L., V. R. Flechtner, and J. R. Johansen. 2001. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in Cyanobacteria. Mol. Biol. Evol. 18:1057-1069. [DOI] [PubMed] [Google Scholar]
- 6.Brusseau, G. A., E. S. Bulygina, and R. S. Hanson. 1994. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl. Environ. Microbiol. 60:626-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpe, W. A., and S. Rheem. 1989. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol. Ecol. 62:243-249. [Google Scholar]
- 9.Costa, R., M. Gotz, N. Mrotzek, J. Lottmann, G. Berg, and K. Smalla. 2006. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56:236-249. [DOI] [PubMed] [Google Scholar]
- 10.Costa, R., J. F. Salles, G. Berg, and K. Smalla. 2006. Cultivation-independent analysis of Pseudomonas species in soil and in the rhizosphere of field-grown Verticillium dahliae host plants. Environ. Microbiol. 8:2136-2149. [DOI] [PubMed] [Google Scholar]
- 11.Danovaro, R., G. M. Luna, A. Dell'Anno, and B. Pietrangeli. 2006. Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Appl. Environ. Microbiol. 72:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbeltagy, A., K. Nishioka, H. Suzuki, T. Sato, Y. I. Sato, H. Morisaki, H. Mitsui, and K. Minamisawa. 2000. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 46:617-629. [Google Scholar]
- 13.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Martinez, J., S. G. Acinas, A. I. Anton, and F. Rodriguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 15.Gourion, B., M. Rossignol, and J. A. Vorholt. 2006. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. USA 103:13186-13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, P. N. 2006. Methylobacterium, p. 257-265. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 5. Springer, New York, NY. [Google Scholar]
- 17.Gürtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 19.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyer, J., V. F. Galchenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 21.Hirano, S. S., and C. D. Upper. 1991. Bacterial community dynamics, p. 271-294. In J. H. Andrews and S. S. Hirano (ed.), Microbial ecology on leaves. Springer-Verlag, New York, NY.
- 22.Hogue, R., M. Graves, S. Moler, and J. M. Janda. 2007. Pink-pigmented non-fermentative gram-negative rods associated with human infections: a clinical and diagnostic challenge. Infection 35:126-133. [DOI] [PubMed] [Google Scholar]
- 23.Idris, R., M. Kuffner, L. Bodrossy, M. Puschenreiter, S. Monchy, W. W. Wenzel, and A. Sessitsch. 2006. Characterization of Ni-tolerant methylobacteria associated with the hyperaccumulating plant Thlaspi goesingense and description of Methylobacterium goesingense sp. nov. Syst. Appl. Microbiol. 29:634-644. [DOI] [PubMed] [Google Scholar]
- 24.Idris, R., R. Trifonova, M. Puschenreiter, W. W. Wenzel, and A. Sessitsch. 2004. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70:2667-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, E. F., H. L. Echlin, and C. R. Jackson. 2006. Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol. Ecol. 58:236-246. [DOI] [PubMed] [Google Scholar]
- 26.Jaftha, J. B., B. W. Strijdom, and P. L. Steyn. 2002. Characterization of pigmented methylotrophic bacteria which nodulate Lotononis bainesii. Syst. Appl. Microbiol. 25:440-449. [DOI] [PubMed] [Google Scholar]
- 27.Jourand, P., E. Giraud, G. Bena, A. Sy, A. Willems, M. Gillis, B. Dreyfus, and P. de Lajudie. 2004. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 54:2269-2273. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Y. S., J. Kim, H. D. Shin, Y. D. Nam, J. W. Bae, C. O. Jeon, and W. Park. 2007. Methylobacterium platani sp. nov., isolated from a leaf of the tree Platanus orientalis. Int. J. Syst. Evol. Microbiol. 57:2849-2853. [DOI] [PubMed] [Google Scholar]
- 29.Kato, Y., M. Asahara, D. Arai, K. Goto, and A. Yokota. 2005. Reclassification of Methylobacterium chloromethanicum and Methylobacterium dichloromethanicum as later subjective synonyms of Methylobacterium extorquens and of Methylobacterium lusitanum as a later subjective synonym of Methylobacterium rhodesianum. J. Gen. Appl. Microbiol. 51:287-299. [DOI] [PubMed] [Google Scholar]
- 30.Kisand, V., and J. Wikner. 2003. Limited resolution of 16S rDNA DGGE caused by melting properties and closely related DNA sequences. J. Microbiol. Methods 54:183-191. [DOI] [PubMed] [Google Scholar]
- 31.Koenig, R. L., R. O. Morris, and J. C. Polacco. 2002. tRNA is the source of low-level trans-zeatin production in Methylobacterium spp. J. Bacteriol. 184:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutschera, U. 2007. Plant-associated methylobacteria as co-evolved phytosymbionts. Plant Signal. Behav. 2:74-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lacava, P. T., W. B. Li, W. L. Araujo, J. L. Azevedo, and J. S. Hartung. 2006. Rapid, specific and quantitative assays for the detection of the endophytic bacterium Methylobacterium mesophilicum in plants. J. Microbiol. Methods 65:535-541. [DOI] [PubMed] [Google Scholar]
- 34.Lambais, M. R., D. E. Crowley, J. C. Cury, R. C. Bull, and R. R. Rodrigues. 2006. Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917. [DOI] [PubMed] [Google Scholar]
