Abstract
To elevate the production level of heterologous polyketide in Streptomyces venezuelae, an additional copy of the positive regulatory gene pikD was introduced into the pikromycin (Pik) polyketide synthase (PKS) deletion mutant of S. venezuelae ATCC 15439 expressing tylosin PKS genes. The resulting mutant strain showed enhanced production of both tylactone (TL) and desosaminyl tylactone (DesTL) of 2.7- and 17.1-fold, respectively. The notable increase in DesTL production strongly suggested that PikD upregulates the expression of the desosamine (des) biosynthetic gene cluster. In addition, two hydroxylated forms of DesTL were newly detected from the extract of this mutant. These hydroxylated forms presumably resulted from a PikD-dependent increase in expression of the pikC gene that encodes P450 hydroxylase. Gene expression analysis by reverse transcriptase PCR and bioconversion experiments of 10-deoxymethynolide, narbonolide, and TL into the corresponding desosaminyl macrolides indicated that PikD is a positive regulator of the des and pikC genes, as well as the Pik PKS genes. These results demonstrate the role of PikD as a pathway-specific positive regulator of the entire Pik biosynthetic pathway and its usefulness in the development of a host-vector system for efficient heterologous production of desosaminyl macrolides and novel hydroxylated compounds.
Polyketides are a group of secondary metabolites that exhibit remarkable diversity in their structures and possess a wealth of pharmacologically important activities (14). As the number of cloned and sequenced genes involved in the biosynthesis of useful polyketide natural products has increased, the production of heterologous polyketide by genetically and physiologically well-characterized hosts has received considerable attention due to the difficulties in genetic manipulation of many original producers (21). The transfer of biosynthetic genes to a more amenable and robust heterologous host has, therefore, emerged as a viable alternative to producing high levels of desired compounds or to providing a basis for subsequent combinatorial biosynthetic approaches. Among several well-known actinomycete heterologous hosts, a pikromycin (Pik)/methymycin-producing strain, Streptomyces venezuelae ATCC 15439, has recently been developed as a promising host in that it is amenable to genetic manipulation and requires a short culture period (3 to 4 days) for metabolite production compared to other Streptomyces species (7, 8, 19, 23, 24). In addition, the presence of two post-polyketide synthase (PKS) tailoring enzymes that have unusual substrate flexibility, DesVII glycosyltransferase (2, 3, 6, 25) and PikC P450 monooxygenase (11, 13, 18), offers it another advantage as a heterologous host for combinatorial biosynthesis of novel compounds. However, one of the key challenges in developing S. venezuelae as a competent heterologous host is the relatively low production of heterologous polyketides (8).
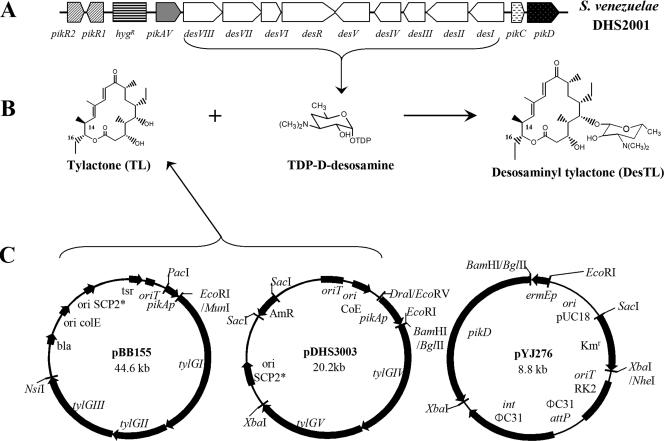
The PikD regulatory factor that originates from S. venezuelae was the sole transcriptional regulator of the Pik biosynthetic cluster (Fig. 1A) and required for the macrolide antibiotic biosynthesis (22). This factor belongs to one of the newly characterized regulatory groups, the large ATP-binding regulators of the LuxR family (LAL), which contains nucleotide triphosphate-binding motifs and a C-terminally located helix-turn-helix motif (22). The other LAL family regulators, such as RapH from the rapamycin biosynthetic pathway of Streptomyces hygroscopicus, have also been reported as positive regulatory elements of their biosynthetic cluster (10). The positive roles of LAL family regulators can be applied to improve the native antibiotic production.
FIG. 1.
Organization of the Pik gene cluster of S. venezuelae mutant DHS2001 in which the hygromycin resistance gene replaced Pik PKS (A), formation of DesTL by attaching TDP-d-desosamine to TL (B), and maps of three plasmids for expressing tylosin PKS (pBB155 and pDHS3003) and the pikD regulatory gene (pYJ276) (C).
