Abstract
We evaluated four priming-boosting vaccine regimens for the highly pathogenic simian human immunodeficiency virus SHIV89.6P in Macaca nemestrina. Each regimen included gene gun delivery of a DNA vaccine expressing all SHIV89.6 genes plus Env gp160 of SHIV89.6P. Additional components were two recombinant vaccinia viruses, expressing SHIV89.6 Gag-Pol or Env gp160, and inactivated SHIV89.6 virus. We compared (i) DNA priming/DNA boosting, (ii) DNA priming/inactivated virus boosting, (iii) DNA priming/vaccinia virus boosting, and (iv) vaccinia virus priming/DNA boosting versus sham vaccines in groups of 6 macaques. Prechallenge antibody responses to Env and Gag were strongest in the groups that received vaccinia virus priming or boosting. Cellular immunity to SHIV89.6 peptides was measured by enzyme-linked immunospot assay; strong responses to Gag and Env were found in 9 of 12 vaccinia virus vaccinees and 1 of 6 DNA-primed/inactivated-virus-boosted animals. Vaccinated macaques were challenged intrarectally with 50 50% animal infectious doses of SHIV89.6P 3 weeks after the last immunization. All animals became infected. Five of six DNA-vaccinated and 5 of 6 DNA-primed/particle-boosted animals, as well as all 6 controls, experienced severe CD4+-T-cell loss in the first 3 weeks after infection. In contrast, DNA priming/vaccinia virus boosting and vaccinia virus priming/DNA boosting vaccines both protected animals from disease: 11 of 12 macaques had no loss of CD4+ T cells or moderate declines. Virus loads in plasma at the set point were significantly lower in vaccinia virus-primed/DNA-boosted animals versus controls (P = 0.03). We conclude that multigene vaccines delivered by a combination of vaccinia virus and gene gun-delivered DNA were effective against SHIV89.6P viral challenge in M. nemestrina.
An effective vaccine to reduce human immunodeficiency virus (HIV) infection and subsequent disease is not yet available. In 2002 alone, an estimated five million people became infected with HIV type 1 (HIV-1) (66). Ninety-five percent of new infections occur in developing countries. Although effective treatments are available, they have a high failure rate and their long-term effects are unknown and, more important, they are unaffordable in the countries hit hardest by the epidemic. Vaccines are likely to be the only viable long-term solution.
A wide variety of HIV vaccine candidates are being tested in small clinical trials with humans; however, to date only one has been evaluated in a phase III efficacy trial (31). As new vaccine concepts are developed, the nonhuman primate models of AIDS allow direct testing of a vaccine followed by a viral challenge. The macaque monkey can be infected with viruses related to HIV. Several strains of simian immunodeficiency virus (SIV), and chimeras of HIV and SIV called simian HIV (SHIV), infect macaques and cause a disease similar to human AIDS (43). Both SIV and SHIV have been used to evaluate protection from infection or from disease. Use of SHIV further allows testing of vaccines that contain HIV Envelope, which could be moved directly into formulations for human trials. Importantly, a number of approaches have shown promise in these models, supporting further development and testing in humans. The predictive value of nonhuman primate models will only be determined after successful human trials.
DNA vaccination is a promising strategy that has yet to be fully exploited. In principle, DNA vaccines share major advantages with live attenuated vaccines: the vaccine antigen is made by transfected host cells, will thus be folded and modified in its native conformation, and can be presented to the immune system by major histocompatibility complex (MHC) class I and II molecules. DNA vaccines are safer than live attenuated viruses because they can be made replication incompetent by carrying only single genes or genomes with large deletions, and a number of animal and human studies have demonstrated their safety. Both cellular and humoral immunity can be generated, and protection from disease has been achieved against a variety of pathogens in animal models (16, 54). Human trials of DNA vaccines for AIDS show induction of antibodies and cellular immune responses to HIV-1 (13, 31, 39), while several macaque studies have shown reduced SIV or SHIV viral load in animals vaccinated with DNA alone (10, 20, 28, 38, 42, 55). However, the relatively low levels of immunity elicited by current methods of DNA vaccine delivery have failed to provide sterilizing immunity against the primate lentiviruses. Thus, efforts have been directed toward enhancing the low-level immune responses elicited by DNA vaccines by combining them with a different type of vaccine modality to boost the priming response (29, 42, 55). The priming-boosting concept showed early promise for SIV and HIV vaccines with recombinant vaccinia virus and subunit protein boosting (33). Several recent studies have demonstrated good protection of macaques from disease induced by SHIV89.6P after priming-boosting vaccines including a DNA priming vaccine (2, 60).
There is increasing evidence that inclusion of multiple immune targets in vaccines is more effective than using a single antigen. Many candidate AIDS vaccines have been made against Env only (reviewed in reference 64), Gag only (20, 60), or Tat only (24). While Env is the sole target of neutralizing antibody, all of the viral genes may be targets for cytotoxic T lymphocyte, and individual infected patients and animals develop cytotoxic-T-lymphocyte responses to multiple viral gene products (12, 44). Different MHC haplotypes are able to present epitopes from different genes; and even in related animals which share some MHC alleles, epitopes from different genes are targeted (23). Given the variable breadth of responses in individuals, single-antigen vaccines may not be effective in all recipients. Furthermore, HIV sequences vary greatly; several antigens in a vaccine increase the chance of providing at least one that is cross-reactive with a virus to which a vaccinee is exposed. Therefore, it is compelling to use multiple antigens in an AIDS vaccine. Indeed, recent studies suggest that multiantigen vaccines are superior to comparable Gag-only or Env-only formulations (1, 47).
We have evaluated the immunogenicity and protective efficacy of several priming-boosting vaccine regimens for SHIV89.6P, a highly virulent challenge virus that causes rapid CD4+-T-cell depletion in unvaccinated Macaca mulatta (rhesus macaques) and in Macaca nemestrina (pigtailed macaques). Each regimen included a multigene DNA vaccine delivered by gene gun. We found significant protection from disease in M. nemestrina macaques given the combination of DNA and recombinant vaccinia virus, with DNA either as the priming vaccine or as the boosting vaccine.
MATERIALS AND METHODS
DNA vaccinations.
The construction of vaccine components is described elsewhere (6, 18, 20). Plasmids for vaccination were prepared with the EndoFree Maxiprep kit (Qiagen, Valencia, Calif.). DNA vaccines were administered with the Helios gene gun (Bio-Rad, Hercules, Calif.) at 400 lb/in2. DNA was precipitated onto 1-μm-diameter gold beads, and bullets were prepared according to the manufacturer's instructions (Bio-Rad). Mice were shaved on the abdomen and received one or three shots of 1 μg of DNA on 0.5 mg of gold. Macaques received 30 shots of gold; each shot had 2 μg of total DNA on 0.5 mg of gold. Skin was shaved on each thigh, each upper arm, and between the shoulder blades; six shots were administered at each site. The choice of the number of shots was based on a dose-response study (18).
Vaccinia virus vaccinations.
Two recombinant vaccinia viruses were used: v-ELgp160(89.6P), which contains the full-length Env gene of SHIV 89.6P clone KB9 (34), and vELgag/pol(mac239)b(2)9.1, which contains gag and pol (up to but not including integrase) of SIVmac239. The construction and propagation of these viruses is described in reference 18. Macaques were inoculated with 108 PFU of each recombinant virus by skin scarification at two or three sites on the back.
Inactivated virus.
SHIV89.6 virus was grown in CEMx174 cells, inactivated with AT-2 as described in reference 57 and purified in sucrose gradients. Characterization of the preparation is described in reference 18. For each dose, 200 μg (by protein) of the inactivated virus was mixed with adjuvant, 0.025% alhydrogel (Cedarlane, Hornby, Ontario, Canada) in acetate buffer (pH 6.2), and 500 μg of CpG oligonucleotides. The latter were phosphorothioate at all linkages, synthesized by Genosys, Inc. (The Woodlands, Tex.), with the sequence 5′-TCGTCGCTGTTGTCGTTTCTT (32). Macaques received 1 ml of vaccine intramuscularly.
Animals.
Juvenile M. nemestrina macaques were housed in the Washington National Primate Research Center under the care of licensed veterinarians. The University of Washington Institutional Animal Care and Use Committee approved all experimental procedures. Euthanasia was performed on the basis of the following criteria: (i) AIDS, (ii) termination of the experiment, or (iii) an unrelated cause. Euthanasia is considered to be AIDS related if the animal exhibits ≤200 CD4+ cells/mm3 in the peripheral blood at two or more consecutive time points, and two or more of the following conditions are present: wasting (loss of >15% of normal body weight), unsupportable diarrhea, opportunistic infection(s), proliferative disease(s) (e.g., lymphoma), and abnormal hematology (most commonly anemia). Mice were housed at the Seattle Biomedical Research Institute in a Food and Drug Administration-approved facility. All work was approved by the Institutional Animal Care and Use Committee.
Virus challenge.
SHIV89.6PMN stock was derived from monkey-passaged SHIV89.P stock (gift of N. Letvin) (50, 51) by two passages in CD8+-depleted peripheral blood mononuclear cells (PBMC) from M. nemestrina (18). The in vivo titer in M. nemestrina was determined to be 25 50% animal infectious doses (AID50)/ml (63). Vaccinated and control macaques were challenged with two doses, 1 h apart, of 1 ml of undiluted virus atraumatically in the rectum (50 AID50 total).
Virus load determinations.
Viral loads in plasma and PBMC were determined by real-time reverse transcription (RT)-PCR and real-time PCR, respectively, as described in reference 18. The limit of detection was 100 copies/ml of plasma. Viral load assays, as well as immunophenotyping, were performed by the Virology Core of the Washington National Primate Research Center.
