Abstract
The identity and ecophysiology of a group of uncultured protein-hydrolyzing epiphytic rods attached to filamentous bacteria in activated sludge from nutrient removal plants were investigated by using the full-cycle rRNA approach combined with microautoradiography and histochemical staining. The epiphytic group consists of three closely related clusters, each containing 11 to 16 clones. The closest related cultured isolate is the type strain Haliscomenobacter hydrossis (ATCC 27775) (<87% similarity) in the family Saprospiraceae of the phylum Bacteroidetes. Oligonucleotide probes at different hierarchical levels were designed for each cluster and used for ecophysiological studies. All three clusters behaved similarly in their physiology and were specialized in protein hydrolysis and used amino acids as energy and carbon sources. They were not involved in denitrification. No storage of polyphosphate and polyhydroxyalkanoates was found. They all colonized probe-defined filamentous bacteria belonging to the phyla Chloroflexi, Proteobacteria, and candidate phylum TM7, with the exception of cluster 1, which did not colonize TM7 filaments. The three epiphytic clusters were all widespread in domestic and industrial wastewater treatment plants with or without biological phosphorus removal, constituting, in total, up to 9% of the bacterial biovolume. A new genus, “Candidatus Epiflobacter,” is proposed for this epiphytic group in activated-sludge treatment plants, where it presumably plays an important role in protein degradation.
Microbial growth on biotic surfaces is widespread in the biosphere (33). Examples are growth on human tissues in relation to diseases (12) and growth on plant surfaces such as the rhizosphere, where interactions between the microorganism and plants are well described (34). However, certain bacteria also attach themselves to other microorganisms. The sheaths of large filamentous bacteria (e.g., Beggiatoa and Thioploca) are often colonized by other bacteria such as sulfate-reducing bacteria or bacteria belonging to the phylum Bacteroidetes, where it is believed that they may use sheath material for growth (16). In activated-sludge wastewater treatment plants (WWTPs), the growth of small, rod-formed microorganisms attached to different filamentous bacteria was noticed more than 30 years ago (9). Some of these filamentous morphotypes with attached growth (epiflora) (Eikelboom types 0041, 1701, and 1851) can cause bulking problems in activated sludge (40), and the presence of epiflora has been used as an important criterion for a morphological classification of these unwanted filamentous bacteria (8, 9).
Limited information exists about the identity and ecophysiology of microorganisms colonizing filamentous bacteria in activated sludge (43). Recently, we found (45) that most epiflora bacteria could hybridize with oligonucleotide probe Sap309, which is designed to target most of the members of the family Saprospiraceae in the phylum Bacteroidetes (38). Sap309 hybridized not only with the epiflora bacteria but also with some filamentous bacteria, indicating the existence of a diverse phylogeny (45). In an attempt to identify undescribed dominant microorganisms in WWTPs, we designed a series of oligonucleotide probes for clones retrieved from an enhanced biological phosphorus removal (EBPR) WWTP (Skagen) (17). Of these, probe Bac111, targeting clones (approximately 50 clones of >1,200 bp) distantly related to Haliscomenobacter hydrossis (ATCC 27775), also hybridized with the epiflora bacteria, confirming the affiliation to the family Saprospiraceae. However, although it is more specific than Sap309, probe Bac111 also hybridized with some filaments and cocci, so the detailed phylogeny of the epiflora bacteria is still unresolved.
The family Saprospiraceae consists of the genera Aureispira, Haliscomenobacter, Lewinella, and Saprospira and is represented by five type strains and approximately 160 16S rRNA gene sequences (>1,200 bp as of June 2007), and they are found in various habitats. Large, filamentous microorganisms hybridizing with probe Sap309 are widely distributed in freshwater lakes (38) and are believed to play an important role there. Molecular evidence demonstrates that members of the family Saprospiraceae are also present in hypersaline mats (26), and three strains of gliding bacteria belonging to the genus Saprospira have been isolated from marine sponges and algae from the southern coastline of Thailand (13). H. hydrossis was isolated from activated sludge (44), and 16S rRNA gene analysis indicated that Haliscomenobacter-related bacteria were also present in a methanol-fed denitrifying sequencing batch reactor (11). However, to our knowledge, the presence of epiphytic Saprospiraceae has not been reported in habitats other than activated sludge.
Members of Bacteroidetes are generally associated with the degradation of complex organic materials (6, 35, 36), but except for H. hydrossis, little is known about the detailed ecophysiology of other Saprospiraceae. Isolates of H. hydrossis can hydrolyze starch and grow aerobically on glucose, N-acetylglucosamine, lactose, and sucrose but not glycerol, lactate, acetate, and succinate (44). Therefore, in activated sludge, H. hydrossis may be involved in the hydrolysis of polysaccharides and utilize the hydrolysates as energy and carbon sources for growth. Interestingly, in our recent efforts to identify microorganisms involved in protein hydrolysis in activated sludge, the epiflora bacteria hybridizing with the probe Sap309 or Bac111 showed up as a candidate. They were abundant in different activated-sludge WWTPs, accounting for up to 8 to 12% of the biomass (45). Thomsen et al. (42) carried out a detailed study of the ecophysiology of filamentous bacterial type 0041 and some of the epiphytic bacteria. However, as they were unable to identify most of the epiflora bacteria with any probes, the physiology of epiflora bacteria belonging to Saprospiraceae is uncertain. In order to understand the function of this large epiflora group in the wastewater treatment process, there is a need to reveal more details about the identity and physiology of the epiflora bacteria.
