Abstract
Carotenoids are structurally diverse pigments of biotechnological interest as natural colorants and in the prevention of human disease. The carotenoids present in 19 strains taxonomically related to the poorly described, nonphotosynthetic bacterial genus Hymenobacter, including 10 novel isolates cultivated from Victoria Upper Glacier, Antarctica, were characterized using high-performance liquid chromatography (HPLC). Nine chemically distinct carotenoids, present in various combinations irresolvable by conventional crude spectrophotometric analyses, were purified by preparative HPLC and characterized using UV-visible light absorption spectroscopy and high-resolution mass spectrometry. All major Hymenobacter carotenoids appear to be derived from a common backbone of 2′-hydroxyflexixanthin and include previously unreported presumptive hexosyl, pentosyl, and methyl derivatives. Their distribution does not, however, correlate perfectly with 16S rRNA gene phylogeny. Carotenoid composition, therefore, may be strain specific and does not follow a strictly homogeneous pattern of vertical evolutionary descent.
Carotenoids are isoprenoid pigments responsible for most natural red, orange, and yellow coloration (21, 30). Over 600 known structurally unique carotenoids are distributed throughout all major lineages of the tree of life (6, 30). Although best known as auxiliary components of the photosynthetic light-harvesting apparatus, many carotenoids are also produced by nonphotosynthetic bacteria and fungi, where they function as antioxidants (24, 40, 44) and in the stabilization of cellular membranes (9, 19). Higher animals lack the ability to synthesize carotenoids de novo and instead assimilate them from their diet (27).
Recently, carotenoids have gained biotechnological interest as natural nutritional supplements (17, 29) and natural pigments (25), with a projected market in 2010 exceeding 1 billion U.S. dollars (13). Carotenoids are particularly well-known scavengers of reactive oxygen species, such as singlet oxygen (21, 35), and in humans contribute both to the maintenance of proper cell function and to disease prevention (21, 27, 34, 35). Carotenoid biosynthesis has been well studied in many microorganisms (10, 32), particularly regarding the creation of recombinant biosynthetic pathways leading to the production of novel pigments (29, 41) with biotechnologically interesting properties, such as improved antioxidant activity (1, 26). These approaches require the identification of novel biosynthetic enzymes having expanded substrate ranges and activities, the discovery of which requires a clear understanding of carotenoid diversity and distribution. Most high-resolution structural studies are limited to the carotenoids present in single representative strains. Whether this approach captures all structural diversity is unknown because systematic, high-resolution studies of carotenoid composition in taxonomically related strains are lacking. The current understanding of the evolution of carotenoid metabolism (and that of other secondary metabolites) suggests a “tree-like” model (41) containing a highly conserved core biosynthetic pathway with terminal “branches” that are freer to evolve. The degree of evolutionary plasticity exhibited by these terminal biosynthetic branches remains unclear due to insufficient study.
This work describes the carotenoids produced by bacteria of the genus Hymenobacter, a poorly studied lineage within the Bacteroidetes, including several novel strains that we have isolated from Victoria Upper Glacier, Antarctica (VUG). An undefined “Taxeobacter” (now Hymenobacter) (8) strain has been previously reported to contain 2′-hydroxyflexixanthin and 3-deoxy-2′-hydroxyflexixanthin as major carotenoids (5), although this identification is considered to be poorly supported (6). In order to obtain sufficient structural resolution, high-performance liquid chromatography (HPLC) was used to separate and compare the carotenoids present in this collection of Hymenobacter and related strains. Because of the previously reported carotenoid structures and taxonomic relatedness of these strains, we hypothesized that systematic examination of their carotenoids would indicate the extent of carotenoid evolution within these related genera and species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Hymenobacter reference strains were purchased from the DSMZ and CCUG culture collections, except for the Hymenobacter norwichensis and Hymenobacter rigui strains and Hymenobacter sp. strain NS/2, which were kindly provided by Hans-Jürgen Busse (University of Vienna). VUG Hymenobacter-like strains were isolated by direct plating of aseptically melted glacier ice onto R2A agar (Difco) and incubation at 4°C, 10°C, or room temperature (approximately 20°C) in the dark, with subsequent restreaking to purity. Because most strains did not grow consistently in liquid culture or at room temperature, cultures were maintained on chilled R2A plates incubated at 10°C for 4 weeks. Except where indicated, growth experiments were conducted at 18°C in the dark for 7 days, using 1-week-old cultures incubated at 10°C as inocula to ensure active growth.
16S rRNA gene sequencing and phylogenetic analysis.