- 35.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, H. S., M. Madhaiyan, C. W. Kim, S. J. Choi, K. Y. Chung, and T. M. Sa. 2006. Physiological enhancement of early growth of rice seedlings (Oryza sativa L.) by production of phytohormone of N2-fixing methylotrophic isolates. Biol. Fertil. Soils 42:402-408. [Google Scholar]
- 37.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckman, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madhaiyan, M., B. Y. Kim, S. Poonguzhali, S. W. Kwon, M. H. Song, J. H. Ryu, S. J. Go, B. S. Koo, and T. M. Sa. 2007. Methylobacterium oryzae sp. nov., an aerobic, pink-pigmented, facultatively methylotrophic, 1-aminocyclopropane-1-carboxylate deaminase-producing bacterium isolated from rice. Int. J. Syst. Evol. Microbiol. 57:326-331. [DOI] [PubMed] [Google Scholar]
- 39.Mano, H., F. Tanaka, C. Nakamura, H. Kaga, and H. Morisaki. 2007. Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ. 22:175-185. [Google Scholar]
- 40.Muyzer, G., T. Brinkoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 2004. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 743-770. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermanns, and D. Van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Nesme, X., and P. Normand. 2004. Easy individual strain and community typing by rDNA ITS1 analysis, p. 671-688. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermanns, and D. Van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 42.Nishio, T., T. Yoshikura, and H. Itoh. 1997. Detection of Methylobacterium species by 16S rRNA gene-targeted PCR. Appl. Environ. Microbiol. 63:1594-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunn, D. N., and M. E. Lidstrom. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J. Bacteriol. 166:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omer, Z. S., R. Tombolini, A. Broberg, and B. Gerhardson. 2004. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Regul. 43:93-96. [Google Scholar]
- 45.Omer, Z. S., R. Tombolini, and B. Gerhardson. 2004. Plant colonization by pink-pigmented facultative methylotrophic bacteria (PPFMs). FEMS Microbiol. Ecol. 47:319-326. [DOI] [PubMed] [Google Scholar]
- 46.Opelt, K., C. Berg, S. Schönmann, L. Eberl, and G. Berg. 2007. High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J. 1:502-516. [DOI] [PubMed] [Google Scholar]
- 47.Pirttilä, A. M., H. Laukkanen, H. Pospiech, R. Myllylä, and A. Hohtola. 2000. Detection of intracellular bacteria in the buds of Scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl. Environ. Microbiol. 66:3073-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasche, F., R. Trondl, C. Naglreiter, T. G. Reichenauer, and A. Sessitsch. 2006. Chilling and cultivar type affect the diversity of bacterial endophytes colonizing sweet pepper (Capsicum anuum L.). Can. J. Microbiol. 52:1036-1045. [DOI] [PubMed] [Google Scholar]
- 49.Ryu, J., M. Madhaiyan, S. Poonguzhali, W. Yim, P. Indiragandhi, K. Kim, R. Anandham, J. Yun, K. H. Kim, and T. Sa. 2006. Plant growth substances produced by Methylobacterium spp. and their effect on tomato (Lycopersicon esculentum L.) and red pepper (Capsicum annuum L.) growth. J. Microbiol. Biotechnol. 16:1622-1628. [Google Scholar]
- 50.Sait, L., M. Galic, R. A. Strugnell, and P. H. Janssen. 2003. Secretory antibodies do not affect the composition of the bacterial microbiota in the terminal ileum of 10-week-old mice. Appl. Environ. Microbiol. 69:2100-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 52.Schöniger, M., and A. von Haeseler. 1999. Toward assigning helical regions in alignments of ribosomal RNA and testing the appropriateness of evolutionary models. J. Mol. Evol. 49:691-698. [DOI] [PubMed] [Google Scholar]
- 53.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sy, A., E. Giraud, P. Jourand, N. Garcia, A. Willems, P. de Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sy, A., A. C. Timmers, C. Knief, and J. A. Vorholt. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 71:7245-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trotsenko, Y. A., E. G. Ivanova, and N. V. Doronina. 2001. Aerobic methylotrophic bacteria as phytosymbionts. Microbiology 70:623-632. [Google Scholar]
- 57.Tsien, H. C., B. J. Bratina, K. Tsuji, and R. S. Hanson. 1990. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl. Environ. Microbiol. 56:2858-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Aken, B., C. M. Peres, S. L. Doty, J. M. Yoon, and J. L. Schnoor. 2004. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides × nigra DN34). Int. J. Syst. Evol. Microbiol. 54:1191-1196. [DOI] [PubMed] [Google Scholar]
- 59.Van Aken, B., J. M. Yoon, and J. L. Schnoor. 2004. Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoides × nigra DN34). Appl. Environ. Microbiol. 70:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weon, H. Y., B. Y. Kim, J. H. Joa, J. A. Son, M. H. Song, S. W. Kwon, S. J. Go, and S. H. Yoon. 2008. Methylobacterium iners sp. nov. and Methylobacterium aerolatum sp. nov., isolated from air samples in Korea. Int. J. Syst. Evol. Microbiol. 58:93-96. [DOI] [PubMed] [Google Scholar]
- 61.Yang, C. H., D. E. Crowley, J. Borneman, and N. T. Keen. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 98:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.