In this study, we investigated the phylogeny of the epiphytic protein-hydrolyzing Saprospiraceae by using the full-cycle rRNA approach. More specific gene probes were designed and used to study their in situ ecophysiology by MAR-FISH (microautoradiography [MAR] combined with fluorescence in situ hybridization [FISH]) and FISH combined with histochemical staining. The identity of the colonized filamentous bacteria and the distribution and abundance of the Saprospiraceae epiflora bacteria in full-scale activated-sludge WWTPs were investigated. Finally, their ecological relationship with the filamentous bacteria and their taxonomy were discussed.
MATERIALS AND METHODS
Sampling and description of WWTPs.
The activated-sludge samples used in this study were collected from eight Danish WWTPs, one American WWTP, and one Swedish WWTP. For the influent characteristics and configurations of the WWTPs, see Table 3. For all experiments, fresh sludge samples were taken from the aerobic tanks and transferred to the laboratory within 0.5 to 1 h.
TABLE 3.
Amino acid uptake by the epiphytic bacteria in cluster 1 (hybridizing with Epi993A), cluster 2 (hybridizing with Epi993B), and cluster 3 (hybridizing with Epi1004) under different electron acceptor conditions
| Plant name | Aerobic (oxygen)
|
Anaerobica
|
Anoxic (nitrate or nitrite)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 1 | Cluster 2 | Cluster 3 | |
| AAV | ++b | ++ | ++ | + | + | + | + | + | + |
| Egaa | ++ | +++ | +++ | + | ++ | + | + | ++ | + |
| Hjorring | ++ | ++ | +++ | ++ | + | ++ | ++ | + | + |
| Aabybro | +++ | ++ | ++ | ++ | + | ++ | + | + | ++ |
Anaerobic indicates that no electron acceptors were added.
The symbols +, ++, and +++ indicate that 10 to 30%, 30 to 50%, and 50 to 70% of a probe-defined epiphytic cluster could take up the radioactive amino acid mixture under the specified electron acceptor condition. In each sample, at least 200 epiphytic bacteria hybridizing with an individual probe were examined. Each counting was repeated with three independent samples, and the results were expressed as a range of percentages.
Micromanipulation and clone library construction.
Fresh sludge samples were collected from the Egaa and Aalborg West (AAV) WWTPs. Filaments with epiflora were micromanipulated and used as PCR templates for clone library construction by following the procedures previously described (43). Ninety clones were picked from each WWTP. Clones with the correct insert were sequenced (see the following section for more details) with primer 8F (4), and the partial sequences obtained were compared in the Ribosomal Database Project (RDP) (5). The sequences having members of Saprospiraceae as their closest relatives were chosen to be sequenced with 1492R (24) as the reverse primer.
Clone library construction using probe Bac111 as a primer.
Community DNA was extracted from fresh sludge samples obtained from the Egaa, AAV, Skagen, Hjorring, and Aabybro WWTPs (15 October 2006) with the Fast Soil DNA Extraction Kit by following the protocol provided by Qbiogene (Carlsbad, CA). The extracted DNA was pooled and used for PCR amplification with the complementary strand (Bac111F, 5′-GGGTGAGTAACGCGTACA-3′) of probe Bac111, targeting the epiflora cells, as the forward primer and bacterial universal primer 1492R as the reverse primer. The PCR cycle used was as follows: initial denaturation at 94°C for 5 min followed by 30 cycles of denaturation (45 s at 94°C), annealing (45 s at 55°C), and extension (1 min at 72°C) before a final extension at 72°C for 5 min. The PCR amplicon was confirmed on a 1% agarose gel before being ligated into the pCRII-TOPO vector provided in the TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands). Clones with the correct insert were sequenced at Macrogen, Inc., Seoul, Korea (http://www.macrogen.com), with an ABI 3730 XL automated sequencer (Applied Biosystems, Foster City, CA).
Phylogenetic analysis.
Partial 16S rRNA gene sequences were retrieved into ARB (28) and aligned. The aligned sequences were checked with the check_chimera tool in the RDP and Bellerophon (14) for chimeric artifacts before being compared in GenBank (32) by using the BLAST program to search for related sequences with high similarity values. A comparative analysis of all retrieved sequences and their closely related sequences was performed in ARB and Mega3 (23) by using different algorithms, including neighbor joining, maximum parsimony, and maximum likelihood and the default setups. Bootstrap values were calculated with the Mega3 program. Sequence similarities were calculated on the basis of the neighbor-joining tree with the function provided in ARB.
Oligonucleotide probe design and specification.