Genomic DNA was extracted from R2A plate-grown cells by using a bead-beating and chemical lysis method described previously (16), and nearly full-length 16S rRNA gene sequences (Escherichia coli positions 8 to 1509) (7) were amplified by PCR using primers PB36F and PB38R (16). PCR products were sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems Instruments [ABI]) and an ABI 3700 DNA sequencer (ABI) with PB36F, PB38R, and internal primers (11). Sequences were assembled using the PREGAP v1.5 and GAP4 v4.10 programs of the Staden package (12), checked for chimeras using PINTAIL (2), and submitted to GenBank. To construct bootstrapped trees, 16S rRNA genes of VUG and reference Hymenobacter strains were aligned using CLUSTAL_X (39) and trimmed using the SEQRET program of the EMBOSS bioinformatics suite (28) to eliminate primer sequences and ensure the comparison of sequences of equal lengths (nucleotides 38 to 1486 [E. coli numbering]) (7). With PHYLIP version 3.6a3 (14), 100 phylogenetic trees were created using the Kimura two-parameter method (20), and a consensus neighbor-joining tree was constructed using superimposed branch lengths generated using the method of Fitch and Margoliash (15). The 16S rRNA gene sequence of E. coli (J01859) was used as an outgroup.
Carotenoid extraction and analysis.
To extract carotenoids, plate-grown Hymenobacter cultures were added to 2 ml of HPLC grade methanol (Fisher Scientific) and extracted at 65°C for 5 min. Cell debris was pelleted at low speed in a clinical centrifuge, the supernatant removed, and the pellet extraction repeated using fresh methanol. Pooled supernatants were dried under a stream of nitrogen and redissolved in a small volume of methanol by gentle heating.
UV-visible light (UV-Vis) absorption spectra of crude methanolic extracts were determined using an Ultraspec 3000 spectrophotometer (Pharmacia Biotech). Wavelengths from 200 nm to 800 nm were recorded at 0.5-nm intervals.
HPLC analyses of concentrated methanolic extracts were conducted using an Agilent 1100 series HPLC (Agilent) equipped with a 125- by 4-mm LiChrospher 100 RP-18 column (5-μm particle size; Agilent). Carotenoids were eluted at a flow rate of 1.5 ml/min by a 10-min linear gradient of 100% 80:20 HPLC grade methanol-MilliQ filtered water to 100% 80:20 HPLC grade methanol-HPLC grade ethyl acetate (BDH), followed by 8 min of elution with isocratic 80:20 methanol-ethyl acetate. All solvents were degassed by vacuum filtration through a 0.45-μm-pore-diameter HVLP filter (Millipore) prior to use. Carotenoids were detected at 470 nm by using an online photodiode array (PDA) detector with a 600-nm reference wavelength.
Peak areas generated by an HPLC auto-integrator were used for carotenoid quantification as all detected carotenoids shared similar absorption maxima (see Table 2) and their absorption coefficients were unknown. For data analysis, peak areas were exported to an EXCEL spreadsheet and peaks with retention times less than 1.2 min or areas less than 100 milli-absorbance units/s were excluded to eliminate injection artifacts or background noise, respectively. Peaks with widths greater than 0.22 min were also excluded, except in cases where a distinct peak was obviously apparent from visual inspection of the HPLC chromatogram. In time course assays, certain peaks below the 100-milli-absorbant-unit/s threshold were included for uniformity based on their presence in more-concentrated parallel samples. Peak areas of cis carotenoids, as determined by their distinct UV spectra (6), were added to those of all trans isomers to further simplify analysis. The identities of these cis peaks were confirmed by the homogeneous matrix-assisted laser desorption/ionization (MALDI) mass spectra of mixtures containing both cis and trans isomers (J. L. Klassen, unpublished results).
TABLE 2.
UV-Vis absorption maxima, high-resolution MALDI m/z values, deduced chemical formulae, and presumptive identifications of carotenoids purified from strains VUG-A141a (carotenoids 1 to 5) and VUG-A42aa (carotenoids 6 to 9)a
| Carotenoid | UV-Vis maximum(a) (nm) | m/z | Deduced chemical formula | Presumptive identity |
|---|---|---|---|---|
| 1 | 478 | ND | ND | Unknown |
| 2 | 478 | ND | ND | Unknown |
| 3 | 484, 502, sh | 760.45449 | C46H64O9 | Hexosyl-2′-hydroxyflexixanthin |
| 4 | 480, 502, sh | 730.44392 | C45H62O8 | Pentosyl-2′-hydroxyflexixanthin |
| 5 | 486, 502, sh | 621.39143 (598.4) | C40H54O4Na C40H54O4 | 2′-Hydroxyflexixanthin |
| 6 | 484, 502, sh | 635.40708 (612.4) | C41H56O4Na C41H56O4 | Methyl-2′-hydroxyflexixanthin |
| 7 | 480, 502, sh | 596.42240 | C41H56O3 | Methyl-3-deoxy-2′-hydroxyflexixanthin, methyl-flexixanthin or methyl-1′,2′-dihydro-2,2′-dihydroxy-3′,4′-didehydro-4-keto-γ-carotene |
| 8 | 446, 472, 502 | 568.42748 | C40H56O2 | Plectaniaxanthin, saproxanthin or 1′,2′-dihydro-2,2′-dihydroxy-3′,4′-didehydro-γ-carotene |
| 9 | 446, 470, 500 | 582.44313 | C41H58O2 | Methyl-plectaniaxanthin, methyl-saproxanthin or methyl-1′,2′-dihydro-2,2′-dihydroxy-3′,4′-didehydro-γ-carotene |
The high-resolution electrospray ionization m/z values of carotenoids 5 and 6 are given as sodiated derivatives, with their nonsodiated, low-resolution m/z values in parentheses. ND, not determined, due to low abundance (Table 1); sh, shoulder peak.