Antibody assays.
Binding antibody responses to SHIV antigens were measured by enzyme-linked immunosorbent assay (ELISA) as described previously (18). Briefly, Immunosorp plates (Nalge Nunc, Rochester, N.Y.) were coated with 2 μg of recombinant gp120/ml. Diluted plasma was incubated for 1 h on the plates and detected with biotin-conjugated anti-human immunoglobulin G (IgG) (ICN Biomedicals, Costa Mesa, Calif.) followed by Extravidin-horseradish peroxidase. Neutralization assays were performed with the cMAGI assay. cMAGI cells (15) were maintained in Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum (FBS) with Geneticin (250 μg/ml), hygromycin (100 μg/ml), and puromycin (1 μg/ml). Cells were seeded in 96-well flat-bottom plates at 104 cells/well in 100 μl of DMEM-10% FBS medium and incubated for 24 h prior to infection. Virus was diluted in DMEM-10% FBS to give 100 to 200 infectious units per well. Heat-inactivated plasma was serially diluted twofold in DMEM-10% FBS, mixed with equal volumes of input virus, and incubated for 1 h at 37°C in 5% CO2. DEAE-dextran was then added to a final concentration of 20 μg/ml. The virus-plasma mixture (110 μl/well) was then added to duplicate aspirated cell monolayers. Plates were incubated for 2 h, after which an additional 200 μl of medium/well was added. After 48 h, cells were fixed with 400 μl of fixing solution (1% formaldehyde, 0.2% glutaraldehyde in phosphate-buffered saline [PBS]) and stained for 50 min with 50 μl of staining solution (5 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal; Sigma]/ml, 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2 in PBS) and washed thoroughly with PBS. A positive reaction for virus infection is denoted by nuclei or syncytia that are stained deep blue. Wells were counted on a Bioreader (Biosys, Heidelberg, Germany) calibrated to a manual count of at least 6 wells. The percent neutralization at a given titer is calculated by the equation (Vo − Vn)/Vo × 100, where Vn is the number of infected cells in the virus-plus-antibody wells and Vo is the number of positive cells in the virus alone wells. Titers were normalized to the titer of a standard HIV-positive human serum pool that was included on each assay plate.
ELISpot assay.
A gamma interferon (IFN-γ) enzyme-linked immunospot (ELISpot) assay was performed on frozen PBMC. The cells were thawed in RPMI and 10% fetal calf serum 1 day before the assay and incubated overnight at 37°C. The assay was carried out with an ELISpot system monkey IFN-γ kit (U-CyTech, Utrecht, The Netherlands) according to the manufacturer's protocol. Duplicate wells of 105, 5 × 104, 2.5 × 104, and 1.25 × 104 PBMC were plated in a 96-well format. Overlapping 15-mer peptides obtained from the National Institutes of Health AIDS Research and Reference Reagent Program were used at between 0.5 and 10 μg/ml (Gag peptides derived from SIVmac239; Env from SHIV89.6; Pol from SIVmac239; the accessory pool contained peptide from HIV Tat and Rev and SIVmac239 Nef, Vpr, and Vpx). Each well received the same amount of peptide. As a positive control, streptococcal enterotoxin B was added to responder cells at 1 μg/ml; as a negative control, an irrelevant peptide was added. After drying, the plates were stripped and read with a Zeiss microscope by using KS ELISpot (Carl Zeiss Vision GmbH, Göttingen, Germany) software. The response to the irrelevant peptide was subtracted from the response to each antigen peptide pool, and a response to antigen was considered positive if it was greater than twice the response to irrelevant peptide and >40 per 106 PBMC (which is the mean plus three standard deviations of the naive animal response to antigen).
RESULTS
Construction and characterization of SHIV89.6-based DNA vaccines.
The DNA vaccine consisted of six plasmids, which together allow high levels of expression of all SHIV genes under the control of the human cytomegalovirus Immediate Early-1 promoter. The plasmids pV1Jns:89.6Penv160OPTcleavable and pVIR SIV GAG OPT have been described elsewhere (6, 20) and are immunogenic in macaques (10). The other plasmids were constructed for this study and were validated in vitro (18) and found to elicit antibodies to the expected viral gene products in DNA-immunized mice (Table 1). pV1Jns:89.6Penv160OPTcleavable contains codon-optimized env gp160 from the animal-passaged virus SHIV89.6P; the other five plasmids correspond to SHIV89.6, the parental construct. Additional env constructs were the rev-env construct pCIneo89.6 and pEMC*896, a gp120 expression construct in which gp120 is fused to the signal sequence of tissue plasminogen activator. pVIR SIV GAG OPT encodes codon-optimized Gag from SIVmac239, which is identical to the gene in SHIV89.6. Nef expression is provided by pC3aNef. Accessory genes tat, rev, vif, vpr, vpx, and vpu, as well as gag, pol, and env, are provided by pC3a896gpe, which carries nucleotides 536 to 8903 in the SHIV89.6 genome (34). We demonstrated that this plasmid can produce virus-like particles (VLP) in COS cells with processed Gag and fully functional Envelope (18), as was shown previously for an SIV plasmid that directed the expression of SIV VLP (42).
TABLE 1.
Plasmids used in DNA vaccine
| Plasmid | Gene product(s) | Source virus | Assay(s) showing in vitro expressionc | Antibody response in micea |
|---|---|---|---|---|
| pV1Jns:89.6Penv160OPTcleavable | Env (gp160) | SHIV89.6P | IFA, Western blotting, sCD4 binding | 5/5 to gp160 |
| pCIneo89.6 | Env (gp160) | SHIV89.6 | IFA, cell fusion, Western blotting, sCD4 binding | 2/20 to gp160 |
| pEMC*896 | Env (gp120) | SHIV89.6 | Western blotting, sCD4 binding | 5/5 to gp160 |
| pC3a896gpe | Gag | SHIV89.6 | Western blotting, ELISA | 2/5 to Gag-Pol |
| Pol | Western blotting | NT | ||
| Tat | cMAGI assay | NT | ||
| Env (gp160) | IFA, cell fusion, sCD4 binding | 2/5 to gp160 | ||
| Vif, Vpr, Vpv, Rev | NTb | NT | ||
| pVIR SIV OPT GAG | Gag | SHIV89.6 | Western blotting, ELISA | NT |
| pC3a Nef | Nef | SHIV89.6 | Western blotting | NT |
| All 6 mixed | 5/5 to gp160 | |||
| 2/5 to Gag-Pol |
Number of mice responding/total number of mice.
NT, not tested.
IFA, immunofluorescence assay; sCD4, soluble CD4.
Boosting agents.
To improve the potency of the DNA vaccine, we used several different boosting agents. The first group of animals received three priming doses and two additional boosting doses of DNA and is referred to as DNA-DNA or DNA alone. The second group was boosted twice with virus particles inactivated with AT-2 (57) and is referred to as DNA priming/particle boosting (DNA-Particle on figures). AT-2 inactivation does not affect the structure of Env on the surface of the virions (57) and may therefore be a better immunogen for the elicitation of conformation-dependent and neutralizing antibodies than purified protein.
Two recombinant vaccinia viruses were constructed for this study: v-ELgp160(89.6P), which contains the full-length Env gp160 gene of SHIV89.6P, and vELgag/pol(mac239)b(2)9.1, which expresses Gag-Pol of SIVmac239 (18). The third group was boosted twice with a mixture of these two recombinant vaccinia viruses and is referred to as DNA priming/vaccinia virus boosting (DNA-Vacc on figures). Previous work by our group showed that vaccinia virus priming/protein boosting vaccines provided sterilizing immunity against SIVmne (33). Therefore, we chose to also include a fourth group in which the regimen was reversed: two priming doses of recombinant vaccinia virus followed by three boosts with DNA. This group is referred to as vaccinia virus priming/DNA boosting (Vacc-DNA on figures).
Vaccination of macaques and prechallenge humoral and cellular antiviral immunity.
Thirty M. nemestrina macaques were vaccinated according to the time line shown in Fig. 1. Prior experience with vaccinia virus indicated that an 8-week interval between doses was optimal for an effective boost of the immune response (S.-L. Hu, unpublished data). Long intervals following the second and third doses were chosen to maximize the effect of boosting doses, as several groups have shown the benefits of such rest periods for both DNA (27) and protein (4) immunizations.
FIG. 1.
Time line of vaccination and viral challenge. Arrows indicate times of vaccinations. DNA, DNA vaccination by gene gun; Particle, inactivated virus particles; Vaccinia, recombinant vaccinia virus.
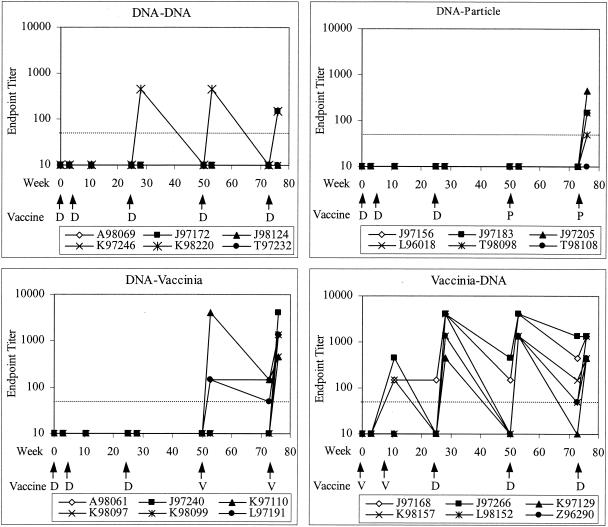
On the day of and 3 weeks after each immunization, blood samples were taken from the animals and tested for binding antibody to HIV-1 Env. The development of the IgG response to Env is shown in Fig. 2. Control animals had endpoint titers of <50 and were considered negative at all time points prior to challenge. DNA alone elicited very low binding antibody responses. In the group that received DNA alone (group 1), only 2 of 6 animals had detectable anti-Env IgG after five doses. Examination of all animals in the three groups primed with three doses of DNA showed that only 1 of 18 animals had detectable antibody after three doses. Boosting with inactivated virus particles greatly improved the antibody response: after the second dose, 4 of 6 animals had binding antibody against Env (Fig. 2) and all had IgG to disrupted SIV, which contains the same Gag-Pol as the vaccine (data not shown).