Oligonucleotide probes were designed with the function provided in the ARB software. The specificity of these probes was further confirmed by using the Check Probe program in RDP. The optimal formamide (FA) concentration of these probes used in FISH was determined and confirmed in different ways. Paraformaldehyde-fixed sludge samples from the AAV, Skagen, Hjorring, Egaa, and Aabybro WWTPs were used as positive controls for all of the probes designed. The FA concentration was increased in 5% increments from 0 to 60%. The last FA concentration before the hybridization signal was lost was used as the optimal concentration. The names, sequences, specificities, and optimal FA concentrations of all of the probes designed in this study are listed in Table 1.
TABLE 1.
New oligonucleotide probes designed to target the three epiphytic clusters belonging to Saprospiraceae
| Probe name | Sequence (5′-3′) | 16S rRNA target site | GC content (%) | Tm (°C)a | Length (nt) | Specificity | FA concn (%) |
|---|---|---|---|---|---|---|---|
| Epi741 (S-S-Sap-0741-a-18) | CA GCG TCA ATC AAG GCC C | 741-759 | 61.1 | 53 | 18 | Clusters 1-3 and a few other Saprospiraceae clones | 35-40 |
| Epi993A (S-S-Sap-0993A-a-21) | CAC CAT CTG TCA CTC ACA TTC | 993-1014 | 47.6 | 51 | 21 | Cluster 1 | 35-40 |
| Epi993B (S-S-Sap-0993B-a-21) | CAC AGW CCG TCA CTC ACA TTC | 993-1014 | 52.4 | 53 | 21 | Cluster 2 | 35 |
| Epi1004 (S-S-Sap-1004-a-18) | GCA CCT TTC AGT GCC GGT | 1004-1022 | 61.1 | 53 | 18 | Cluster 3 | 35-40 |
Tm, melting temperature (according to the manufacturer).
No pure cultures with zero to two mismatches with the probes designed in this study were available to be used as negative controls, so the clone-FISH technique (39) was adopted to find the optimal FA concentration for probes Epi741 and Epi993A, as clones with zero and one mismatch are available in this study. Clones Epr107 and Epr117, which have zero and one mismatch with probe Epi741, respectively, were used to specify probe Epi741. Similarly, clones Epr8 and Epr97, having zero and one mismatch with probe Epi993A, respectively, were used to specify probe Epi993A. The clone-FISH procedure and calculation of the optimal FA concentration for each probe are described elsewhere (18).
No negative controls were used for probes Epi993B and Epi1004, as they have at least three and two mismatches with all of the available sequences, respectively. The optimal FA concentration determined for each of the probes designed in this study was also checked by FISH probing with a combination of different hierarchical level probes Sap309, Bac111, Epi741, Epi993A, Epi993B, and Epi1004 on a series of sludge samples from the AAV, Egaa, Hjorring, and Aabybro WWTPs. The coverage ratio of probe EpiMix (a mixture of Epi993A, Epi993B, and Epi1004) was estimated by examining at least 500 epiphytic cells hybridized with probe Sap309, Bac111, or Epi741.
FISH.
FISH was carried out according to Amann et al. (2). Besides the new oligonucleotide probes described in this paper, the following were included: EUBmix, equimolar concentrations of EUB338 (1), EUB338II, and EUB338III (7) targeting most bacteria; a mixture of GFX1223 (3) and GNSB941 (10) targeting most of the members of the phylum Chloroflexi; TM7-905 (15), targeting candidate phylum TM7; Aqs997 (42), targeting Aquaspirillum-related bacteria in the class Betaproteobacteria; Sap309 (38), targeting most of the members of the family Saprospiraceae; and Bac111 (17), targeting activated-sludge clones in Saprospiraceae. All of these probes were labeled with either Cy3 or FLUOS. Detailed information about most of these probes is given in probeBase (27).
Production of exoenzymes and storage compounds.
Sludge samples used to detect the presence of exoenzymes and storage compounds were collected from the AAV, Hesingborg, Egaa, Skagen, Hjorring, Horsens, Middelfart, Kerteminde, and Aabybro WWTPs. The ability of the probe-defined epiflora bacteria to produce the exoenzymes was detected by using enzyme-labeled fluorescence (ELF 97; Molecular Probes, Eugene, OR) combined with FISH and following the procedure described previously (22). The exoenzymes included esterase (ELF 97 acetate), lipase (ELF 97 palmitate), β-d-galactosidase (ELF 97 β-d-galactopyranoside), β-d-glucuronidase (ELF 97 β-d-glucuronide), chitinase/N-acetylglucosaminidase (ELF 97 N-acetylglucosaminide; ELF 97 NAG), and phosphatase (ELF endogenous phosphatase detection kit). The activity of extracellular proteases was determined by using BODIPY enzyme staining combined with FISH (45). Neisser staining and Nile blue staining combined with FISH were used to investigate the ability of a probe-defined cluster to store polyphosphate and polyhydroxyalkanoates (PHA) (20).