Carotenoid purification and mass spectrometry.
To isolate large volumes of carotenoids, cell biomasses from 25 to 30 replicate R2A plates of isolates VUG-A42aa and VUG-A141a were separately extracted twice in 100 ml of HPLC grade methanol at 65°C for 5 min and gravity filtered through Whatman no. 1 filter paper (Whatman), which was finally washed twice with 25 ml of methanol. The pooled filtrate was concentrated to near dryness by rotary evaporation, the carotenoid extract removed, and the flask washed several times with small volumes of methanol which were added to the concentrate and dried under a nitrogen stream until visual precipitation occurred. After warming to room temperature, carotenoid precipitates were dissolved by adding a minimal volume of methanol.
Individual carotenoids were purified by repeated preparative HPLC using the conditions described above or slight modifications thereof. Pooled fractions were concentrated by rotary evaporation and/or drying under nitrogen gas, redissolved in methanol, and repurified as described above until no further contaminants were visible with the PDA detector. High-resolution mass spectra were determined by electrospray ionization as sodiated derivatives with a Mariner biospectrometry workstation (ABI) in positive-ionization mode (carotenoids 5 and 6) or by high-resolution MALDI in a trans-2-[3-{4-tert-butylphenyl}-2-methyl-2-propenylidene]malononitrile matrix with a Bruker Daltonics 9.4T Apex-Qe Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics) in positive-ionization mode (carotenoids 3, 4, 7, 8, and 9).
Nucleotide sequence accession numbers.
The sequences obtained in this study were deposited in GenBank under accession numbers EU155008 to EU155017 and EU159489.
RESULTS
Phylogenetic analysis of the genus Hymenobacter and related VUG strains.
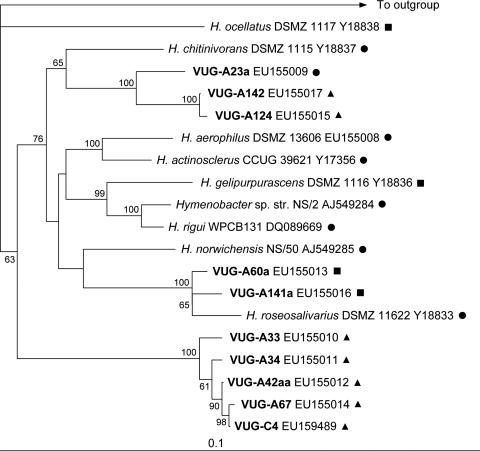
An updated phylogenetic tree of the genus Hymenobacter (Fig. 1) was constructed using the nearly full-length 16S rRNA gene sequences of all currently described Hymenobacter species plus 10 novel VUG strains. Isolates VUG-A60a and VUG-A141a were closely related to Hymenobacter roseosalivarius, and isolates VUG-A23a, VUG-A142, and VUG-A124 were loosely affiliated with Hymenobacter chitinivorans. Isolates VUG-A33, VUG-A34, VUG-A42aa, VUG-A67, and VUG-C4 formed a cluster (hereafter referred to as the “novel VUG clade”) distinct from all other strains and may comprise a novel genus based on <95% 16S rRNA gene sequence similarity (33) to all other described Hymenobacter species.
FIG. 1.
Nearly full-length 16S rRNA gene phylogeny of cultured Hymenobacter and Hymenobacter-like VUG strains (in boldface) constructed by neighbor joining. Only bootstrap values of >50 are shown. The scale bar shown represents a 10% difference in nucleotide composition. The 16S rRNA gene sequence of E. coli was used as an outgroup. GenBank accession numbers are indicated to the right of the sequence names. The major carotenoid content of strains is indicated as follows: circles, carotenoid 5 only; triangles, carotenoids 5 and 6; squares, carotenoids 3, 4, and 5.
Carotenoid content of Hymenobacter and related strains.
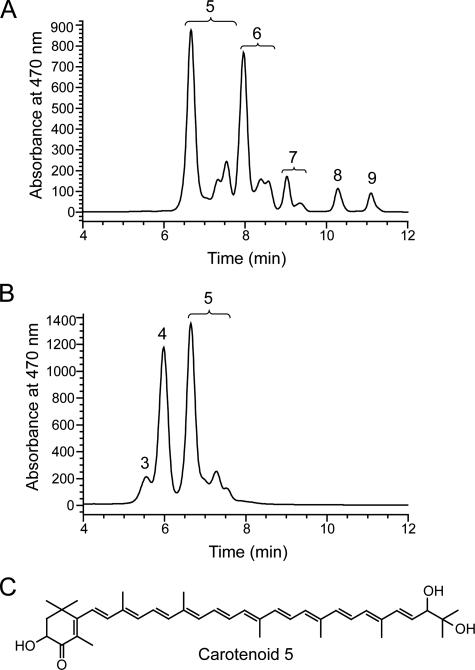
One feature common to all Hymenobacter and VUG strains is their bright red-pink pigmentation. Analyses of crude methanolic extracts of Hymenobacter and VUG strains by UV-Vis spectroscopy revealed the common presence of a single broad peak with an absorbance maximum at 500 nm and a shoulder at 517 nm (results not shown). In contrast, HPLC analyses revealed a total of nine unique carotenoids, the distribution of which differed between strains (Table 1 and Fig. 2). Stereoisomers of carotenoids 5 to 7 (bracketed in Fig. 2) were detected and resolved by this HPLC method and identified by their characteristic UV absorption spectra (6, 30). No stereoisomers of any other carotenoids were observed.