FIG. 2.
Development of prechallenge anti-Env binding antibody (IgG) in vaccinated macaques. The dotted line indicates the limit of detection (1:50 dilution); values below this limit were assigned values of 10. Arrows indicate times of vaccinations. D, DNA; P, particle; V, recombinant vaccinia virus. Endpoint titer is the reciprocal of the highest dilution at which the optical density is twice that of preimmune sera at the corresponding dilution.
Recombinant vaccinia virus expressing Gag-Pol and Env was an effective boosting agent for DNA. After three doses of DNA, no animals in the DNA-Vacc group had antibodies to Env (Fig. 2); however, after one boosting dose of vaccinia, 3 of 6 animals had detectable anti-Env antibodies. After a second vaccinia dose, 6 of 6 animals had antibody to Env, with a geometric mean titer of 1:1,100. Conversely, DNA served as an effective boosting agent for vaccinia virus in the Vacc-DNA group. Animals primed with two doses of vaccinia virus had low but detectable responses, with 3 of 6 animals having antibodies to Env. After the first DNA boosting dose, 6 of 6 animals responded, with a geometric mean titer of 1:1,900.
We measured cellular immunity to SHIV89.6 by ELISpot assay and compared samples after priming and after full vaccination (Table 2). PBMC isolated after the last priming dose (dose 2 or 3) (Fig. 1) or the last boosting dose (day of challenge) were stimulated with one of four pools of peptides, corresponding to Gag, Pol, Env, or accessory proteins of SHIV89.6. Responses ranged from 70 to 1,595 spot-forming units/million PBMC and were found in 9 of 24 vaccinated animals and none of the controls on the day of challenge. After three DNA priming immunizations, 17 of the 18 DNA-vaccinated animals were negative for cellular responses while 1 animal was very weakly positive for the Env and accessory gene peptides. In contrast, after two priming doses of recombinant vaccinia virus, 2 of 6 animals were positive by ELISpot assay for Gag and Env peptides. After the boosting immunizations, 1 of 6 DNA priming/particle boosting animals was positive for Gag while 5 of 6 DNA priming/vaccinia virus boosting animals and 4 of 6 vaccinia virus priming/DNA boosting animals were positive for Gag, Env, or both. Three macaques had low-level prechallenge responses to Pol or accessory proteins that were above the background; one vaccinia virus Gag and Env-primed macaque (Z96290) showed an unexpected response to Pol and accessory genes that did not persist with boosting. These data indicate that, as seen for antibody, DNA was an effective priming or boosting agent for cellular immune responses elicited by the vaccinia virus recombinants.
TABLE 2.
Cellular immunity measured by IFN-γ ELISpot assay
| Group | Animal | Resulta for viral antigen:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag
|
Env
|
Pol
|
Accessory peptide
|
||||||||||||||
| Primeb | Wk 0c | Wk 2 | Wk 4 | Prime | Wk 0 | Wk 2 | Wk 4 | Prime | Wk 0 | Wk 2 | Wk 4 | Prime | Wk 0 | Wk 2 | Wk 4 | ||
| Control | J96258 | —d | 2 | NDe | 50 | — | 0 | ND | 90 | — | 2 | ND | 0 | — | 2 | ND | 0 |
| J97167 | — | 5 | 10 | 25 | — | 5 | 0 | 40 | — | 5 | 35 | 5 | — | 15 | 0 | 25 | |
| J97200 | — | 0 | 0 | 5 | — | 15 | 5 | 5 | — | 5 | 0 | 0 | — | 0 | 0 | 0 | |
| J98071 | — | 0 | 5 | 10 | — | 0 | 10 | 0 | — | 0 | 0 | 10 | — | 0 | 0 | 0 | |
| K96166 | — | 20 | 120f | 70 | — | 0 | 30 | 0 | — | 0 | 0 | 5 | — | 0 | 5 | 150 | |
| K97107 | — | 0 | 100 | 55 | — | 5 | 0 | 10 | — | 0 | 5 | 10 | — | 0 | 20 | 0 | |
| DNA-DNA | A98069 | 0 | 5 | 320 | 160 | 10 | 5 | 0 | 60 | 0 | 0 | ND | 0 | 5 | 10 | ND | 30 |
| J97172 | 0 | 0 | 25 | 50 | 0 | 5 | 75 | 90 | 0 | 5 | 430 | 0 | 0 | 5 | 145 | 0 | |
| J98124 | 0 | 0 | 75 | 100 | 0 | 0 | 560 | 0 | 0 | 0 | 0 | 20 | 5 | 0 | 5 | 5 | |
| K97246 | 5 | 0 | 225 | 90 | 15 | 5 | 890 | 75 | 0 | 0 | 35 | 0 | 0 | 0 | 55 | 100 | |
| K98220 | 0 | 0 | 60 | 0 | 5 | 0 | 345 | 25 | 0 | 5 | 25 | 0 | 0 | 0 | 630 | 0 | |
| T97232 | 0 | 0 | 300 | 0 | 0 | 20 | 15 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 0 | |
| DNA-particle | J97156 | 0 | 10 | ND | 0 | 0 | 0 | ND | 0 | 0 | 0 | ND | 0 | 0 | 0 | ND | 0 |
| J97183 | 0 | 10 | 5 | 5 | 0 | 10 | 70 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 25 | 0 | |
| J97205 | 0 | 15 | 595 | 120 | 0 | 0 | 180 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 35 | 0 | |
| L96018 | 10 | 160 | 60 | 595 | 15 | 30 | 345 | 45 | 0 | 35 | 25 | 70 | 10 | 10 | 630 | 30 | |
| T98098 | 5 | 0 | ND | 30 | 0 | 0 | ND | 10 | 10 | 0 | ND | 10 | 5 | 0 | ND | 15 | |
| T98108 | 0 | 0 | 55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 10 | 0 | 0 | 305 | 20 | |
| DNA-vaccinia virus | A98061 | 5 | 398 | 1,090 | 230 | 20 | 362 | 500 | 220 | 15 | 0 | 1,300 | 0 | 0 | 0 | 15 | 70 |
| J97240 | 0 | 90 | 910 | 180 | 0 | 300 | 130 | 1,485 | 0 | 0 | 10 | 15 | 0 | 0 | 0 | 0 | |
| K97110 | 0 | 1,018 | 0 | 20 | 0 | 1,315 | 110 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 170 | 235 | |
| K98097 | 0 | 10 | 705 | 445 | 0 | 15 | 670 | 520 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | |
| K98099 | 40 | 40 | ND | 10 | 65 | 145 | ND | 1,595 | 10 | 0 | ND | 0 | 55 | 0 | ND | 10 | |
| L97191 | 10 | 325 | 875 | 30 | 0 | 255 | 70 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 205 | 35 | |
| Vaccinia virus-DNA | J97168 | 0 | 25 | 100 | 145 | 10 | 15 | 0 | 25 | 10 | 0 | 0 | 25 | 10 | 10 | 5 | 65 |
| J97266 | 0 | 20 | 440 | 235 | 55 | 440 | 205 | 230 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | |
| K97129 | 10 | 210 | 430 | 185 | 10 | 635 | 5 | 925 | 0 | 22 | 0 | 5 | 0 | 10 | 0 | 110 | |
| K98157 | 0 | 10 | 230 | 150 | 10 | 15 | 1,425 | 10 | 0 | 0 | 15 | 10 | 0 | 0 | 95 | 0 | |
| L98152 | 215 | 85 | 815 | 280 | 35 | 35 | 455 | 10 | 5 | 5 | 15 | 0 | 0 | 0 | 10 | 10 | |
| Z96290 | 310 | 90 | 70 | 5 | 210 | 575 | 470 | 15 | 90 | 0 | 0 | 5 | 135 | 0 | 5 | 5 | |
Spot-forming cells/106 PBMC measured by IFN-γ ELISpot with pools of overlapping peptides.
Prime, samples taken after 3 doses of DNA or 2 doses of recombinant vaccinia virus.
Wk 0, day of challenge (3 weeks after the last boosting dose).
—, Not applicable.
ND, not determined.
Numbers in boldface type are >2 times the background for that animal and >40 spot-forming cells/106 PBMC.
Lymphocyte depletion following viral challenge.