Investigation of possible PHA production from amino acids was carried out with 10-ml serum bottles under aerobic and anaerobic conditions. The anaerobic condition was achieved as described for microautoradiographic incubations (19). To fresh sludge samples in serum bottles (2 ml sludge, approximately 4 g liter−1 mixed liquor suspended solids), a mixture of the amino acids (2 mM final concentration each) from a stock solution was added. The composition of the amino acid mixture was identical (but unlabeled) to that of the mixture of labeled amino acids used in MAR incubations (18) (see the following section for further details). All of the incubations were carried out on a shaking disk (250 rpm) kept at 20 ± 1°C for 3 h before being fixed and stained for PHA (20).
MAR-FISH.
MAR-FISH was carried out according to Lee et al. (25). All of the sludge samples used in MAR-FISH were obtained from the Egaa, AAV, Hjorring, and Aabybro WWTPs. All of the MAR incubation conditions and the labeled chemicals used are described elsewhere (18, 19, 21 [for N-acetylglucosamine]). Briefly, biomass samples were incubated with a radioactively labeled compound under different well-defined electron acceptor and electron donor conditions before fixation with freshly prepared paraformaldehyde in a phosphate-buffered saline buffer. All of the incubations were carried out on the same shaking disk as mentioned above.
Identification and enumeration of filamentous bacteria colonized with epiflora bacteria.
FISH probing was used to identify and enumerate the filaments colonized with the probe-defined epiflora bacteria. Sludge samples used for FISH probing were obtained from the Hjorring, Egaa, Skagen, AAV, and Aabybro WWTPs. Two probes (both Cy3 labeled), one targeting a group (group or phylum level) of filamentous bacteria and the other targeting an epiflora cluster, were used for FISH probing.
Biovolume measurement by quantitative FISH.
Quantitative FISH was carried out as described previously (20). For descriptions of all of the sludge samples used for FISH probing, see Table 5. FISH probing was carried out on gelatin-coated cover glasses (24 by 60 mm). Activated-sludge flocs were efficiently homogenized by rubbing two glass slides with a 20-μl sample (4 to 5 g mixed liquor suspended solids liter−1) to form a thin and evenly dispersed biomass layer to ensure a reliable biovolume measurement. At least 40 microscopic fields (magnification, ×1,000) were analyzed for each enumeration. Within each field, cells hybridized with a given probe were expressed as a percentage of the total area of bacteria hybridized by the EUBmix by using the functions provided in Meta Vue (Universal Image Corporation, Downingtown, PA). In some experiments, the ratio of epiflora bacteria hybridized with probe EpiMix to those hybridized with Bac111 was determined. Sludge samples were obtained from the Hjorring, Egaa, AAV, and Aabybro WWTPs. Biomass was double stained with probe EpiMix labeled with Cy3 and probe Bac111 labeled with FLUOS.
TABLE 5.
Distribution and biovolume of probe-defined epiflora bacteria in full-scale wastewater treatment plants with different influent characteristics and configurations
| Plant name | Influenta | Configurationb | Population equivalent | Sampling datec | Biovolume of probe-defined microbial group (%)d
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Bac111 | Epi741 | Cluster 1 | Cluster 2 | Cluster 3 | |||||
| Helsingborg (Sweden) | M | C, N, DN, EBPR, CP | 120,000 | 16/03/05 | 11 ± 2 | 4 ± 1 | 1 ± 1 | <1 | 2 ± 1 |
| Helsingborg (Sweden) | M | C, N, DN, EBPR, CP | 120,000 | 31/08/06 | 13 ± 2 | 5 ± 1 | 1 ± 1 | <1 | 4 ± 1 |
| AAV (Denmark) | M | C, N, DN, EBPR, CP | 300,000 | 06/02/05 | 15 ± 3 | 6 ± 2 | 3 ± 1 | 1 ± 1 | 2 ± 1 |
| Skagen (Denmark) | I | C, N, DN, EBPR, CP | 280,000 | 08/07/03 | 13 ± 3 | 4 ± 1 | 2 ± 1 | 2 ± 1 | 1 ± 1 |
| Skagen (Denmark) | I | C, N, DN, EBPR, CP | 280,000 | 22/03/05 | 11 ± 2 | 5 ± 2 | 1 ± 1 | 2 ± 1 | 1 ± 0 |