TABLE 1.
Percents abundance of carotenoids present in VUG and reference Hymenobacter strains grown on R2A plates for 1 week at 18°C in the darka
| Strain | % of total peak area for indicated cartenoid no.
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| H. ocellatus DSMZ 1117 | − | − | 11 ± 0.7 | 9.3 ± 0.4 | 80 ± 0.8 | Tr | Tr | − | − |
| H. chitinivorans DSMZ 1116 | − | − | − | − | 100 ± 0 | − | − | − | − |
| VUG-A23a | − | − | − | Tr | 95 ± 0.7 | 3.7 ± 0.4 | − | Tr | − |
| VUG-A142 | − | − | − | − | 16 ± 0.6 | 84 ± 0.6 | − | − | Tr |
| VUG-A124 | − | − | − | − | 15 ± 2 | 84 ± 2 | − | − | Tr |
| H. aerophilus DSMZ 13606 | − | − | − | − | 93 ± 0.7 | 2.9 ± 0.7 | − | 1.5 ± 0.2 | 2.2 ± 0.1 |
| H. actinosclerus CCUG 39621 | − | − | − | − | 99 ± 0.01 | 1.1 ± 0.01 | − | − | − |
| H. gelipurpurascens DSMZ 1116 | − | − | 8.8 ± 0.1 | 8.2 ± 0.1 | 83 ± 0.2 | − | − | − | − |
| Hymenobacter strain NS/2 | − | − | − | Tr | 100 ± 0.3 | − | − | − | − |
| H. rigui WPCB131 | − | − | − | − | 100 ± 0.2 | − | − | − | − |
| H. norwichensis NS/50 | − | − | − | Tr | 98 ± 0.1 | 1.1 ± 0.2 | − | − | − |
| VUG-A60a | 2.7 ± 0.6 | 4.0 ± 0.1 | 14 ± 0.5 | 23 ± 1 | 49 ± 2 | Tr | − | − | − |
| VUG-A141a | Tr | 2.0 ± 0.1 | 9.6 ± 0.2 | 30 ± 1 | 56 ± 2 | 1.1 ± 0.03 | − | − | − |
| H. roseosalivarius DSMZ 11622 | − | − | − | Tr | 99 ± 0.2 | Tr | − | − | − |
| VUG-A33 | − | − | − | − | 62 ± 2 | 37 ± 2 | Tr | Tr | Tr |
| VUG-A34 | − | − | − | − | 79 ± 2 | 15 ± 2 | Tr | 3.6 ± 0.3 | 2.1 ± 0.2 |
| VUG-A42aa | − | − | − | − | 49 ± 2 | 40 ± 0.9 | 5.1 ± 0.6 | 3.3 ± 0.5 | 2.8 ± 0.3 |
| VUG-A67 | − | − | − | − | 27 ± 0.3 | 68 ± 1 | 2.9 ± 0.6 | Tr | 1.7 ± 0.1 |
| VUG-C4 | − | − | − | − | 86 ± 0.3 | 6.9 ± 0.2 | − | 6.8 ± 0.3 | − |
Values represent the means of three independent replicates and their standard deviations. Strains are presented in the same order as in Fig. 1, and carotenoid abundances in boldface correspond to the groupings in the figure. −, below detection level; Tr, trace amounts (<1% of the total carotenoids) detected.
FIG. 2.
Representative HPLC chromatograms for VUG-A42aa (A) and VUG-A141a (B). Carotenoids were detected using the HPLC PDA detector at 470 nm and numbered in accordance with Tables 1 and 2. Carotenoids 1 and 2 were present at concentrations below the detection level in this sample and are therefore not labeled. Brackets indicate stereoisomer groups as determined by the presence of cis peaks in their UV absorption spectra. An abbreviated timescale showing only minutes 4 to 12 is shown as no carotenoids were detected outside this range. Panel C shows the proposed structure of the trans isomer of carotenoid 5, 2′-hydroxyflexixanthin.