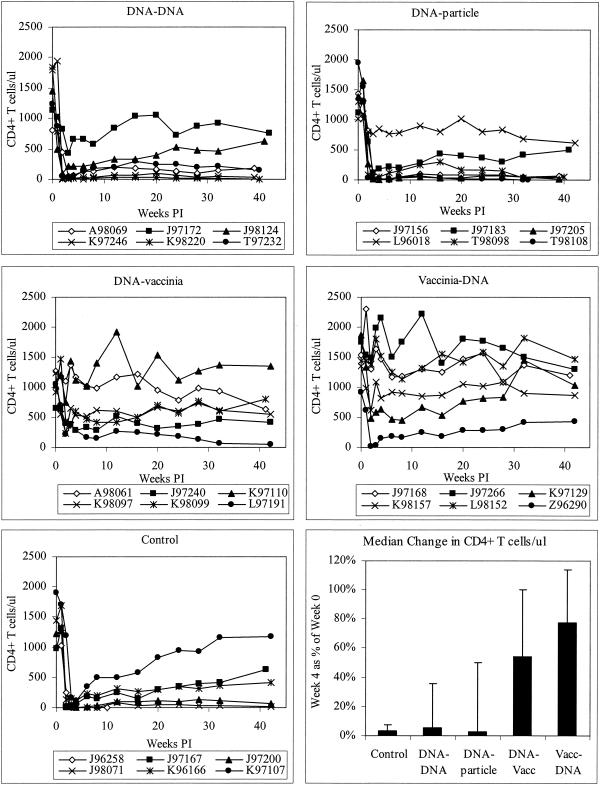
Three weeks after the fifth immunization, all animals were challenged intrarectally with 50 AID50 of SHIV89.6P. We chose the mucosal route to model sexual transmission, the dominant form of HIV transmission worldwide. Lymphocyte subsets in the blood of all macaques were measured over the course of the experiment for 42 weeks after challenge. As shown in Fig. 3, naive animals experienced dramatic and rapid CD4+-T-cell loss in the first 4 weeks postchallenge, similar to that documented for M. mulatta. One animal died of simian AIDS at week 10, 2 animals gradually recovered some CD4+ T cells, and the remaining 3 control animals had counts of less than 500 cells/μl for 42 weeks. The animals vaccinated with DNA alone or DNA priming/particle boosting were not protected from this pathological effect; in each group, 5 of 6 animals experienced dramatic loss and sustained low levels of CD4+ T cells, and 1 of 6 animals showed slow CD4+-T-cell decline over several months.
FIG. 3.
CD4+-T-cell counts of infected macaques. The bottom right panel shows the median for each group of each animal's CD4+-T-cell count at week 4 compared to week 0. Error bars indicate interquartile ranges. Vacc, vaccinia virus; PI, postinfection.
Animals vaccinated with DNA and vaccinia virus, in either regimen, fared much better. DNA priming/vaccinia virus boosting animals experienced slow or moderate declines in CD4+-T-cell counts; 1 of 6 animals experienced no immunodepletion at all (Fig. 3). Macaques given vaccinia virus priming/DNA boosting were also partially protected from disease. Three of 6 animals showed no effect of infection on the level of CD4+ T cells. Two of 6 animals had slow, moderate declines but maintained CD4+-T-cell counts of over 500 for the 42 weeks of the study, and 1 animal initially had a severe loss but gradually recovered to 50% of its baseline CD4+-T-cell level.
Since the prechallenge CD4+-T-cell counts were quite variable from animal to animal, we analyzed CD4+-T-cell loss by comparing each animal's counts at weeks 0 and 4. This parameter is shown in Fig. 3, lower right panel. The median loss was much more severe in control animals and the DNA-only and DNA/particle groups than in the DNA/vaccinia virus and vaccinia virus/DNA groups. CD4+-T-cell loss is significantly greater in controls than in the last two groups (P = 0.002 for each group compared to controls) (Table 3). Additionally, we looked at the levels of CD4+ T cells in the chronic phase of infection. Again, the vaccinia virus/DNA group showed significantly better maintenance of CD4+ T cells (P = 0.03); however, the DNA/vaccinia virus group did not achieve statistically significant maintenance of CD4+ T cells in the chronic phase of infection.
TABLE 3.
Analysis of CD4+-T-cell counts in infected macaques
| Group | Median CD4+-T-cell count as % of wk 0 for wk:
|
P value for wka:
|
||
|---|---|---|---|---|
| 4 | 24 | 4 | 24 | |
| Control | 3.2 | 23.7 | ||
| DNA-DNA | 5.7 | 19.0 | 0.240 | 0.931 |
| DNA-particle | 2.6 | 9.1 | 0.485 | 0.662 |
| DNA-vaccinia virus | 53.9 | 57.3 | 0.002b | 0.052 |
| Vaccinia virus-DNA | 77.9 | 88.2 | 0.002 | 0.030 |
P values from Mann-Whitney test of each vaccine group compared to controls.
Boldface type indicates P < 0.05.
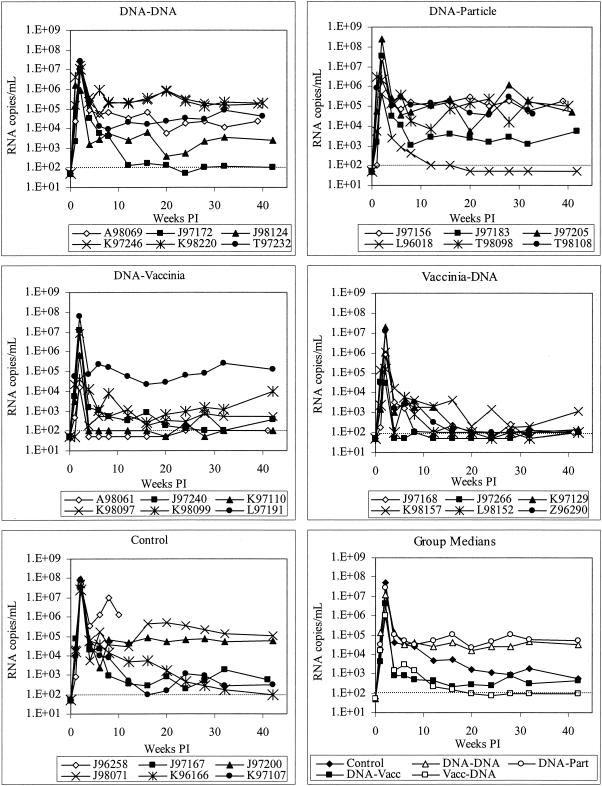
Virus loads in vaccinated and control macaques.
Virus loads were measured by real-time RT-PCR of viral RNA in plasma. This analysis showed that all animals in the study were infected, and this was confirmed by PCR of PBMC DNA and lymph node DNA (data not shown). As shown in Fig. 4, controls had consistently high peak viremia, up to 108 copies/ml. All vaccine groups except DNA priming/particle boosting had small but significant reductions in peak viremia (Table 4). Set point viremia was highly peak variable in controls and in animals vaccinated with DNA alone or primed with DNA and boosted with particles. One control animal (J96258) sustained very high viremia, progressed rapidly to AIDS, and was euthanized at week 10. In concordance with the effects on CD4+ T cells, we found that animals receiving DNA and recombinant vaccinia virus, in either regimen, had consistently lower viral loads in the chronic phase of infection. In macaques that were primed with DNA and boosted with vaccinia virus, 5 of 6 animals controlled viremia to less than 104 copies/ml by week 6 postinfection, and these low levels were stable through week 42. Among animals primed with vaccinia virus and boosted with DNA, all 6 animals controlled viremia below 104 copies/ml, with 5 of 6 having virus at or below the limit of detection (100 copies/ml) at multiple time points. The animals vaccinated with vaccinia virus priming/DNA boosting had significantly lower viremia at week 24 than the controls (P = 0.03); this was the only group that reached a level statistically different from the controls.
FIG. 4.
Virus load in plasma in infected macaques. Viremia was quantified by real-time RT-PCR. The dotted line indicates the limit of detection (100 copy equivalents/ml); values below this limit were assigned values of 50 copies/ml. Vacc, vaccinia virus; Part, particle; PI, postinfection.
TABLE 4.
Analysis of plasma viral load in infected macaques
| Group | Median viral load (RNA copies/ml of plasma) for wk:
|
P value for wka:
|
||
|---|---|---|---|---|
| 4 | 24 | 4 | 24 | |
| Control | 5.2 × 107 | 1.2 × 103 | ||
| DNA-DNA | 1.2 × 107 | 2.6 × 104 | 0.004b | 0.931 |
| DNA-particle | 2.9 × 107 | 4.5 × 104 | 0.310 | 0.792 |
| DNA-vaccinia virus | 4.3 × 106 | 2.6 × 102 | 0.026 | 0.178 |
| Vaccinia virus-DNA | 9.8 × 105 | 1.0 × 102 | 0.002 | 0.030 |
P values from Mann-Whitney test of each vaccine group compared to controls.
Boldface type indicates P value of <0.05.
Quantification of postchallenge cellular immunity.
Cellular immunity was measured at weeks 2 and 4 postchallenge by IFN-γ ELISpot assay (Table 2). All 12 of the animals in the DNA-vaccinia virus and vaccinia virus-DNA groups mounted robust cellular responses to Gag and/or Env by week 2, and 11 of 12 had sustained responses through week 4. All 6 animals vaccinated with DNA alone had responses at week 2, and 4 animals sustained the responses at week 4. In contrast, only 2 of 6 controls and 2 of 6 DNA-primed/particle-boosted animals mounted cellular responses. Additionally, at least 1 animal in each group developed responses to additional gene products, as shown by positive ELISpot assays for Pol and accessory gene peptides, including 3 of 6 animals in the DNA-only group. The level of cellular immunity on the day of challenge correlated strongly with protection from disease (see below). The level of response at weeks 2 and 4, however, did not correlate with the set point viral load or protection from CD4+-T-cell decline. ELISpot responses at these early time points were higher and more frequent in vaccinees relative to controls.