In the present study, we exploited the PikD positive regulator for enhancing heterologously produced tylactone (TL) and desosaminyl tylactone (DesTL) in S. venezuelae (Fig. 1). The promotion of TL and DesTL production, as well as the generation of novel hydroxylated DesTL compound, seemed to result from the activation of the entire Pik biosynthetic pathway by PikD, which was verified by gene expression analysis and bioconversion experiments.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and genetic manipulation.
S. venezuelae DHS2001, in which the pikAI-VI genes were deleted (8), was propagated on SPA medium (1 g of yeast extract, 1 g of beef extract, 2 g of tryptose, 10 g of glucose, trace amount of ferrous sulfate, and 15 g of agar/liter). Transformants of S. venezuelae were selected on R2YE agar plates (9). Genetic manipulations were carried out in Escherichia coli DH10B by using standard protocols (16). Transformation of DNA into protoplasts of S. venezuelae was performed according to standard protocols (9). Southern blot hybridization was performed according to the protocol recommended by a digoxigenin labeling and detection kit (Roche).
Construction of pikD expression vector pYJ276 and S. venezuelae mutant strains YJ112 and YJ113.
A derivative of pSET152 (1), in which the apramycin resistance gene [aac(3)IV] was replaced by the kanamycin resistance gene (aphII), was constructed. The aphII gene (20) was amplified from plasmid pYJ100 (6) by PCR using the following primers: 5′-AAAGAGCTCCCCGAACCCCAGAGTCCC-3′ and 5′-AAATCTAGACCTGATACCGCTCGCCGC-3′ (restriction sites are underlined). The PCR product of aphII was cut by SacI/XbaI and ligated to SacI/NheI-digested pSET152, generating pYJ205. A 2.9-kb DNA fragment of pikD was amplified from the genomic DNA of S. venezuelae by PCR using a forward primer, 5′-ACGACTAGATCTCACGGAAGCCCCGGATCG-3′, and a reverse primer, CGGGGACACTCTAGATGGCTCAGGCCGTGA (the restriction sites are underlined). The pikD PCR product was cloned into pGEM-T Easy vector (Promega) and sequenced. This plasmid was digested with BglII/XbaI, and the resulting pikD-containing fragment was placed into BamHI/XbaI sites of pYJ205, yielding pYJ276. Plasmid pYJ276 was integrated into the chromosomal DNA of S. venezuelae mutant DHS2001 (8), thus generating mutant strain YJ112. The two previously constructed replicating plasmids, pBB155 containing tylGI-III and pDHS3003 containing tylGIV-V (8), were subsequently transformed into the pikD-integrated mutant YJ112, generating YJ113.
Production and analysis of heterologous polyketides.
All mutants were cultured and extracted as previously reported (8). The extracts were analyzed by the combination of liquid chromatography-electrospray ionization mass spectrometry (LC/ESI-MS) and electrospray ionization mass spectrometry-mass spectrometry (ESI-MS/MS) using a Waters/Micromass Quattro micro/MS apparatus. Separation was performed on a Nova-Pak C18 (150 by 3.9 mm, 4.0 μm, Waters) reversed-phase column. The analytes were eluted at a flow rate of 180 μl/min with a gradient of 5 mM (wt/vol) ammonium acetate-0.05% acetic acid (vol/vol) in water (solution A) and 80% (vol/vol) acetonitrile with the same additive concentrations (solution B) at 20 to 70% solution B for 25 min, to 90% solution B for 20 min, maintained at 90% solution B for 10 min, and then to 20% solution B for another 10 min for column re-equilibration. Authentic TL and 5-O-mycaminosyl tylonolide standards (provided by Eric Cundliffe at University of Leicester) were used to generate calibration curves for the determination of TL and DesTL titer, respectively. The production levels of TL and DesTL were determined in triplicates.
Extraction of total mRNA from S. venezuelae grown on an R2YE plate.
At day 2 and day 3, total RNA was isolated from S. venezuelae mutants grown on R2YE plates. In order to prevent the degradation of mRNA, 2 volumes of RNA protect reagent (Qiagen) were mixed with 1 volume of phosphate-buffered saline (8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, 0.24 g of KH2PO4, and 800 ml of distilled H2O; pH adjusted to 7.4 with HCl). The resulting solution was added to the plate, and the bacterial mycelia were obtained by using a sterile spreader. The cell pellets were harvested by centrifugation (5,000 × g for 5 min at 4°C) and were then suspended in 1 ml of TRIzol reagent (Invitrogen), followed by incubation at room temperature for 5 min. The mixture was added to 200 μl of chloroform, vigorously vortex mixed for 15 s, and centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was transferred into a new tube and mixed with 1 volume of 70% ethyl alcohol. The mixture was moved to an RNeasy Mini spin column (Qiagen) and centrifuged at 8,000 × g for 15 s at room temperature. The spin column was washed two times with RPE washing buffer (Qiagen), and the resulting total RNAs were dissolved in 50 μl of RNase-free water. Nucleic acid preparations were treated with DNase I (New England Biolabs) as recommended by the manufacturer.