Carotenoid 5 was present in all Hymenobacter and VUG strains and in most cases was the major pigment observed (Table 1). In isolates VUG-A142 and VUG-A124, carotenoid 6 formed the major pigment, with carotenoid 5 present in smaller amounts (Table 1). Carotenoid 6 was also present in members of the novel VUG clade (Table 1) despite their lack of close phylogenetic relationship with strains VUG-A124 and VUG-A142 (Fig. 1). Carotenoids 7 to 9 were present in Hymenobacter aerophilus and strains of the novel VUG clade (Table 1). Carotenoids 1 to 4 were detected in only two strains, VUG-A60a and VUG-A141a, whereas the phylogenetically close species H. roseosalivarius (Fig. 1) contained solely carotenoid 5 (Table 1). Carotenoids 3 and 4, but not carotenoids 1 and 2, were also present in Hymenobacter ocellatus and Hymenobacter gelipurpurascens (Table 1).
Chemical characterization of carotenoids.
In order to chemically characterize all detected carotenoids from VUG-A141a and VUG-A42aa, which together possessed all carotenoids from 1 to 9, UV-Vis absorption spectra of each carotenoid were obtained using the HPLC online PDA detector (Table 2). Additionally, carotenoids 3 to 9 were isolated by preparative HPLC and characterized by high-resolution mass spectrometry (Table 2). The UV-Vis absorption spectra of carotenoids 1 and 2 featured one broad peak at 478 nm, consistent with a ketolated photochrome containing 11 or 12 double bonds (6, 30). Insufficient amounts of these carotenoids were recovered for their mass spectra to be determined. Carotenoids 3 to 7 exhibited identical UV-Vis absorption spectra, with a single peak centered at 480 to 486 nm and a shoulder at 502 nm, suggesting a common ketolated photochrome backbone containing 12 double bonds (6, 30). The chemical formula inferred from the high-resolution mass spectrum of the sodiated ion of carotenoid 5, C40H54O4, is consistent with that of 2′-hydroxyflexixanthin (Fig. 2C), the previously determined major carotenoid of a “Taxeobacter” (now Hymenobacter) (8) strain (5). The inferred chemical formulae of carotenoids 3 (C46H64O9), 4 (C45H62O8), and 6 (C41H56O4, derived from a sodiated ion) are consistent with those of the hexosyl, pentosyl, and methyl derivatives of carotenoid 5, respectively. None of these carotenoids have been reported previously. The inferred chemical formula of carotenoid 7, C41H56O3, is consistent with that of carotenoid 6 minus a single hydroxyl group. Although three structural isomers are possible (methyl-3-deoxy-2′-hydroxyflexixanthin, methyl-flexixanthin, and methyl-1′,2′-dihydro-2,2′-dihydroxy-3′,4′-didehydro-4-keto-γ-carotene) (see Fig. S1 in the supplemental material), methyl-3-deoxy-2′-hydroxyflexixanthin may be the most plausible, given the prior report (5) of 3-deoxy-2′-hydroxyflexixanthin as a minor carotenoid present in the previously studied “Taxeobacter” strain; the latter compound was not detected in the current study. Carotenoids 8 and 9 exhibited UV-Vis absorption spectra with maxima at 446, 470 to 472, and 500 to 502 nm, suggesting a common nonketolated photochrome containing 12 double bonds (6, 30). The mass spectra of carotenoids 8 (C40H56O2) and 9 (C41H58O2) are consistent with that of either plectaniaxanthin, saproxanthin, or 1′,2′-dihydro-2,2′-dihydroxy-3′,4′-didehydro-γ-carotene (see Fig. S1 in the supplemental material) and its methyl derivative, respectively. Plectaniaxanthin and methyl-plectaniaxanthin are the most likely, based on the structures described previously (5), assuming a shared biosynthetic pathway.
The effect of culture age on carotenoid composition in Hymenobacter and VUG strains.
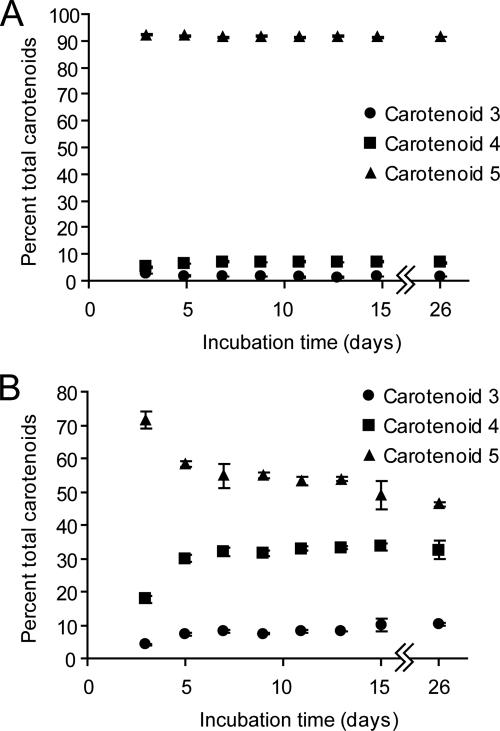
Because of our inability to culture the VUG strains reliably in liquid media, we were unable to standardize carotenoid production based on optical density or growth phase. Previous studies of other genera have determined that carotenoid content is maximal and biosynthesis most complete during stationary phase (4, 43). All Hymenobacter and VUG strains were therefore incubated on plates under the standard conditions of 1 week at 18°C in the dark since these incubation conditions uniformly resulted in visually robust growth, presumed to correspond to maximal cell density. To determine the effects of these growth conditions on the absolute quantifications of carotenoid content in the Hymenobacter and VUG strains, we examined the carotenoid compositions of parallel cultures grown on R2A plates and harvested at different time points. Some strains, such as the H. gelipurpurascens strain (Fig. 3A) and VUG-A124 (data not shown), maintained constant carotenoid compositions throughout incubation for 26 days. Others, such as VUG-A141a (Fig. 3B), the H. aerophilus strain, and VUG-A33 (data not shown), required 7 to 9 days for carotenoid composition to stabilize. In these cases, presumptively glycosylated or methylated carotenoids accumulated, with concomitant reduction of their precursor carotenoid 5 (Fig. 3B).