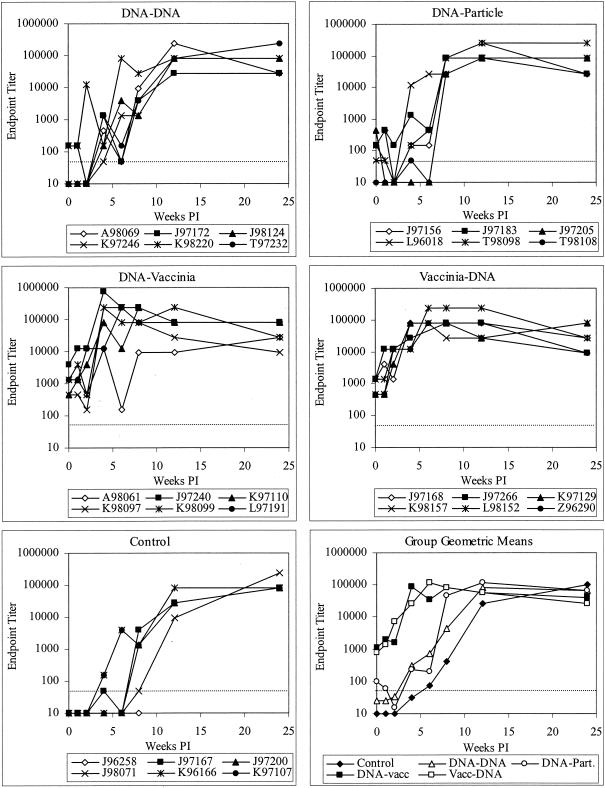
Quantification of postchallenge antiviral humoral responses.
Postchallenge antibody responses were measured for binding to Envelope gp120 (Fig. 5) and neutralizing activity to SHIV89.6 and SHIV89.6P (Fig. 6). Binding antibody increased rapidly in vaccinees that received both vaccinia virus and DNA and peaked by weeks 4 to 6 postinfection, remaining at a high level throughout the study. In contrast, while controls and animals in the first two vaccine groups generated similarly high titers of antibodies, the development was slower, plateauing at weeks 8 to 12. This observation indicates a strong memory response in the DNA priming/vaccinia virus boosting and vaccinia virus priming/DNA boosting groups.
FIG. 5.
Development of postchallenge anti-Env binding antibody (IgG) in vaccinated and control macaques. The dotted line indicates the limit of detection (1:50 dilution); values below this limit were assigned values of 10. Vacc, vaccinia virus; Part, particle; PI, postinfection.
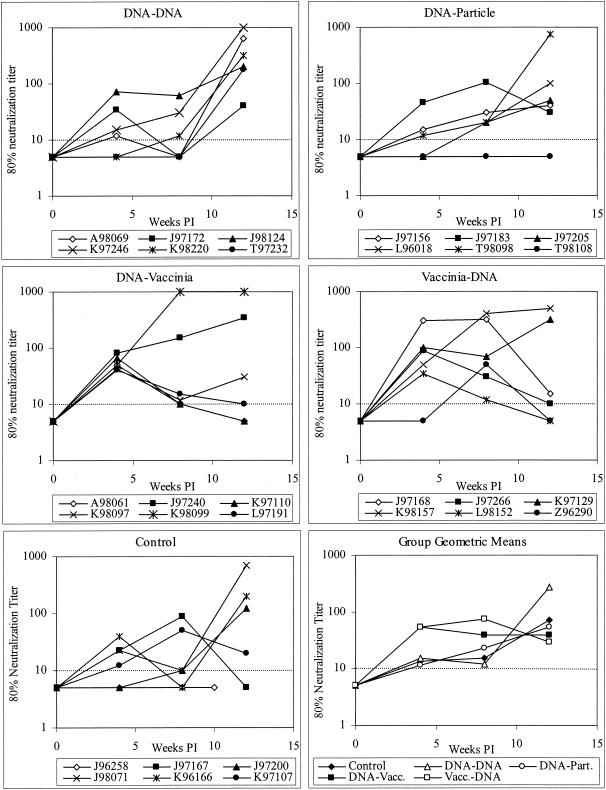
FIG. 6.
Neutralizing antibody against SHIV89.6P. Neutralization was measured by the cMAGI assay. The dotted line indicates the limit of detection (1:10 dilution). Values below this limit were assigned a value of 5. Vacc, vaccinia virus; Part, particle; PI, postinfection.
Neutralizing antibody against the challenge strain was measured by cMAGI assay (15). No neutralizing activity was detected on the day of challenge. However, most animals developed neutralizing antibodies in the first 12 weeks postinfection. The development of neutralizing antibody differed among groups. As shown in Fig. 6, animals immunized with DNA/vaccinia virus or vaccinia virus/DNA rapidly developed neutralizing antibody, with detectable neutralization in 11 of 12 animals and a geometric mean titer of 1:53 (limit of detection, 1:10) at week 4. In contrast, control animals and those immunized with DNA alone or DNA priming/particle boosting had lower and less consistent responses, with only 12 of 18 responding and a geometric mean titer of 1:13 (limit of detection, 1:10) at week 4. The same pattern was seen for neutralization of the vaccine strain SHIV89.6, assayed in GHOST-CCR5 cells (14) at 8 weeks postchallenge (data not shown). These data indicate a memory response or faster maturation of the antibody response in animals immunized with DNA/vaccinia virus or vaccinia virus/DNA.
Correlation of immune responses and outcome of infection.
We analyzed the relationship of preexisting immune responses to the outcome of viral infection. Protection from rapid immunodepletion (defined as reduction of CD4+ T cells to <20% of baseline in the first 4 weeks) correlated with detection of anti-Env IgG on the day of challenge (P = 0.003, Fisher's exact test) and detection of cellular immunity to Gag or Env by IFN-γ ELISpot assay on the day of challenge (P = 0.004). The magnitude of the immune response also correlated with protection (P values for the Spearman rank correlation test are shown in Table 5). The total ELISpot response on the day of challenge, the anti-Env IgG titer on the day of challenge, and the neutralizing antibody titers at week 4 postinfection all correlated positively with the CD4+-T-cell count (as a percentage of the levels on the day of challenge) at weeks 4 and 24 postinfection and correlated negatively with viral load at week 24.
TABLE 5.
Spearman rank correlation test for immune responses compared to outcomes
| Immune response |
P value for comparison to:
|
||
|---|---|---|---|
| Viral load, wk 24 | CD4+ T cells, wk 4 | CD4+ T cells, wk 24 | |
| SFCb/106 PBMC, wk 0 | <0.0001a | 0.0003 | 0.0001 |
| Anti-Env IgG titer, wk 0 | <0.0001 | 0.001 | 0.005 |
| Neutralizing titer, wk 4 | 0.0043 | <0.0001 | 0.0001 |
Boldface type indicates P value of <0.05.
SFC, spot-forming cells.
Clinical outcome in challenged macaques.
One animal in the control group progressed rapidly to AIDS and was euthanized at week 10 postchallenge. This animal did not seroconvert. One animal in the DNA priming/particle boosting group was euthanized at week 33. All other animals were still alive at the conclusion of the study (weeks 39 to 42), despite the fact that many had extremely low CD4+-T-cell counts for >9 months. Only minor illnesses, such as diarrhea which resolved, were noted.
DISCUSSION
In this study, we found that priming vaccinations with gene gun-delivered DNA followed by boosting with vaccinia virus, or vaccinia virus priming/DNA boosting, provided significant protection from disease caused by SHIV89.6P in M. nemestrina. Neither DNA alone nor DNA boosted with inactivated virus particles provided protection from disease. While these two groups, as well as controls, experienced severe and rapid loss of CD4+ T cells, most animals that received DNA priming/vaccinia virus boosting or vaccinia virus priming/DNA boosting remained healthy for more than 39 weeks. Statistically significant differences were seen for these two groups versus controls in the first 4 weeks after infection, and long-term maintenance of CD4+ T cells was significant for the vaccinia virus priming/DNA boosting group. These two regimens also resulted in reduced viral load in the chronic phase of infection. Set point viremia is predictive of disease progression in HIV-1-infected humans (41) and in macaque models with SIV (61, 69). In the highly pathogenic SHIV89.6P model, the relationship is less clear; however, lower viremia generally correlates with preservation of CD4+ T cells (22, 52, 65). We found a statistically significant reduction of set point viremia in the animals primed with vaccinia virus and boosted with DNA relative to controls. The animals that received DNA priming/vaccinia virus boosting also had reduced viral loads, but the reduction was not statistically significant. The median viral load for controls was lower than that in the groups vaccinated with DNA or DNA with particles; this may be due to the early death (week 10) of the control animal with the highest viral load (J96258).
In most previous vaccine studies which used DNA with a viral vector, DNA was used as priming agent but not as a boosting agent (reviewed in references 17 and 48). In a landmark study of malaria in mice, DNA priming/modified vaccinia virus Ankara (MVA) boosting provided sterilizing immunity while MVA priming/DNA boosting was ineffective (58). It has been suggested that DNA can only be effective as a priming agent and that its success may be due to low-dose priming of high-affinity T cells which can be expanded by a viral vector boosting agent (21). Conversely, several groups have suggested that poxviruses are only effective as boosting agents (21, 59) because they prime low-avidity T cells. However, other studies imply that this order may not be required, at least for lentiviruses. A study of SIV in macaques with DNA and vaccinia virus did not show clear differences when the order of priming and boosting was reversed (29). Several experiments by our group show complete (33) or partial (46) protection from SIV challenge after priming with vaccinia virus and boosting with recombinant protein; thus, we expected that vaccinia virus would be effective in priming a DNA vaccine.
Here we present two lines of evidence to show that a SHIV DNA vaccine is effective as a boosting agent for recombinant vaccinia virus. First, the animals immunized with recombinant vaccinia virus priming/DNA boosting had the best outcomes of any group in this study. Second, the time course of development of cellular and humoral immunity reveals a boosting effect of DNA. A comparison of the antibody titers (Fig. 2) illustrates this point. The responses in the vaccinia virus priming/DNA boosting group increase from 3 of 6 responders after vaccinia virus alone to 6 of 6 responders after the DNA inoculations; thus, DNA works as a boosting agent. Furthermore, all 6 macaques primed with DNA and boosted with vaccinia virus developed antibody after the boost, with higher titers than the animals given vaccinia virus first. Thus, DNA primes the antibody responses elicited by the vaccinia virus, as expected. Likewise, ELISpot assays for cellular immunity show that, while vaccinia virus alone elicited strong responses in 2 of 6 macaques, boosting these animals with DNA increased the number of responders to 4 of 6, and although DNA alone did not elicit cellular responses, the animals primed with DNA and boosted with vaccinia virus had the highest responses, with 5 of 6 animals responding (Table 2).