Detection of mRNA transcripts by RT-PCR.
Analysis of semiquantitative reverse transcriptase PCR (RT-PCR) was performed by using a One-Step RT-PCR kit (Qiagen) as recommended by the manufacturer. Dimethyl sulfoxide (5% [vol/vol]) was added to all PCRs together with RNAguard RNase inhibitor (Amersham Pharmacia). A total of 5 ng of DNase I-treated total RNA was used as a template for reverse transcription and PCR amplification under the following conditions: 50°C for 30 min, followed by 95°C for 15 min and then 32 to 35 cycles of 94°C for 1 min (tylGI, tylGV, pikC, and pikD, 32 cycles; desVI and desIV, 35 cycles), 55°C for 1 min, and 72°C for 45 s. Negative controls were carried out with each experimental reaction using Taq DNA polymerase (New England Biolabs) to confirm that amplified products were not derived from the chromosomal DNA.
Primer design for RT-PCR analysis.
Primers (20-mers) were designed to possess a similar Tm value at 60°C and to generate PCR products of approximately 500 bp. Five primer pairs specific for Tyl PKS genes (tylGI, tylGII, tylGIII, tylGIV, and tylGV) and four primer pairs specific for des genes (desVII, desVI, desIV, and desII) were designed to detect the transcripts of Tyl PKS and the des cluster, respectively. Among these primer pairs, one pair was selected per expected mRNA transcript based on the visibility of the RT-PCR band. Consequently, two primer pairs for Tyl PKS genes (tylGI and tylGIII) and two primer pairs for the des cluster (desVI and desIV) were selected. The specific primer pairs for the pikC and pikD genes were also designed. The oligonucleotide primers and the target genes are summarized in Table 1.
TABLE 1.
Primers for RT-PCR
| Primer | Sequence (5′-3′) | Description |
|---|---|---|
| TylGIF | TTCGTCCTCTTCTCCTCCGT | Forward primer for tylGI |
| TylGIR | TTCGGATGCCGCAGCACAAA | Reverse primer for tylGI |
| TylGVF | ATCTGCTCTTCGACCGGGAC | Forward primer for tylGV |
| TylGVR | ACTCGTTCTCCCGGCAGAAA | Reverse primer for tylGV |
| DesVIF | TTCACCAAGGAGTTCGGCGA | Forward primer for desVI |
| DesVIR | AGATGGACGTCGGAGAAGTG | Reverse primer for desVI |
| DesIVF | TTGTTGCAGCAGCGGGTGAT | Forward primer for desIV |
| DesIVR | TGAGGTGATCGTCCTGGACA | Reverse primer for desIV |
| PikCF | CGCTCAACCACAACATGCTG | Forward primer for pikC |
| PikCR | TGGCGATCAGATTGACCGTG | Reverse primer for pikC |
| PikDF | CGCGATGAATCTGGTGGAAC | Forward primer for pikD |
| PikDR | GCAGTACAGGAGGAACCTCA | Reverse primer for pikD |
Semiquantification of RT-PCR products.
Image analysis of agarose gel electrophoresis was carried out to achieve the relative quantification of RT-PCR products by using Quantity One software (version 4.1.1). After RT-PCR amplification was complete, 20-μl aliquots of the reaction product (total volume of 50 μl) were electrophoresed onto a 1% agarose gel containing 0.5 μg of ethidium bromide/ml and photographed under UV light by using Gel Doc 2000 (Bio-Rad). The relative amounts of amplified products were determined by calculating the average signal intensity of each DNA band in relation to that of a known quantity of the standard. All data were calculated from at least three replicates.
Bioconversion experiments of polyketide aglycones using S. venezuelae mutant strains DHS2001 and YJ112.
The native aglycones produced from S. venezuelae, 10-deoxymethynolide (10-DML) and narbonlide (NL), were obtained from the des deletion mutant YJ003 (6) by the previously described purification steps (12). These macrolactones (10-DML, 0.9 mg; NL, 1.5 mg; TL, 0.06 mg) suspended in ethanol were added to 1 ml of double-distilled water. The suitable feeding concentrations of the exogenous macrolactones for bioconversion experiments, which included 10-DML, NL, and TL, were determined on the basis of the production yields of wild-type S. venezuelae (7) and the yield of TL from the YJ005 mutant strain (8). After the mutant strains, DHS2001 and YJ112, were cultured on R2YE solid medium at 30°C for 2 days, 1 ml of each solution was spread onto the solid culture medium, followed by incubation for four additional days. The resulting agar-grown culture was extracted as described above. The calibration curves using the purified 10-DML, NL, and TL were used for determining the conversion yields of each aglycone.