FIG. 3.
Relative carotenoid compositions of H. gelipurpurascens (A) and VUG-A141a (B) during incubation for 26 days on parallel R2A plates incubated in the dark at 18°C. Percentages were determined from online PDA detection of absorption at 470 nm relative to a 600-nm reference as quantified using the HPLC auto-integrator. Values are the means of three independent replicates, and error bars (where visible) show 1 standard deviation.
The standard culture conditions used in these experiments were chosen without prior quantification of the effect of culture age on carotenoid composition. Although in hindsight we recognize that this could have compromised the quantification of the carotenoid content in the strains (Table 1), the results demonstrate that whereas absolute quantification of carotenoid composition may not be precise without standardization of growth phase (Fig. 3B), this method is capable of identifying the major carotenoids present and discriminating between the carotenoid compositions of related organisms (Table 1). Furthermore, absolute quantification of carotenoid mixtures by use of coupled HPLC and UV-Vis absorption spectroscopy is technically difficult due to the variability of their absorbance maxima and extinction coefficients. We suggest that the method applied here is sufficient to accomplish the purpose of this study, namely, to systematically identify the carotenoids present in a related group of organisms for comparison with their 16S rRNA gene phylogeny.
DISCUSSION
Carotenoid diversity is important because of both its biotechnological potential (29) and its role in understanding the evolution of secondary metabolism (41). Ascertaining the extent of carotenoid diversity from the literature is problematic due to the focus of most high-resolution studies on a single strain and/or the utilization of methods having insufficient resolution to differentiate between related compounds. For example, the organisms in the Bacteroidetes division for which high-resolution carotenoid structures have been determined (5, 22, 23, 31) are all taxonomically distantly related. From these data, it cannot be deduced whether the carotenoids described are representative of the taxa studied. In contrast, when taxonomically close organisms have been compared (e.g., reference 3 and references within) the methods used could only characterize pigments as carotenoids or, at best, flexirubin-like pigments. These methods are clearly insufficient to accurately analyze carotenoid distribution and diversity in these organisms. In contrast, the current study used an HPLC-based method to detect nine different carotenoids, including some previously unreported in the literature, and to estimate their abundance in a taxonomically related group of 19 strains. Comparison of the carotenoid distribution in these strains with their 16S rRNA gene phylogeny (Fig. 1) indicates that carotenoid composition differs between genera (i.e., the novel VUG clade versus all other described Hymenobacter spp.) and between some species (e.g., H. gelipurpurascens versus H. rigui). Furthermore, the erratic phylogenetic distribution of certain carotenoids (e.g., carotenoids 3 and 4) strongly suggests differences in the evolution of their cognate biosynthetic pathways relative to that of their phylogenetic neighbors.
In nearly all organisms, carotenoid biosynthesis proceeds through a conserved central pathway leading from geranylgeranyl pyrophosphate to lycopene (32). At this point, the pathway branches by further cyclization, ketolation, hydroxylation, esterification, desaturation, or epoxidation reactions (10, 32); the differential presence and/or regulation of the responsible enzymes results in the formation of distinct carotenoids in different taxa. As terminal “branches” in a tree-like model of carotenoid evolution (41), these later biosynthetic steps are the most evolutionarily plastic due to their modulation (as opposed to gain or loss) of carotenoid structure and, therefore, function.
Based on the carotenoids identified in this study, we infer a biosynthetic pathway in Hymenobacter leading from geranylgeranyl pyrophosphate to lycopene, typical of most other bacteria (32). Lycopene is subsequently cyclized, ketolated, and hydroxylated at one end of the molecule and hydroxylated and desaturated at the other, in accordance with other related studies (36, 37, 38). It is unclear from the current results whether carotenoids 7 to 9 represent biosynthetic intermediates of this pathway or the products of a separate, parallel one (e.g., reference 18). The inferred terminal biosynthetic step in the Hymenobacter carotenoid biosynthetic pathway is glycosylation or methylation leading to the production of carotenoids 3 and 4 or 6, 7, and 9, respectively. The erratic distribution of these carotenoids (Table 2) clearly indicates differences in the distribution or regulation of the cognate enzymes as a consequence of differential gene gain, gene loss, or evolution of regulatory mechanisms in these organisms. This inference is in accordance with the increased evolutionary plasticity of these terminal biosynthetic “branches.”