| Skagen (Denmark) | I | C, N, DN, EBPR, CP | 280,000 | 17/01/06 | 18 ± 3 | 6 ± 2 | 3 ± 1 | 2 ± 1 | 1 ± 0 |
| Egaa (Denmark) | M | C, N, DN, EBPR | 160,000 | 10/01/06 | 16 ± 2 | 9 ± 2 | 3 ± 1 | 3 ± 1 | 3 ± 1 |
| Hjorring (Denmark) | M | C, N, DN, EBPR | 100,000 | 15/12/05 | 15 ± 4 | 9 ± 3 | 4 ± 2 | 1 ± 0 | 3 ± 2 |
| Hjorring (Denmark) | M | C, N, DN, EBPR | 100,000 | 30/10/06 | 12 ± 2 | 6 ± 2 | 3 ± 1 | 1 ± 0 | 2 ± 1 |
| Hjorring (Denmark) | M | C, N, DN, EBPR | 100,000 | 24/01/07 | 11 ± 2 | 6 ± 2 | 3 ± 1 | 1 ± 1 | 2 ± 1 |
| Aabybro (Denmark) | M | C, N, DN, CP | 9,800 | 12/10/05 | 10 ± 2 | 3 ± 1 | 1 ± 1 | 1 ± 1 | 1 ± 0 |
| Aabybro (Denmark) | M | C, N, DN, CP | 9,800 | 10/01/06 | 17 ± 3 | 3 ± 1 | 1 ± 1 | 1 ± 1 | 1 ± 1 |
| Aabybro (Denmark) | M | C, N, DN, CP | 9,800 | 31/10/07 | 12 ± 3 | 4 ± 1 | 1 ± 1 | 1 ± 1 | 1 ± 0 |
| Horsens (Denmark) | M | C, NDN, CP | 140,000 | 07/02/05 | 6 ± 1 | 4 ± 2 | 1 ± 1 | 1 ± 1 | 1 ± 1 |
| Horsens (Denmark) | M | C, NDN, CP | 140,000 | 07/08/06 | 11 ± 2 | 3 ± 1 | 1 ± 1 | 1 ± 1 | 1 ± 1 |
| Horsens (Denmark) | M | C, NDN, CP | 140,000 | 07/02/07 | 8 ± 1 | 5 ± 2 | 1 ± 1 | 2 ± 1 | 2 ± 1 |
| Kerteminde (Denmark) | M | C, N, DN | 16,000 | 07/08/06 | 10 ± 2 | 4 ± 2 | 2 ± 1 | 1 ± 0 | 1 ± 0 |
| Middelfart (Denmark) | M | C, N, DN | 20,000 | 14/11/06 | 16 ± 4 | 5 ± 2 | 2 ± 1 | 1 ± 0 | 2 ± 1 |
| Districts N (United States) | M/I | C, N, DN | 180,000 | 15/12/06 | 14 ± 3 | 4 ± 2 | 3 ± 1 | <1 | <1 |
M and I represent primarily municipal and industrial wastewaters, respectively.
C, carbon removal; N, nitrification; DN, denitrification; CP, chemical phosphorus removal.
The dates the samples were taken (day/month/year) are shown.
Percentage of biovolume calculated from all bacteria fluorescing with EUBmix. The values are expressed as averages ± standard deviations based on at least 40 measurements.
Microscopy.
All of the FISH and MAR-FISH images were taken with an epifluorescence microscope as previously described (19).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database under accession numbers EF523437 to EF523471 and EU177672 to EU177766.
RESULTS
Phylogenetic analysis of the epiphytic Saprospiraceae.
To determine the identities of the epiphytic Saprospiraceae hybridizing with probes Sap309 and Bac111, micromanipulation of filamentous bacteria (from the AAV and Egaa WWTPs) carrying many epiphytic bacteria was carried out. Two clone libraries were constructed, and a total of 180 clones were partially or fully sequenced. Phylogenetic analysis showed that 32 clones fell into Saprospiraceae, but only 4 of them, after being fully sequenced, grouped with 16S rRNA gene sequences close to the putative epiflora sequences targeted by probe Bac111. Therefore, another clone library was constructed by using the complementary strand of probe Bac111 (Bac111F) as the forward primer and universal primer 1492R as the reverse primer. Community DNA from five WWTPs, where the epiflora bacteria were abundant, was extracted, pooled, and used as the template. One hundred fifty clones were obtained, and 130 were affiliated to members in Saprospiraceae.
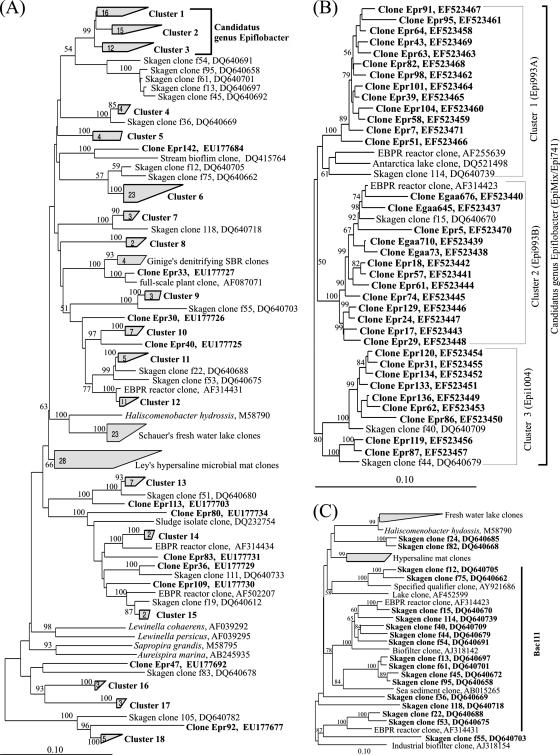
The 130 clones retrieved were distributed widely in the family Saprospiraceae (Fig. 1A). Most of the clones could be included in 18 clusters, each consisting of 2 to 16 clones with >90% similarity. Most of the clones clustered with clones from full-scale WWTPs with a similarity of 85 to 99% and particularly with those from the Skagen WWTP (17). None of the clones were closely related to any type strains in the family Saprospiraceae, as they had less than 87% similarity. Of all of the clusters, only clusters 1, 2, and 3 are closely related to each other, containing a total of 35 clones from this study and 7 clones from the GenBank database (32) (Fig. 1B). Other clusters are diversely distributed in Saprospiraceae and relatively far away from each other.