RESULTS AND DISCUSSION
Construction of the S. venezuelae mutant strain YJ113 expressing pikD and Tyl PKS genes.
We recently reported that the expression of Tyl PKS using two SCP2*-type replicating plasmids, pBB155 and pDHS3003 (Fig. 1C), under the control of pikAI promoter in the Pik PKS deletion mutant DHS2001 led to the successful production of heterologous polyketides, TL and DesTL (8). To improve the production level of these compounds, we introduced an additional copy(ies) of pikD regulatory gene into the YJ005 mutant strain (expressing Tyl PKS in DHS2001), in which the native pikD gene remained intact. The ΦC31-based integrating plasmid pYJ276 (Fig. 1C) containing pikD regulatory gene under the control of a strong constitutive ermEp1 ΔTGG (ermE*) promoter (17), which carries a 3-bp deletion (TGG) in the −35 region of ermEp1 promoter of Saccharopolyspora erythraea, was transformed into the DHS2001 mutant strain, generating YJ112. The integration of an extra copy(ies) of pikD into the chromosome of DHS2001 was confirmed by Southern hybridization. Chromosomal DNA of YJ112 was isolated and digested with EcoRI/XbaI. When the digested genomic DNA was probed with the digoxigenin-labeled 280-bp EcoRI-BamHI fragment of the ermE* promoter region, a 3.1-kb DNA fragment was observed, whereas no DNA band was detected from the genomic DNA of DHS2001 (data not shown). The ΦC31 integrase of pYJ276 often yields cointegrates of several copies of the introduced gene, and these results showed that at least one extra copy of pikD was integrated into the chromosomal DNA of the YJ112 strain. A mutant strain YJ113 was constructed by introduction of the two plasmids pBB155 and pDHS3003 into the pikD-integrated mutant YJ112.
Production analysis of S. venezuelae YJ005 (expressing Tyl PKS) and YJ113 (expressing Tyl PKS and PikD).
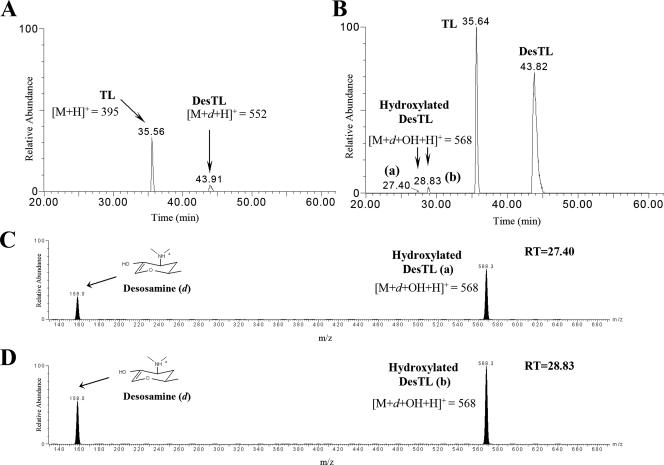
To compare the production of heterologous polyketides, TL and DesTL, between YJ005 and YJ113, both mutant strains were cultured on R2YE plates, extracted, and then analyzed by LC/ESI-MS and ESI-MS/MS. The peaks with the calculated molecular mass for TL (m/z = 395) and DesTL (m/z = 552) were observed at retention times of 35.6 and 43.9 min (Fig. 2A), respectively, from the LC/MS analysis of the YJ005 extract. MS/MS fragmentations of these compounds produced patterns identical to those of TL and DesTL, respectively, as reported previously (8). The titers of TL and DesTL from YJ005 were approximately 0.5 and 0.1 mg/liter, respectively. Approximately 1.4 mg of TL/liter and 1.8 mg of DesTL/liter were produced from the pikD-integrated mutant YJ113 (Fig. 2B), showing the 2.7- and 17.1-fold enhanced production of TL and DesTL, respectively, compared to those produced from YJ005. These results strongly indicated that the overproduction of PikD regulator resulted in upregulated expression of Tyl PKS and the des cluster by binding to pikAI promoters on two plasmids used to express Tyl PKS genes (pBB155 and pDHS3003), as well as promoters acting on the des cluster in the chromosomal DNA of S. venezuelae.
FIG. 2.
LC/ESI-MS/MS chromatograms of TL and DesTL produced by YJ005 (A) and YJ113 (B). The ESI-MS/MS analysis of a hydroxylated DesTL (a) from the peak at 27.4 min (C) and the other hydroxylated DesTL (b) from the peak at 28.8 min (D).