The presence of preexisting antibody or cellular immunity on the day of challenge correlated with protection. Overall, a positive measurement by ELISpot or binding antibody assay on the day of challenge correlated with protection from CD4+-T-cell loss, and the magnitude of the immune response correlated with higher CD4+-T-cell counts and lower viremia. The two regimens that were protective were those that elicited measurable antibody and cellular responses in most animals prior to challenge. Even within the groups that fared poorly overall, the few animals that responded to the vaccines were more likely to remain healthy. For example, the one animal in the DNA/particle group (L96018) that had cellular immunity, as measured by ELISpot, on the day of challenge was the only animal in its group that maintained a normal CD4+-T-cell count. Likewise, the animal in the DNA-only group (J97172) that had the lowest set point viral load and highest CD4+-T-cell count was one of the two animals that had detectable binding antibody to Env on the day of challenge. Furthermore, strong memory antibody responses were noted in the protected groups; these indicate the presence of memory B cells and are likely also related to the maintenance of CD4+ T cells which were able to provide help for a vigorous memory response. Although neutralizing antibody was not detected on the day of challenge in any animals, those immunized with both DNA and recombinant vaccinia virus rapidly developed modest neutralizing antibody titers after infection. At week 4, neutralizing antibody titers were threefold higher in the DNA/vaccinia virus or vaccinia virus/DNA groups than in the other animals. Since most animals developed titers of at least 1:40 by weeks 8 to 12, even those with severe immunodepletion, we infer that the earlier response in animals receiving DNA and vaccinia virus was not due solely to the preservation of CD4+ T cells in those animals. The earlier development of a neutralizing response may have helped protect those animals from CD4+-T-cell depletion. Priming of a rapid neutralizing response, in the absence of detectable neutralizing activity on the day of challenge, has been seen in other studies with SHIV89.6P (1, 10). In addition, binding antibody that was present on the day of challenge may itself have played a role in protection. Binding antibody may have protective effects that are not measured by our neutralization assay, such as antibody-dependent cell-mediated cytotoxicity (5); it has also been speculated that binding of shed gp120 glycoprotein may reduce a proapoptotic effect of gp120 on uninfected cells (1).
Several recent vaccine studies have used the highly pathogenic virus SHIV89.6P and shown protection from CD4+-T-cell loss and disease. The levels of protection from SHIV89.6P provided by DNA/vaccinia virus and vaccinia virus/DNA in this study are similar to published results of vaccination of rhesus macaques M. mulatta with recombinant MVA (rMVA) (9), recombinant vesicular stomatitis virus (56), DNA with recombinant Semliki Forest virus (40), low doses of DNA given intramuscularly and boosted with rMVA (2), and DNA vaccines given intramuscularly (10). The latter study used two of the same plasmids used here (pV1Jns:89.6Penv160OPTcleavable and pVIR SIV GAG OPT). A few studies performed with M. mulatta have shown better protection from SHIV89.6P-induced disease, notably by using recombinant adenovirus (60), rMVA primed with a high dose of DNA (2), or DNA with interleukin-2/Ig (10); however, none have elicited sterilizing immunity. The major difference between these studies and the one reported here is our use of M. nemestrina. We found that M. nemestrina macaques are susceptible to SHIV89.6P infection and that naive animals have levels of peak and set point viremia similar to those of M. mulatta macaques (50). Furthermore, the hallmark of SHIV89.6P infection in naive rhesus monkeys is the very rapid and severe depletion of circulating CD4+ T cells, and we observed this effect in naive M. nemestrina. Therefore, we conclude that M. nemestrina is an appropriate species for evaluating vaccines against this virus and that comparisons to other studies are valid.
We noted several differences in the long-term outcome of infection, notably the survival (>9 months) of animals with extremely low CD4+-T-cell counts. This contrasts with several studies of SHIV89.6P in rhesus macaques in which high mortality was noted in nonvaccinated animals (10, 51). However, more recent data indicate that survival among infected rhesus macaques is highly variable, with better survival in Mamu-A01+ animals (35, 72) and some naive animals partially recovering CD4+ T cells (60). M. nemestrina macaques may be better able to survive with very low levels of CD4+ T cells than M. mulatta. Indeed, there is precedent for this in the HIV-2-287 model, where rapid irreversible loss of CD4+ T cells by M. nemestrina macaques is uniformly observed, and macaques can survive for long periods following HIV-2-287-induced CD4+-T-cell depletion (68).
Several recent studies have compared viral vectors alone to the same vectors primed with a DNA vaccine. Macaques immunized with rMVA, or primed with DNA and boosted with rMVA, all had good control of a pathogenic SHIV challenge; however, the regimens elicited different responses (3). The combination elicited higher levels of IFN-γ-producing T cells but lower antibody titers relative to rMVA alone. Similarly, an adenovirus-based vaccine alone or as a boost after DNA vaccination protected macaques from the same virus, but the latter regimen elicited a higher cellular response (60). These studies utilized needle injection of DNA. The promise of gene gun delivery to provide equivalent immunity with much lower doses of DNA than needle injection (30) has been called into question by studies that indicate poor protection from virus challenge. No protection was seen in a study of a gene gun-delivered gp120 DNA vaccine with an SHIVsf13 challenge (67) or gene gun delivery of gag, pol, env, and nef and challenge with SHIV-IIIb (55). However, in the SIV model, protection from disease at a level similar to that indicated by the data presented here has been obtained by using the gene gun to protect against SIVdeltaB670 challenge (28). This study is the first example of gene gun-based DNA delivery protecting against disease caused by any SHIV, including the highly virulent SHIV89.6P. Although our DNA vaccine alone was not effective in eliciting strong immunity, the DNA clearly improved the responses elicited by recombinant vaccinia virus. It is possible that the effectiveness of the DNA as a priming or boosting agent was due in part to the ability of one of the plasmids to direct expression of VLP in vivo. Additionally, these gene gun-delivered vaccines primed and boosted cellular immune responses. Some previous studies have found that gene gun delivery elicited a primarily Th2 response (26), although others have shown a balanced Th1/Th2 response (11, 45) and cellular immunity (55).
Boosting with inactivated virus particles was not as effective as we had anticipated, based on the prior experience of our group (73) with autologous macaque dendritic cells for delivery of inactivated SIV, and others which used virus particles as the sole antigen (37) or as a boosting agent for vaccinia virus (70). We used alum plus CpG oligonucleotides as adjuvants in an effort to maintain the overall virion architecture and the Envelope conformation present in the original preparation. It is possible that the low immunogenicity of the virions can be overcome by the use of more effective adjuvants or higher doses of particles.
The vaccines tested here included multiple viral genes: the inactivated virus particles and the recombinant vaccinia viruses provided Gag, Pol, and Env, while the DNA vaccine provided these plus all accessory genes. This design was chosen based on the hypothesis that a multiantigen vaccine would improve the outcome of the challenge, and it differs from the many studies which have used single antigens (7, 20, 24, 60, 64). We measured prechallenge antibody and cellular responses to multiple gene products. Some animals responded to both Gag and Env while others responded only to one or the other. Thus, including more than one antigen successfully increased the total number of animals responding to the vaccine. We were disappointed to find only limited responses to Pol or accessory genes prior to challenge. Sporadic cellular responses to Pol and Env also were noted after challenge; however, these did not correlate with any one vaccine regimen.
The utility of the SHIV89.6P model has recently come into question (25). The very rapid loss of CD4+ T cells is unlike the course of HIV disease progression in humans. Furthermore, while the parental HIV89.6 is dualtropic, SHIV89.6P uses CXCR4 as a coreceptor (71), again differing from HIV infection, in which most transmitted viruses are CCR5 users. However, there are advantages to this virus as well (53). SHIVs are invaluable for testing vaccine strategies aimed at eliciting immune responses to Env, particularly neutralizing antibody, because they contain HIV-1 Env. Furthermore, while many SHIV strains are highly attenuated, the pathogenicity of SHIV89.6P allows assessment of protection from both infection and disease. In addition, a large number of recent vaccine studies have used SHIV89.6P as a challenge virus (1, 2, 10, 19, 36, 40, 49, 56); thus, these studies can all be easily compared to each other, as noted above.
Because low viral load is associated with slow disease progression, control of viral load rather than sterilizing immunity may be considered as an endpoint for vaccine-induced protection (31, 62). Most of the encouraging recent vaccine studies with macaques have used this endpoint (2, 60). Recently, however, vaccinated M. mulatta macaques were shown to advance to disease despite very low virus loads early after challenge with SHIV89.6P (8) or SIV (7), casting doubt on the durability of vaccine-induced virus control. It is thus desirable to design vaccines that are even more effective in limiting virus load for the long term. To most effectively improve upon current approaches, it is valuable to understand what types of immunity each vaccine component provides, alone and in combination with other components. We have found that comparative studies can provide information on which regimen(s) are most effective in preventing disease. In conclusion, we show that multigene vaccines in priming-boosting regimens comprised of gene gun-delivered DNA and recombinant vaccinia virus, in either order, are effective in preventing SHIV89.6P disease. Further modifications to the vaccines, possibly including recombinant protein to boost neutralizing antibody responses, will be needed to provide complete protection from disease or sterilizing immunity.