From a taxonomic perspective, these results support the application of carotenoid composition to discriminate between organisms of different genera, as typically applied in polyphasic taxonomic studies (42). In some cases, carotenoid composition can differ between species (Table 2); this property must be properly supported by other taxonomic and/or genetic tests to achieve specific identification. The results of this study do, however, support the increased application of HPLC-based methods (or others with similar structural resolution) in the systematic study of carotenoid distribution, such as that compiled by Bernardet et al. for the Flavobacteriaceae (3). This will allow the identification of novel carotenoids, the cognate biosynthetic enzymes of which may be applicable in the design of recombinant biosynthesis pathways (29, 41). Of the 213 papers published in 2006 in the International Journal of Systematic and Evolutionary Microbiology reporting pigmented strains, only 2 used HPLC to precisely determine the pigments present. This suggests that increased adoption of these methods by the systematics community will greatly enhance our currently limited knowledge of carotenoid diversity and distribution.
Supplementary Material
Acknowledgments
This work was supported by an NSERC Discovery grant (J.M.F.). Funding to J.L.K. was provided by an NSERC Postgraduate Scholarship and a Province of Alberta Graduate Scholarship.
Thanks to Joel Barker for providing VUG ice samples. DNA sequencing was conducted at the University of Alberta Department of Biological Sciences Molecular Biology Service Unit, and mass spectrometry was conducted at the University of Alberta Department of Chemistry Mass Spectrometry Laboratory. We thank Hans-Jürgen Busse (University of Vienna) for the provision of Hymenobacter reference strains and Phillip Fedorak (University of Alberta) for access to equipment and helpful discussion.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albrecht, M., S. Takaichi, S. Steiger, Z.-Y. Wang, and G. Sandmann. 2000. Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat. Biotechnol. 18:843-846. [DOI] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardet, J. F., Y. Nakagawa, and B. Holmes. 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52:1049-1070. [DOI] [PubMed] [Google Scholar]
- 4.Bhosale, P., and P. S. Bernstein. 2004. β-Carotene production by Flavobacterium multivorum in the presence of inorganic salts and urea. J. Ind. Microbiol. Biotechnol. 31:565-571. [DOI] [PubMed] [Google Scholar]
- 5.Bircher, C., and H. Pfander. 1997. Synthesis of carotenoids in the gliding bacteria Taxeobacter: (all-E,2′R)-3-deoxy-2′-hydroxyflexixanthin. Helv. Chim. Acta 80:832-837. [Google Scholar]
- 6.Britton, G., S. Liaaen-Jensen, and H. Pfander. 2004. Carotenoids handbook. Birkhauser Verlag, Basel, Switzerland.
- 7.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 8.Buczolits, S., E. B. M. Denner, P. Kämpfer, and H.-J. Busse. 2006. Proposal of Hymenobacter norwichensis sp. nov., classification of ‘Taxeobacter ocellatus’, ‘Taxeobacter gelipurpurascens’ and ‘Taxeobacter chitinivorans’ as Hymenobacter ocellatus sp. nov., Hymenobacter gelipurpurascens sp. nov. and Hymenobacter chitinivorans sp. nov., respectively, and emended description of the genus Hymenobacter Hirsch et al. 1999. Int. J. Syst. Evol. Microbiol. 56:2071-2078. [DOI] [PubMed] [Google Scholar]
- 9.Chattopadhyay, M. K., M. V. Jagannadham, M. Vairamani, and S. Shivaji. 1997. Carotenoid pigments of an Antarctic psychrotrophic bacterium, Micrococcus roseus: temperature dependent biosynthesis, structure, and interaction with synthetic membranes. Biochem. Biophys. Res. Commun. 239:85-90. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Q. 2006. Structural diversity and functional novelty of new carotenoid biosynthesis genes. J. Ind. Microbiol. Biotechnol. 33:552-559. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, S. M., and J. M. Foght. 2007. Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol. Ecol. 59:318-330. [DOI] [PubMed] [Google Scholar]
- 12.Dear, S., and R. Staden. 1991. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 19:3907-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Campo, J. A., M. García-González, and M. G. Guerrero. 2007. Outdoor cultivation of microalgae for carotenoid production: current state and perspectives. Appl. Microbiol. Biotechnol. 74:1163-1174. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 15.Fitch, W. M., and E. Margoliash. 1967. The construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 16.Foght, J., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, P. D., and P. M. Bramley. 2004. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43:228-265. [DOI] [PubMed] [Google Scholar]