Two small peaks with m/z = 568 were also detected from the extract of YJ113 at retention times of 27.4 and 28.8 min (Fig. 2B), which might correspond to the hydroxylated forms of DesTL. The structures of those two compounds were predicted by ESI-MS/MS analysis. The two compounds with identical m/z = 568 values were fragmented into peaks at m/z = 158 and 568, which corresponded to the oxonium ion fragment from desosamine (4) and the hydrogen adduct of hydroxylated DesTL, respectively (Fig. 2C and D). Two newly detected hydroxylated forms of DesTL were presumably synthesized by the action of PikC P450 hydroxylase. Therefore, the generation of hydroxylated DesTL in the pikD overexpression mutant YJ113 suggested that the overproduced PikD also activated the pikC gene expression. The two novel hydroxylated compounds seem to be synthesized by the introduction of a hydroxyl group into position C-14 or C-16 of DesTL, respectively (Fig. 1B), according to the intrinsic catalytic specificity of PikC. However, the exact hydroxylation positions could not be characterized due to their low concentrations. Further scale-up separation and structural analyses, such as nuclear magnetic resonance spectroscopy, are needed to confirm the hydroxylated site of DesTL.
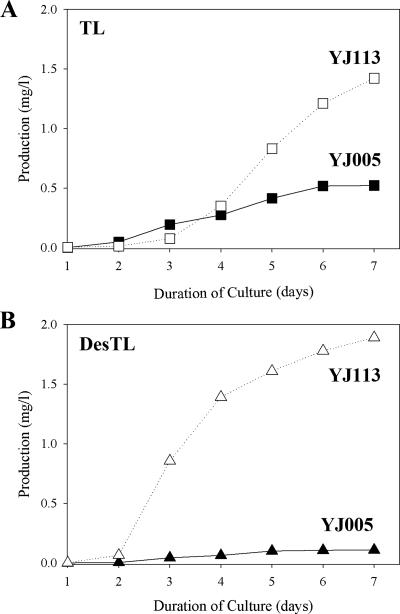
Monitoring the production of TL and DesTL by YJ005 or YJ113 strains.
The production of TL and DesTL in the YJ005 or YJ113 strain was monitored throughout a 7-day incubation period (Fig. 3). The TL production level of the mutant YJ113 showed a notable increase between 3 and 4 days compared to that of YJ005 (Fig. 3A). In the case of DesTL, the YJ005 strain produced only a small amount of DesTL (0.1 mg/liter) until day 7, while a significantly enhanced production of DesTL was observed in the mutant YJ113 between 2 and 3 days and was maintained up through day 7 (Fig. 3B). The total sum of the 16-membered ring heterologous polyketides (TL plus DesTL) produced by the YJ113 strain was greatly increased between day 2 and day 3, and a wider gap was exhibited between YJ005 and YJ113 after 3 days (Fig. 3).
FIG. 3.
Time courses of TL (A) and DesTL (B) production by the YJ005 and YJ113 strains. Incubation was carried out on R2YE solid medium for 7 days.
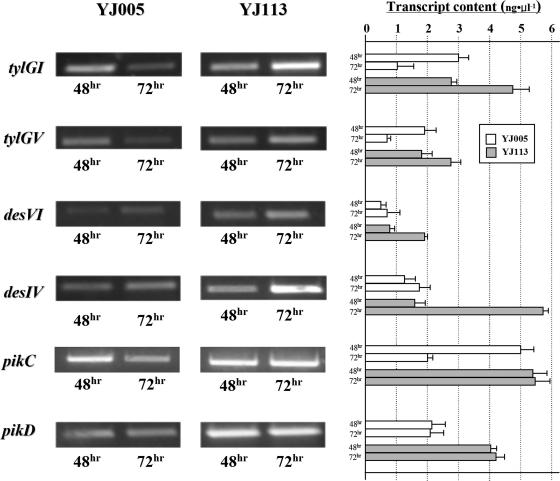
Gene expression analysis in S. venezuelae YJ005 and YJ113 strains.