Acknowledgments
This work was supported by Public Health Service grants P01-AI-23566 (to D.A., P.D.G., N.L.H., and S.-L.H.) and P30-AI27577 (to P.D.G. and N.L.H.) and by grants T32A107509 and T32CA0922925 to N.A.D.-R.
We thank V. Planelles for pEMC*, N. L. Letvin for the SHIV89.6P seed stock, J. Shiver for pV1Jns:89.6Penv160OPTcleavable and pVIR SIV GAG OPT, and the Chiron Corp. for recombinant gp120. We thank P. S. Choi, L. Fink, D. Lauman, S. Reymond, and W. F. Sutton for expert technical assistance and B. Richardson for assistance with statistical analysis.
REFERENCES
- 1.Amara, R. R., J. M. Smith, S. I. Staprans, D. C. Montefiori, F. Villinger, J. D. Altman, S. P. O'Neil, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, J. M. McNicholl, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 76:6138-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, S. I. Staprans, J. D. Altman, D. C. Montefiori, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Different patterns of immune responses but similar control of a simian-human immunodeficiency virus 89.6P mucosal challenge by modified vaccinia virus Ankara (MVA) and DNA/MVA vaccines. J. Virol. 76:7625-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, K. P., C. Lucas, C. V. Hanson, H. F. Londe, A. Izu, T. Gregory, A. Ammann, P. W. Berman, and J. W. Eichberg. 1989. Effect of dose and immunization schedule on immune response of baboons to recombinant glycoprotein 120 of HIV-1. J. Infect. Dis. 160:960-969. [DOI] [PubMed] [Google Scholar]
- 5.Banks, N. D., N. Kinsey, J. Clements, and J. E. Hildreth. 2002. Sustained antibody-dependent cell-mediated cytotoxicity (ADCC) in SIV-infected macaques correlates with delayed progression to AIDS. AIDS Res. Hum. Retrovir. 18:1197-1205. [DOI] [PubMed] [Google Scholar]
- 6.Barouch, D. H., A. Craiu, M. J. Kuroda, J. E. Schmitz, X. X. Zheng, S. Santra, J. D. Frost, G. R. Krivulka, M. A. Lifton, C. L. Crabbs, G. Heidecker, H. C. Perry, M. E. Davies, H. Xie, C. E. Nickerson, T. D. Steenbeke, C. I. Lord, D. C. Montefiori, T. B. Strom, J. W. Shiver, M. G. Lewis, and N. L. Letvin. 2000. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc. Natl. Acad. Sci. USA 97:4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 9.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6p viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 11.Belperron, A. A., D. Feltquate, B. A. Fox, T. Horii, and D. J. Bzik. 1999. Immune responses induced by gene gun or intramuscular injection of DNA vaccines that express immunogenic regions of the serine repeat antigen from Plasmodium falciparum. Infect. Immun. 67:5163-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer, J. D., A. D. Cohen, S. Vogt, K. Schumann, B. Nath, L. Ahn, K. Lacy, M. L. Bagarazzi, T. J. Higgins, Y. Baine, R. B. Ciccarelli, R. S. Ginsberg, R. R. MacGregor, and D. B. Weiner. 2000. Vaccination of seronegative volunteers with a human immunodeficiency virus type 1 env/rev DNA vaccine induces antigen-specific proliferation and lymphocyte production of beta-chemokines. J. Infect. Dis. 181:476-483. [DOI] [PubMed] [Google Scholar]
- 14.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 17.Doria-Rose, N. A., and N. L. Haigwood. DNA vaccine strategies: candidates for immune modulation and immunization regimens. Methods, in press. [DOI] [PubMed]
- 18.Doria-Rose, N. A., C. C. Pierce, S. L. Hu, Y. D. Zhu, W. F. Sutton, M. T. Hensel, L. Kuller, D. Anderson, P. S. Polacino, N. Sheikh, and N. L. Haigwood. 2003. Multi-gene DNA prime-boost vaccines for SHIV89.6P. J. Med. Primatol. 32:218-228. [DOI] [PubMed]
- 19.Earl, P. L., L. S. Wyatt, D. C. Montefiori, M. Bilska, R. Woodward, P. D. Markham, J. D. Malley, T. U. Vogel, T. M. Allen, D. I. Watkins, N. Miller, and B. Moss. 2002. Comparison of vaccine strategies using recombinant env-gag-pol MVA with or without an oligomeric Env protein boost in the SHIV rhesus macaque model. Virology 294:270-281. [DOI] [PubMed] [Google Scholar]
- 20.Egan, M. A., W. A. Charini, M. J. Kuroda, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estcourt, M. J., A. J. Ramsay, A. Brooks, S. A. Thomson, C. J. Medveckzy, and I. A. Ramshaw. 2002. Prime-boost immunization generates a high frequency, high-avidity CD8(+) cytotoxic T lymphocyte population. Int. Immunol. 14:31-37. [DOI] [PubMed] [Google Scholar]
- 22.Etemad-Moghadam, B., D. Rhone, T. Steenbeke, Y. Sun, J. Manola, R. Gelman, J. W. Fanton, P. Racz, K. Tenner-Racz, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 2002. Understanding the basis of CD4(+) T-cell depletion in macaques infected by a simian-human immunodeficiency virus. Vaccine 20:1934-1937. [DOI] [PubMed] [Google Scholar]
- 23.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 24.Fanales-Belasio, E., A. Cafaro, A. Cara, D. R. Negri, V. Fiorelli, S. Butto, S. Moretti, M. T. Maggiorella, S. Baroncelli, Z. Michelini, A. Tripiciano, L. Sernicola, A. Scoglio, A. Borsetti, B. Ridolfi, R. Bona, P. Ten Haaft, I. Macchia, P. Leone, M. R. Pavone-Cossut, F. Nappi, E. Vardas, M. Magnani, E. Laguardia, A. Caputo, F. Titti, and B. Ensoli. 2002. HIV-1 Tat-based vaccines: from basic science to clinical trials. DNA Cell Biol. 21:599-610. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 26.Feltquate, D. M., S. Heaney, R. G. Webster, and H. L. Robinson. 1997. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J. Immunol. 158:2278-2284. [PubMed] [Google Scholar]
- 27.Fuller, D. H., M. M. Corb, S. Barnett, K. Steimer, and J. R. Haynes. 1997. Enhancement of immunodeficiency virus-specific immune responses in DNA-immunized rhesus macaques. Vaccine 15:924-926. [DOI] [PubMed] [Google Scholar]
- 28.Fuller, D. H., P. A. Rajakumar, L. A. Wilson, A. M. Trichel, J. T. Fuller, T. Shipley, M. S. Wu, K. Weis, C. R. Rinaldo, J. R. Haynes, and M. Murphey-Corb. 2002. Induction of mucosal protection against primary, heterologous simian immunodeficiency virus by a DNA vaccine. J. Virol. 76:3309-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuller, D. H., L. Simpson, K. S. Cole, J. E. Clements, D. L. Panicali, R. C. Montelaro, M. Murphey-Corb, and J. R. Haynes. 1997. Gene gun-based nucleic acid immunization alone or in combination with recombinant vaccinia vectors suppresses virus burden in rhesus macaques challenged with a heterologous SIV. Immunol. Cell Biol. 75:389-396. [DOI] [PubMed] [Google Scholar]
- 30.Fynan, E. F., R. G. Webster, D. H. Fuller, J. R. Haynes, J. C. Santoro, and H. L. Robinson. 1993. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. USA 90:11478-11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham, B. S. 2002. Clinical trials of HIV vaccines. Annu. Rev. Med. 53:207-221. [DOI] [PubMed] [Google Scholar]
- 32.Hartmann, G., R. D. Weeratna, Z. K. Ballas, P. Payette, S. Blackwell, I. Suparto, W. L. Rasmussen, M. Waldschmidt, D. Sajuthi, R. H. Purcell, H. L. Davis, and A. M. Krieg. 2000. Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J. Immunol. 164:1617-1624. [DOI] [PubMed] [Google Scholar]
- 33.Hu, S. L., K. Abrams, G. N. Barber, P. Moran, J. M. Zarling, A. J. Langlois, L. Kuller, W. R. Morton, and R. E. Benveniste. 1992. Protection of macaques against SIV infection by subunit vaccines of SIV envelope glycoprotein gp160. Science 255:456-459. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson, G. B., M. Halloran, J. Li, I. W. Park, R. Gomila, K. A. Reimann, M. K. Axthelm, S. A. Iliff, N. L. Letvin, and J. Sodroski. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71:4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kourtis, A. P., C. C. Ibegbu, F. Scinicariello, C. Y. Oh, and H. M. McClure. 2002. SHIV-KB9 infection of rhesus monkeys does not always cause disease-contribution of host immune factors and thymic output. Virology 303:47-57. [DOI] [PubMed] [Google Scholar]
- 36.Letvin, N. L., S. Robinson, D. Rohne, M. K. Axthelm, J. W. Fanton, M. Bilska, T. J. Palker, H. X. Liao, B. F. Haynes, and D. C. Montefiori. 2001. Vaccine-elicited V3 loop-specific antibodies in rhesus monkeys and control of a simian-human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate envelope. J. Virol. 75:4165-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lifson, J. D., M. Piatak, Jr., J. L. Rossio, J. Bess, Jr., E. Chertova, D. Schneider, R. Kiser, V. Coalter, B. Poore, R. Imming, R. C. Desrosiers, L. E. Henderson, and L. O. Arthur. 2002. Whole inactivated SIV virion vaccines with functional envelope glycoproteins: safety, immunogenicity, and activity against intrarectal challenge. J. Med. Primatol. 31:205-216. [DOI] [PubMed] [Google Scholar]