- 18.Giraud, E., L. Hannibal, J. Fardoux, M. Jaubert, P. Jourand, B. Dreyfus, J. N. Sturgis, and A. Verméglio. 2004. Two distinct crt gene clusters for two different functional classes of carotenoid in Bradyrhizobium. J. Biol. Chem. 279:15076-15083. [DOI] [PubMed] [Google Scholar]
- 19.Gruszecki, W. I., and K. Strzałka. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740:108-115. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Krinsky, N. I., and E. J. Johnson. 2005. Carotenoid actions and their relation to health and disease. Mol. Aspects Med. 26:459-516. [DOI] [PubMed] [Google Scholar]
- 22.Lutnaes, B. F., A. Oren, and S. Liaaen-Jensen. 2002. New C40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 65:1340-1343. [DOI] [PubMed] [Google Scholar]
- 23.Lutnaes, B. F., Å. Strand, S. K. Pétursdóttir, and S. Liaaen-Jensen. 2004. Carotenoids of thermophilic bacteria—Rhodothermus marinus from submarine Icelandic hot springs. Biochem. Syst. Ecol. 32:455-468. [Google Scholar]
- 24.Mathews, M. M., and W. R. Sistrom. 1959. Function of carotenoid pigments in non-photosynthetic bacteria. Nature 184:1892-1893. [DOI] [PubMed] [Google Scholar]
- 25.Mortensen, A. 2006. Carotenoids and other pigments as natural colorants. Pure Appl. Chem. 78:1477-1491. [Google Scholar]
- 26.Nishida, Y., K. Adachi, H. Kasai, Y. Shizuri, K. Shindo, A. Sawabe, S. Komemushi, W. Miki, and N. Misawa. 2005. Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl. Environ. Microbiol. 71:4286-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, A. V., and L. G. Rao. 2007. Carotenoids and human health. Pharmacol. Res. 55:207-216. [DOI] [PubMed] [Google Scholar]
- 28.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 29.Sandmann, G., M. Albrecht, G. Schnurr, O. Knörzer, and P. Böger. 1999. The biotechnological potential and design of novel carotenoids by gene combination in Escherichia coli. Trends Biotechnol. 17:233-237. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, K., A. Connor, and G. Britton. 1994. Analysis of pigments: carotenoids and related polyenes, p. 403-461. In M. Goodfellow and A. G. O'Donnell (ed.), Chemical methods in prokaryotic systematics. John Wiley & Sons, Chichester, England.
- 31.Shindo, K., K. Kikuta, A. Suzuki, A. Katsuta, H. Kasai, M. Yasumoto-Hirose, Y. Matsuo, N. Misawa, and S. Takaichi. 2007. Rare carotenoids, (3R)-saproxanthin and (3R,2′S)-myxol, isolated from novel marine bacteria (Flavobacteriaceae) and their antioxidative activities. Appl. Microbiol. Biotechnol. 74:1350-1357. [DOI] [PubMed] [Google Scholar]
- 32.Sieiro, C., M. Poza, T. de Miguel, and T. G. Villa. 2003. Genetic basis of microbial carotenogenesis. Int. Microbiol. 6:11-16. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Stahl, W., N. Ale-Agha, and M. C. Polidori. 2002. Non-antioxidant properties of carotenoids. Biol. Chem. 383:553-558. [DOI] [PubMed] [Google Scholar]
- 35.Stahl, W., and H. Sies. 2003. Antioxidant activity of carotenoids. Mol. Aspects Med. 24:345-351. [DOI] [PubMed] [Google Scholar]
- 36.Tao, L., H. Yao, H. Kasai, N. Misawa, and Q. Cheng. 2006. A carotenoid synthesis gene cluster from Algoriphagus sp. KK10202C with a novel fusion-type lycopene β-cyclase gene. Mol. Gen. Genomics 276:79-86. [DOI] [PubMed] [Google Scholar]
- 37.Teramoto, M., N. Rählert, N. Misawa, and G. Sandmann. 2004. 1-Hydroxy monocyclic carotenoid 3,4-dehydrogenase from a marine bacterium that produces myxol. FEBS Lett. 570:184-188. [DOI] [PubMed] [Google Scholar]
- 38.Teramoto, M., S. Takaichi, Y. Inomata, H. Ikenaga, and N. Misawa. 2003. Structural and functional analysis of a lycopene β-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett. 545:120-126. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian, B., Z. Xu, Z. Sun, J. Lin, and Y. Hua. 2007. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim. Biophys. Acta 1770:902-911. [DOI] [PubMed] [Google Scholar]
- 41.Umeno, D., A. V. Tobias, and F. H. Arnold. 2005. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 69:51-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandamme, P., B. Pot, M. Gillis, P. De Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veiga-Crespo, P., L. Blasco, F. R. dos Santos, M. Poza, and T. G. Villa. 2005. Influence of culture conditions of Gordonia jacobaea MV-26 on canthaxanthin production. Int. Microbiol. 8:55-58. [PubMed] [Google Scholar]
- 44.Zhang, L., Q. Yang, X. Luo, C. Fang, Q. Zhang, and Y. Tang. 2007. Knockout of crtB or crtI gene blocks the carotenoid biosynthetic pathway in Deinococcus radiodurans R1 and influences its resistance to oxidative DNA-damaging agents due to change of free radicals scavenging ability. Arch. Microbiol. 188:411-419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.