A previous genetic analysis of PikD using the xylE reporter system showed that the PikD regulator activates the pikAI promoter, whereas it directly or indirectly represses desI promoter, and does not participate in pikC regulation in wild-type S. venezuelae (23). Our observations that the overexpression of the pikD regulatory gene triggered a remarkable enhancement of DesTL production (17.1-fold), as well as the generation of novel hydroxylated DesTL compounds, were inconsistent with the previous report. To determine the effect of PikD on the expressions of Tyl PKS genes, which are under the control of the pikAI promoter, and especially on the des cluster and pikC, transcriptional analysis was performed by using semiquantitative RT-PCR. The total RNA was isolated from both YJ005 and YJ113 mutant strains after 48 or 72 h growth (when the production of DesTL notably increased) to compare the expression levels of the Tyl PKS genes, the des gene cluster, and pikC between the two strains. After an agarose gel electrophoresis, the band intensities of the PCR products of YJ005 were compared to those of YJ113 (Fig. 4). In RT-PCR analysis of the expressions of the Tyl PKS genes, there was an observable decrease in the expression levels of Tyl PKS genes (tylGI, 0.3-fold; tylGV, 0.4-fold) in the YJ005 strain at 72 h compared to 48 h, whereas the expression levels of those genes in the YJ113 were increased (tylGI, 1.7-fold; tylGV, 1.5-fold) during the same culture period. These results showed the positive role of PikD on the pikAI promoter of the two Tyl PKS expression vectors (pBB155 and pDHS3003). In the case of the des cluster, slightly enhanced amounts of desVI (1.4-fold) and desIV (1.3-fold) transcripts were observed at 72 h compared to 48 h in the YJ005 strain, whereas the expression levels of those genes in YJ113 were remarkably increased (desVI, 2.7-fold; desIV, 3.6-fold) between 48 and 72 h. These results showed that the desIV gene selected as a representative of the desI-desV gene set was more highly expressed than the desVI gene chosen among the oppositely directed des gene set (desVIII-desR) in both the YJ005 strain and the YJ113 strain (Fig. 1, Fig. 4). This result also indicated that two apparent operons of the des gene cluster that are transcribed in the convergent direction are separately controlled by a PikD regulator. In addition, the remarkably enhanced transcriptions of both desVI and desIV at 72 h were observed compared to those detected at 48 h in the YJ113 strain (Fig. 4), which showed that both parts of the des cluster (desI-desV and desVIII-desR) were positively affected by the overexpression of pikD.
FIG. 4.
Gene expression analyses by RT-PCR applied to the messenger RNAs isolated from S. venezuelae YJ005 and YJ113 strains at two different time intervals. The expression levels of those genes were analyzed by semiquantitative RT-PCR experiments.
The transcriptional level of the pikC P450 hydroxylase gene in the YJ005 strain was decreased at 72 h compared to 48 h (0.4-fold), whereas the amount of pikC transcript in YJ113 at 48 h was similar to that in YJ005 at 48 h and was maintained for up to 72 h. Therefore, after 72 h of cultivation, the pikC gene of the YJ113 strain was expressed 2.7-fold more than that of YJ005, suggesting the overexpression of pikD stimulates pikC expression. The pikD regulatory gene was constantly expressed in both the YJ005 strain and the YJ113 strain during the culture time from 48 to 72 h. However, the amount of pikD gene transcript in YJ113 was 2.0-fold higher than that in YJ005.
Bioconversion of polyketide aglycones using S. venezuleae DHS2001 (pikA-deleted mutant) and YJ112 (pikD-overexpressed DHS2001).
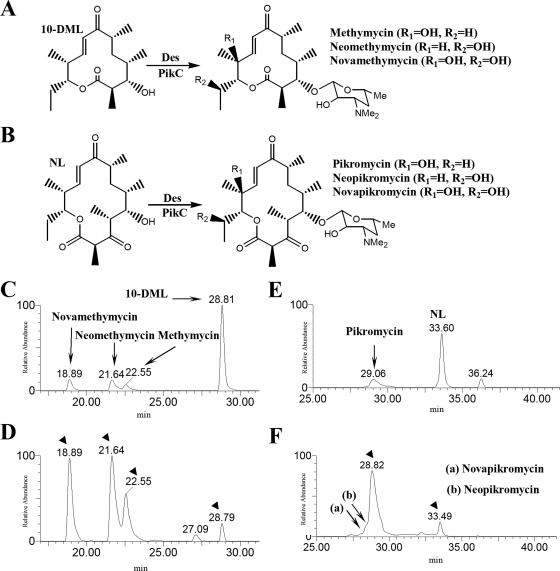
In order to investigate the desosaminylation and hydroxylation activity in the pikD overexpression mutant, aglycone bioconversion experiments were conducted using DHS2001 and YJ112. At first, the purified TL (0.06 mg) was fed onto 30 ml of solid medium containing DHS2001 and YJ112. Approximately 0.1 and 1.7 mg of DesTL/liter was biosynthesized by the DHS2001 and YJ112 strains, respectively (data not shown), and their amounts were similar to those of YJ005 (0.1 mg/liter) and YJ113 (1.8 mg/liter), respectively (Fig. 3). In the extract of YJ112 fed with TL, two novel hydroxylated forms of DesTL were also found, as was the case for the YJ113 strain. When the larger amount of TL (>0.06 mg) was used for the bioconversion experiment, an extra amount of TL remained unconverted (data not shown). In addition, purified 10-DML (0.9 mg) and NL (1.5 mg) were supplemented onto the 30 ml of growing medium containing DHS2001 and YJ112 (Fig. 5A and B). Both DHS2001 and YJ112 converted 10-DML to 12-membered ring macrolides including methymycin, neomethymycin, and novamethycin (26). The conversion yield of 10-DML to the total 12-membered ring macrolides represented ca. 91% in YJ112, showing a 5.1-fold improvement (17%) over that in DHS2001 (Fig. 5C and D). Approximately 14% of the NL was successfully bioconverted to the Pik, together with a small amount of neopikromycin and novapikromycin (12) in DHS2001 fed with NL (Fig. 5E), whereas a 91% conversion of NL to the 14-membered ring macrolides, including Pik, neopikromycin, and novapikromycin, was observed in YJ112 (Fig. 5F). Overexpression of pikD in the DHS2001 strain resulted in 6.3-fold-enhanced bioconversion of NL to 14-membered ring macrolides. These results also supported our hypothesis that PikD activates the expression of the des cluster and pikC gene.