FIG. 1.
(A) Simplified distance tree (neighbor joining) built for all of the genera and the main microbial groups in the family Saprospiraceae of the phylum Bacteroidetes. (B) Distance tree built for the three clusters of “Candidatus genus Epiflobacter” as shown in panel A. (C) Distance tree showing the Skagen clones targeted by probe Bac111. The sequences with bold names in panels A and B were obtained in this study. The values in the quadrangles of panel A represent the numbers of clones in the clone clusters. The bootstrap values in all of the panels (only those >50% are shown) were calculated on the basis of 1,000 resamplings. The scale bars in all panels represent 1 substitution per 10 nucleotides.
To further clarify the phylogeny of the epiflora bacteria, the clones putatively representing the epiflora clusters were screened by using clones targeted by probe Bac111 (mainly Skagen clones, as shown in Fig. 1C) as “internal markers.” It is believed that they contain representatives of the epiflora bacteria. Clusters 1, 2, and 3 each contains at least one marker clone and were chosen for the screening.
Clusters 1, 2, and 3 were further analyzed, and different treeing algorithms (maximum parsimony, maximum likelihood, and neighbor joining) were adopted to calculate the phylogenetic trees, and the topology of the trees obtained did not significantly change (Fig. 1B). Clusters 1, 2, and 3 consist of 16, 15, and 11 sequences, respectively, and within each cluster the similarity is 93 to 99%. The similarity is 90 to 95% between clusters 1 and 2, 90 to 95% between clusters 2 and 3, and 91 to 94% between clusters 1 and 3. The type strain closest to the putative epiflora group is H. hydrossis (similarity, ≤87.0%), and the similarity to other Saprospiraceae type strains, including Aureispira marina, Saprospira grandis, and Lewinella persica, is <82%. The epiflora group also has <87% similarity to other clones of uncultured bacteria, including those from freshwater lakes (38), hypersaline microbial mats (26), and a methanol-fed denitrifying reactor (11) (Fig. 1A).
Gene probe design and FISH of activated sludge.
Gene probes at two hierarchical levels were designed to target clusters 1, 2, and 3 and the Skagen marker cluster consisting of the four marker clones. The probe designed for the marker cluster hybridized with only a few cocci (<1% of the total bacterial biovolume, naked-eye estimation) and is not included in this report. The sequences, specificities, and optimal FA concentrations of the probes designed for clusters 1 to 3 are listed in Table 1. Probes Epi993A, Epi993B, and Epi1004 were designed to target clusters 1, 2, and 3, respectively. Probe Epi741 targets all three clusters and a few other clones. The optimal FA concentration for probes Epi741 and Epi993A by the clone-FISH technique generally agreed with those determined by using FISH probing of sludge samples, although a slightly higher optimal FA concentration was found by clone-FISH (1.1 times). As a weak FISH signal was observed from the epiflora cells at the FA concentrations obtained from clone-FISH, the FA concentrations (Table 1) determined in FISH probing of sludge samples were used.
The specificity of the newly designed gene probes was checked by FISH probing with sludge samples from the AAV, Egaa, Hjorring, and Aabybro WWTPs (Table 2). Under the optimal FA concentrations determined, probes Epi993A, Epi993B, and Epi1004 hybridized only with epiflora bacteria consisting of single rods or two or three rods connected end to end (Fig. 2A and B). Epi741 hybridized mainly with epiflora bacteria but occasionally with short filaments, too. All of the epiflora bacteria hybridizing with Epi993A, Epi993B, or Epi1004, targeting the individual clusters, also hybridized with Epi741, Bac111, or Sap309. Application of probes Epi993A, Epi993B, and Epi1004 in a mixture (EpiMix) yielded the same result. No cross hybridization was found among probes Epi993A, Epi993B, and Epi1004. Most (>95%) of the epiflora bacteria hybridizing with probe Epi741 also hybridized with EpiMix. All of these tests confirm the specificity of the new cluster level probes. Not all of the epiflora bacteria hybridizing with probe Bac111 or Sap309 (both match well in coverage of the epiflora bacteria; data not shown) also hybridized with EpiMix. In the Egaa and Hjorring WWTPs, 80 to 90% of the epiphytic cells hybridized with Sap309 or Bac111 hybridized with EpiMix. The ratio was 70 to 80% in AAV and 60 to 70% in Aabybro.
TABLE 2.