- 38.Lu, S., J. Arthos, D. C. Montefiori, Y. Yasutomi, K. Manson, F. Mustafa, E. Johnson, J. C. Santoro, J. Wissink, J. I. Mullins, J. R. Haynes, N. L. Letvin, M. Wyand, and H. L. Robinson. 1996. Simian immunodeficiency virus DNA vaccine trial in macaques. J. Virol. 70:3978-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacGregor, R. R., J. D. Boyer, K. E. Ugen, K. E. Lacy, S. J. Gluckman, M. L. Bagarazzi, M. A. Chattergoon, Y. Baine, T. J. Higgins, R. B. Ciccarelli, L. R. Coney, R. S. Ginsberg, and D. B. Weiner. 1998. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J. Infect. Dis. 178:92-100. [DOI] [PubMed] [Google Scholar]
- 40.Matano, T., M. Kano, H. Nakamura, A. Takeda, and Y. Nagai. 2001. Rapid appearance of secondary immune responses and protection from acute CD4 depletion after a highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA prime/Sendai virus vector boost regimen. J. Virol. 75:11891-11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 42.Mossman, S. P., C. C. Pierce, M. N. Robertson, A. J. Watson, D. C. Montefiori, M. Rabin, L. Kuller, J. Thompson, J. B. Lynch, W. R. Morton, R. E. Benveniste, R. Munn, S.-L. Hu, P. Greenberg, and N. L. Haigwood. 1999. Immunization against SIVmne in macaques using multigenic DNA vaccines. J. Med. Primatol. 28:206-213. [DOI] [PubMed] [Google Scholar]
- 43.Nathanson, N., V. M. Hirsch, and B. J. Mathieson. 1999. The role of nonhuman primates in the development of an AIDS vaccine. Aids 13(Suppl. A):S113-S120. [PubMed] [Google Scholar]
- 44.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 45.Oliveira, S. C., G. M. Rosinha, C. F. de-Brito, C. T. Fonseca, R. R. Afonso, M. C. Costa, A. M. Goes, E. L. Rech, and V. Azevedo. 1999. Immunological properties of gene vaccines delivered by different routes. Braz. J. Med. Biol. Res. 32:207-214. [DOI] [PubMed] [Google Scholar]
- 46.Polacino, P., V. Stallard, D. C. Montefiori, C. R. Brown, B. A. Richardson, W. R. Morton, R. E. Benveniste, and S. L. Hu. 1999. Protection of macaques against intrarectal infection by a combination immunization regimen with recombinant simian immunodeficiency virus SIVmne gp160 vaccines. J. Virol. 73:3134-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polacino, P. S., V. Stallard, J. E. Klaniecki, S. Pennathur, D. C. Montefiori, A. J. Langlois, B. A. Richardson, W. R. Morton, R. E. Benveniste, and S. L. Hu. 1999. Role of immune responses against the envelope and the core antigens of simian immunodeficiency virus SIVmne in protection against homologous cloned and uncloned virus challenge in macaques. J. Virol. 73:8201-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramshaw, I. A., and A. J. Ramsay. 2000. The prime-boost strategy: exciting prospects for improved vaccination. Immunol. Today 21:163-165. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen, R. A., R. Hofmann-Lehman, D. C. Montefiori, P. L. Li, V. Liska, J. Vlasak, T. W. Baba, J. E. Schmitz, M. J. Kuroda, H. L. Robinson, H. M. McClure, S. Lu, S. L. Hu, T. A. Rizvi, and R. M. Ruprecht. 2002. DNA prime/protein boost vaccine strategy in neonatal macaques against simian human immunodeficiency virus. J. Med. Primatol. 31:40-60. [DOI] [PubMed] [Google Scholar]
- 50.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I. W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reimann, K. A., J. T. Li, G. Voss, C. Lekutis, K. Tenner-Racz, P. Racz, W. Lin, D. C. Montefiori, D. E. Lee-Parritz, Y. Lu, R. G. Collman, J. Sodroski, and N. L. Letvin. 1996. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J. Virol. 70:3198-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimann, K. A., A. Watson, P. J. Dailey, W. Lin, C. I. Lord, T. D. Steenbeke, R. A. Parker, M. K. Axthelm, and G. B. Karlsson. 1999. Viral burden and disease progression in rhesus monkeys infected with chimeric simian-human immunodeficiency viruses. Virology 256:15-21. [DOI] [PubMed] [Google Scholar]
- 53.Robinson, H. L. 2002. New hope for an AIDS vaccine. Nat. Rev. Immunol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 54.Robinson, H. L., S. Lu, D. M. Feltquate, C. T. Torres, J. Richmond, C. M. Boyle, M. J. Morin, J. C. Santoro, R. G. Webster, D. Montefiori, Y. Yasutomi, N. L. Letvin, K. Manson, M. Wyand, and J. R. Haynes. 1996. DNA vaccines. AIDS Res. Hum. Retrovir. 12:455-457. [DOI] [PubMed] [Google Scholar]
- 55.Robinson, H. L., D. C. Montefiori, R. P. Johnson, K. H. Manson, M. L. Kalish, J. D. Lifson, T. A. Rizvi, S. Lu, S. L. Hu, G. P. Mazzara, D. L. Panicali, J. G. Herndon, R. Glickman, M. A. Candido, S. L. Lydy, M. S. Wyand, and H. M. McClure. 1999. Neutralizing antibody-independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat. Med. 5:526-534. [DOI] [PubMed] [Google Scholar]
- 56.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 57.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider, J., S. C. Gilbert, T. J. Blanchard, T. Hanke, K. J. Robson, C. M. Hannan, M. Becker, R. Sinden, G. L. Smith, and A. V. Hill. 1998. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat. Med. 4:397-402. [DOI] [PubMed] [Google Scholar]
- 59.Schneider, J., S. C. Gilbert, C. M. Hannan, P. Degano, E. Prieur, E. G. Sheu, M. Plebanski, and A. V. Hill. 1999. Induction of CD8+ T cells using heterologous prime-boost immunisation strategies. Immunol. Rev. 170:29-38. [DOI] [PubMed] [Google Scholar]
- 60.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 61.Smith, S. M., B. Holland, C. Russo, P. J. Dailey, P. A. Marx, and R. I. Connor. 1999. Retrospective analysis of viral load and SIV antibody responses in rhesus macaques infected with pathogenic SIV: predictive value for disease progression. AIDS Res. Hum. Retrovir. 15:1691-1701. [DOI] [PubMed] [Google Scholar]
- 62.Spearman, P. W. 2003. HIV vaccine research: lessons from the past and promise for the future. Curr. HIV Res. 1:101-120. [DOI] [PubMed] [Google Scholar]
- 63.Spouge, J. L. 1992. Statistical analysis of sparse infection data and its implications for retroviral treatment trials in primates. Proc. Natl. Acad. Sci. USA 89:7581-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stamatatos, L., and D. Davis. 2001. New insights into protective humoral responses and HIV vaccines. AIDS 15(Suppl. 5):S105-S115. [DOI] [PubMed] [Google Scholar]
- 65.Ten Haaft, P., B. Verstrepen, K. Uberla, B. Rosenwirth, and J. Heeney. 1998. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J. Virol. 72:10281-10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.UNAIDS. 2002. AIDS epidemic update: December 2002. Joint United Nations Programme on HIV/AIDS. [Online.] http://www.unaids.org.
- 67.Verschoor, E. J., P. Mooij, H. Oostermeijer, M. van der Kolk, P. ten Haaft, B. Verstrepen, Y. Sun, B. Morein, L. Akerblom, D. H. Fuller, S. W. Barnett, and J. L. Heeney. 1999. Comparison of immunity generated by nucleic acid-, MF59-, and ISCOM-formulated human immunodeficiency virus type 1 vaccines in rhesus macaques: evidence for viral clearance. J. Virol. 73:3292-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson, A., J. McClure, J. Ranchalis, M. Scheibel, A. Schmidt, B. Kennedy, W. R. Morton, N. L. Haigwood, and S. L. Hu. 1997. Early postinfection antiviral treatment reduces viral load and prevents CD4+ cell decline in HIV type 2-infected macaques. AIDS Res. Hum. Retrovir. 13:1375-1381. [DOI] [PubMed] [Google Scholar]
- 69.Watson, A., J. Ranchalis, B. Travis, J. McClure, W. Sutton, P. R. Johnson, S. L. Hu, and N. L. Haigwood. 1997. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J. Virol. 71:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willey, R. L., R. Byrum, M. Piatak, Y. B. Kim, M. W. Cho, J. L. Rossio, Jr., J. Bess, Jr., T. Igarashi, Y. Endo, L. O. Arthur, J. D. Lifson, and M. A. Martin. 2003. Control of viremia and prevention of simian-human immunodeficiency virus-induced disease in rhesus macaques immunized with recombinant vaccinia viruses plus inactivated simian immunodeficiency virus and human immunodeficiency virus type 1 particles. J. Virol. 77:1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Z. Q., T. M. Fu, D. R. Casimiro, M. E. Davies, X. Liang, W. A. Schleif, L. Handt, L. Tussey, M. Chen, A. Tang, K. A. Wilson, W. L. Trigona, D. C. Freed, C. Y. Tan, M. Horton, E. A. Emini, and J. W. Shiver. 2002. Mamu-a*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 76:12845-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, Y., K. Koo, J. D. Bradshaw, W. F. Sutton, L. R. Kuller, R. Bucala, D. Anderson, S. P. Mossman, F. Villinger, and N. L. Haigwood. 2000. Macaque blood-derived antigen-presenting cells elicit SIV-specific immune responses. J. Med. Primatol. 29:182-192. [DOI] [PubMed] [Google Scholar]