FIG. 5.
The conversion schemes of 10-DML to 12-membered ring series (methymycin, neomethymycin, and novamethymycin) (A) and NL to 14-membered ring series (Pik, neopikromycin, and novapikromycin) (B). LC/ESI-MS chromatograms of 12-membered ring macrolides (C) and 14-membered ring macrolides (D) generated from DHS2001 fed with 10-DML and NL, respectively, and 12-membered ring macrolides (E) and 14-membered ring macrolides (F) generated from YJ113 fed with 10-DML and NL, respectively.
As mentioned above, these results are not consistent with the previous analysis using the xylE reporter system, which determined that the PikD regulatory element downregulates the des cluster and does not take part in regulation of the pikC expression (22). However, in the same report, the results obtained using the intermediate feeding experiment to wild-type S. venezuelae and pikD deletion mutant showed that the desosaminylation activity of 10-DML and NL disappeared in the pikD deletion mutant, which supports our hypothesis. In addition, although the xylE assay used in the previous report has been widely used as a reporter system, it has been suggested to use caution in interpreting data derived from xylE reporter system under aerobic conditions (5). The exact reason for these discrepancies has yet to be further revealed; however, our results using RT-PCR and bioconversion experiment would provide more direct evidence for the positive role of PikD regulator on the des cluster and pikC hydroxylase gene.
The present study shows that improved production of heterologous polyketides can be achieved by the overexpression of a pikD regulatory gene in S. venezuelae. The positive role of PikD toward the des cluster, as well as pikC, can be also harnessed for the combinatorial biosynthesis of a variety of desosaminyl macrolides and their hydroxylated forms. Our efforts are currently being focused on enhancing the production levels of heterologous polyketides by genetic engineering, as well as a metabolic strategy including direct supplementation with limiting coenzyme A ester (15), which will facilitate the development of S. venezuelae as an efficient heterologous host and combinatorial biosynthetic system.
Acknowledgments
We are grateful to Eric Cundliffe (University of Leicester) for kindly providing TL and 5-O-mycaminosyl tylonolide.
This study was supported by grant 20050401-034-682-006-02-00 from the BioGreen 21 Program, grants from the National R&D Program for Cancer Control (0620300-1), the Korea Science and Engineering Foundation grant funded by the Korea government (MOST, M10749000201-07N4900-20110), and the Marine and Extreme Genome Research Center Program, Ministry of Maritime Affairs and Fisheries, Republic of Korea. B.C.P. is grateful for the MOCIE grants from the Intelligence Bioinformatics and Application Center (TGW10011459), KRIBB. We are grateful to the Ministry of Education for the Brain Korea 21 fellowship.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Borisova, S. A., C. Zhang, H. Takahashi, H. Zhang, A. W. Wong, J. S. Thorson, and H. W. Liu. 2006. Substrate specificity of the macrolide-glycosylating enzyme pair DesVII/DesVIII: opportunity, limitations, and mechanistic hypotheses. Angew. Chem. Int. Ed. 45:2748-2753. [DOI] [PubMed] [Google Scholar]
- 3.Borisova, S. A., L. Zhao, C. E. Melançon III, C. L. Kao, and H. W. Liu. 2004. Characterization of the glycosyltransferase activity of DesVII: analysis and implication for the biosynthesis of macrolide antibiotics. J. Am. Chem. Soc. 126:6534-6535. [DOI] [PubMed] [Google Scholar]
- 4.Butler, A. R., N. Bate, D. E. Kiehl, H. A. Kirst, and E. Cundliffe. 2002. Genetic engineering of aminodeoxyhexose biosynthesis in Streptomyces fradiae. Nat. Biotechnol. 20:713-716. [DOI] [PubMed] [Google Scholar]
- 5.Hassett, D. J., U. A. Ochsner, S. L. Groce, K. Parvatiyar, J. F. Ma, and J. D. Lipscomb. 2000. Hydrogen peroxide sensitivity of catechol-2,3-dioxygenase: a cautionary note on use of xylE reporter fusions under aerobic conditions. Appl. Environ. Microbiol. 66:4119-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong, J. S. J., S. H. Park, C. Y. Choi, J. K. Sohng, and Y. J. Yoon. 2004. New olivosyl derivatives of methymycin/pikromycin from an engineered strain of Streptomyces venezuelae. FEMS Microbiol. Lett. 238:391-399. [DOI] [PubMed] [Google Scholar]