Evaluation of cross hybridization and coverage ratios of gene probes targeting cluster 1 (Epi993A), cluster 2 (Epi993B), and cluster 3 (Epi1004) of epiphytic Saprospiraceae
| Plant name | Hybridizationa with following probe(s) in FISH analysis:
|
||||
|---|---|---|---|---|---|
| Sap309 or Bac111 (EpiMix) | Epi741 (EpiMix) | Epi993A (Epi993B + Epi1004) | Epi993B (Epi993A + Epi1004) | Epi1004 (Epi993A + Epi993B) | |
| AAV | Y, ++ | Y, ++++ | N, − | N, − | N, − |
| Egaa | Y, +++ | Y, ++++ | N, − | N, − | N, − |
| Hjorring | Y, +++ | Y, ++++ | N, − | N, − | N, − |
| Aabybro | Y, + | Y, ++++ | N, − | N, − | N, − |
The symbols −, +, ++, +++, and ++++ indicate that 0%, 60 to 70%, 70 to 80%, 80 to 90%, and >95% of the epiphytic bacteria that hybridized with a probe not in parentheses also hybridized with the probe(s) in parentheses. In each sample, at least 500 epiphytic bacteria hybridizing with each of the probes not in parentheses were counted. The results from three independent countings are expressed as a range of percentages. Y indicates that the epiphytic bacteria hybridizing with the probe not in parentheses also hybridized with the probe(s) in parentheses. N indicates that the epiphytic bacteria hybridizing with the probe not in parentheses did not hybridize with the probe(s) in parentheses.
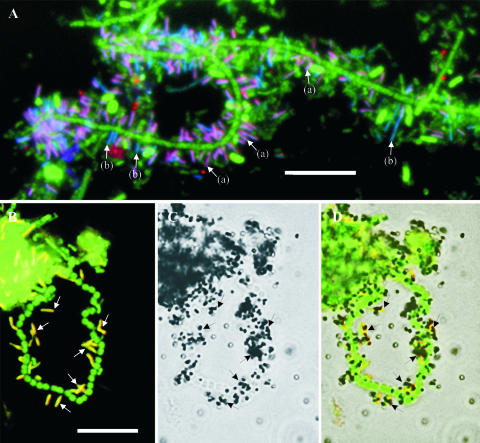
FIG. 2.
(A) FISH image of activated sludge after color combination showing Bacteria hybridized with EUBmix (Cy5 labeled, set as green) as cells labeled with different colors. Epiflora hybridized with Bac111 (FLUOS labeled, set as blue) as cyan-labeled cells (mixture of green and blue, indicated by arrows b), and epiflora hybridized with EpiMix (Cy3 labeled, set as red) as purple-labeled cells (mixture of blue, red, and green, indicated by arrows a). (B, C, and D) MAR-FISH image of activated sludge with epiflora. (B) FISH image of epiflora hybridized with EpiMix (Cy3 labeled, set as red) and Bacteria hybridized with EUBmix (FLUOS labeled, set as green). (C) Bright-field image of MAR of the same field as panel B. (D) Overlay of images D and C. Images B, C, and D show that the epiflora hybridized with EpiMix (e.g., those indicated by arrows) take up a mixture of labeled amino acids under aerobic conditions. The bars in panels A and B represent 10 μm.
Morphological observations and attachment style.
In all of the sludge samples examined, the epiflora bacteria hybridizing with probes Epi993A, Epi993B, and Epi1004 were rods 1.3 to 1.9 μm in length and 0.3 to 0.4 μm in width and all only attached to different filamentous bacteria (Fig. 2A and B) and not to any other microbial morphotypes. The density of epiflora bacteria on the filamentous bacteria varied widely from very dense to sparse colonization. Normally, two or three rods connected end to end attached themselves to the surfaces of filamentous bacteria and they were usually attached perpendicularly to the filament.
Presence of exoenzymes and storage compounds.
The ability of the three probe-defined epiflora clusters to produce different exoenzymes and intracellular storage compounds was investigated in five EBPR and four nitrogen removal WWTPs. In the nine WWTPs investigated, all of the epiflora clusters had exoprotease activity, as revealed by in situ protease staining (BODIPY FL casein and bovine serum albumin) combined with FISH. No cell surface-associated activities of esterase, lipase, β-d-galactosidase, β-d-glucuronidase, chitinase, and phosphatase were found in any of the clusters by using ELF 97 combined with FISH. Intracellular accumulation of polyphosphate or PHA was not detected by FISH in combination with histochemical staining (Neisser staining and Nile blue staining). The potential effect of incubation with amino acids (substrates they can utilize; see the following section) on their intracellular storage of PHA was also investigated. Incubation of activated-sludge samples from AAV, Hjorring, Egaa, and Aabybro with an amino acid mixture for 3 to 5 h under aerobic or anaerobic conditions did not lead to the formation of intracellular PHA.
Uptake of organic substrates.