- 7.Jung, W. S., A. R. Han, J. S. J. Hong, S. R. Park, C. Y. Choi, J. W. Park, and Y. J. Yoon. 2007. Bioconversion of 12-, 14-, and 16-membered ring aglycones to glycosylated macrolides in an engineered strain of Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 76:1373-1381. [DOI] [PubMed] [Google Scholar]
- 8.Jung, W. S., S. K. Lee, J. S. J. Hong, S. R. Park, S. J. Jeong, A. R. Han, J. K. Sohng, B. G. Kim, C. Y. Choi, D. H. Sherman, and Y. J. Yoon. 2006. Heterologous expression of tylosin polyketide synthase and production of a hybrid macrolide in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 72:763-769. [DOI] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hoopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 10.Kuscer, E., N. Coates, I. Challis, M. Gregory, B. Wilkinson, R. Sheridan, and H. Petković. 2007. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 189:4756-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. K., D. B. Basnet, J. S. J. Hong, W. S. Jung, C. Y. Choi, H. C. Lee, J. K. Sohng, K. G. Ryu, D. J. Kim, J. S. Ahn, B. S. Kim, H. C. Oh, D. H. Sherman, and Y. J. Yoon. 2005. Structural diversification of macrolactones by substrate-flexible cytochrome P450 monooxygenase. Adv. Synth. Catal. 347:1369-1378. [Google Scholar]
- 12.Lee, S. K., J. W. Park, J. W. Kim, W. S. Jung, S. R. Park, C. Y. Choi, E. S. Kim, B. S. Kim, J. S. Ahn, D. H. Sherman, and Y. J. Yoon. 2006. Neopikromycin and novapikromycin from the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Nat. Prod. 69:847-849. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. K., J. W. Park, S. R. Park, J. S. Ahn, C. Y. Choi, and Y. J. Yoon. 2006. Hydroxylation of indole by PikC cytochrome P450 from Streptomyces venezuelae and engineering its catalytic activity by site-directed mutagenesis. J. Microbiol. Biotechnol. 16:974-978. [Google Scholar]
- 14.O'Hagan, D. 1991. The polyketide metabolites. Ellis Horwood, Chichester, United Kingdom.
- 15.Park, J. W., W. S. Jung, S. R. Park, B. C. Park, and Y. J. Yoon. 2007. Analysis of intracellular short organic acid-coenzyme A esters from actinomycetes using liquid chromatography-electrospray ionization-mass spectrometry. J. Mass Spectrom. 42:1136-1147. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Schmitt-John, T., and J. W. Engels. 1992. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 36:493-498. [DOI] [PubMed] [Google Scholar]
- 18.Sherman, D. H., S. Li, L. V. Yermalitskaya, Y. Kim, J. A. Smith, M. R. Waterman, and L. M. Podust. 2006. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J. Biol. Chem. 281:26289-26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thapa, L. P., T. J. Oh, H. C. Lee, K. Liou, J. W. Park, Y. J. Yoon, and J. K. Sohng. 2007. Heterologous expression of the kanamycin biosynthetic gene cluster (pSKC2) in Streptomyces venezuelae YJ003. Appl. Microbiol. Biotechnol. 76:1357-1364. [DOI] [PubMed] [Google Scholar]
- 20.Ward, J. M., G. R. Janssen, T. Kieser, M. J. Bibb, and M. J. Buttner. 1986. Construction and characterization of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 21.Wenzel, S. C., and R. Müller. 2005. Recent developments toward the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 16:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, D. J., Y. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue, Y., and D. H. Sherman. 2001. Biosynthesis and combinatorial biosynthesis of pikromycin-related macrolides in Streptomyces venezuelae. Metab. Eng. 3:15-26. [DOI] [PubMed] [Google Scholar]
- 24.Xue, Y., L. Zao, H. Y. Liu, and D. H. Sherman. 1999. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon, Y. J., J. B. Beck, B. S. Kim, H. Y. Kang, K. A. Reynolds, and D. H. Sherman. 2002. Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem. Biol. 9:203-214. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Q., and D. H. Sherman. 2001. Isolation and structure determination of novamethymycin, a new bioactive metabolite of the methymycin biosynthetic pathway in Streptomyces venezuelae. J. Nat. Prod. 64:1447-1450. [DOI] [PubMed] [Google Scholar]