The substrate utilization pattern of the individual epiflora clusters was investigated by using MAR-FISH under different electron acceptor conditions. Organic substrates, including sugars (glucose, mannose, N-acetylglucosamine), short-chain fatty acids (formate, acetate, pyruvate, and propionate), ethanol, a long-chain organic acid (oleic acid), individual amino acids (leucine, aspartic acid, glutamic acid, and glycine), and a mixture of 14 amino acids were tested in biomass samples from the AAV, Egaa, Hjorring, and Aabybro WWTPs. The three epiflora clusters could only take up the mixture of amino acids, and the uptake was observed under aerobic (Fig. 2C and D) and, to some extent, anaerobic conditions (Table 3). However, in each cluster, not all of the epiflora cells took up amino acids. The fractions of the epiflora bacteria taking up amino acids were very similar in the AAV, Egaa, Hjorring, and Aabybro WWTPs, being 50 to 70% under aerobic conditions and 10 to 50% under anaerobic conditions. The amino acid uptake of the epiflora bacteria was also investigated under anoxic conditions by adding NO3− or NO2− to the anaerobic MAR incubations. In all of the WWTPs examined, the ratios (10 to 50%) of the three clusters taking up amino acids did not change, being in the same ranges as observed under anaerobic conditions, indicating that they were not involved in denitrification.
The potential growth ability of the three clusters was also investigated under anaerobic and aerobic conditions by using MAR-FISH with labeled amino acids. This was carried out by preincubating the sludge samples with an unlabeled amino acid mixture (with the same composition as the labeled amino acid mixture they took up) for 3, 6, or 9 h before the labeled amino acid mixture was added. No anaerobic uptake was observed after 3 h of anaerobic preincubation with an unlabeled amino acid mixture, whereas 50 to 70% of the three epiflora clusters could still take up amino acids after 9 h of aerobic preincubation in the AAV, Egaa, Hjorring, and Aabybro WWTPs. This indicates that the epiflora bacteria were obligately aerobic but able to take up amino acids under short-term anaerobic conditions.
Identification of the filamentous bacteria colonized by epiflora bacteria.
The identities of the filamentous bacteria colonized by the different epiflora clusters were investigated in five activated-sludge WWTPs by using FISH. The results are listed in Table 4. Epiphytic colonization by the three epiflora clusters was detected on three probe-defined filamentous bacterial groups (or phyla), which were all abundant in the WWTPs investigated. These were Aquaspirillum-related filaments (defined by probe Aqs997), Chloroflexi-related filaments (probe mixture GNSB941-CFX1223), and uncultured TM7-related filaments (probe TM7-905). Except for cluster 1 (Epi993A), which did not grow on the filaments defined by probe TM7-905, all of the epiflora clusters colonized all of the probe-defined filamentous groups. They were universally present in the WWTPs investigated but showed some differences in their colonization behavior.
TABLE 4.
Identities of filamentous bacteria colonized by the three epiphytic Saprospiraceae clusters found in five wastewater treatment plantsa
| Plant name | Cluster 1 (Epi993A)
|
Cluster 2 (Epi993B)
|
Cluster 3 (Epi1004)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Aqs997b | GNSB941 + CFX1223c | TM7-905d | Aqs997 | GNSB941 + CFX1223 | TM7-905 | Aqs997 | GNSB941 + CFX1223 | TM7-905 | |
| AAV | + | +++ | − | + | + | ++ | + | + | + |
| Egaa | +++ | ++ | − | ++ | + | + | +++ | + | − |
| Hjorring | ++ | +++ | − | ++ | ++ | − | +++ | + | + |
| Aabybro | + | ++ | − | + | + | − | + | + | + |
| Skagen | + | + | − | +++ | +++ | ++ | ++ | ++ | − |
The probe-defined filamentous bacteria were present in all of the plants investigated. The symbols +, ++, and +++ indicate that 1 to 10%, 10 to 20%, and 20 to 30% of the probe-defined filaments were colonized with the indicated epiflora cluster, and a minus sign indicates that the probe-defined epiflora did not colonize the probe-defined filaments. In each sample, at least 100 filaments hybridizing with a probe were counted. The results from three independent countings are expressed as a range of percentages.
Probe targeting genus Aquaspirillum-related bacteria.
Probe mixture targeting most of the members of the phylum Chloroflexi.
Probe targeting most of the members of candidate phylum TM7.
Among the filaments hybridizing with Aqs997 and GNSB941-CFX1223, 10 to 30% were colonized by clusters 1, 2, and/or 3 (Table 4). Clusters 2 and 3 colonized TM7 filaments in some WWTPs but not in others. In the Egaa, Skagen, and AAV WWTPs, 10 to 20% of the TM7 filaments were colonized by cluster 2 but none were colonized in the Hjorring and Aabybro WWTPs. Cluster 3 colonized 1 to 10% of the TM7 filaments in the Hjorring, AAV, and Aabybro WWTPs but none in the Egaa and Skagen WWTPs. Colonization of the epiflora clusters on a few other unidentified filamentous bacteria was also observed.
Distribution and abundance of the individual epiflora clusters in full-scale activated-sludge WWTPs.
The biovolumes of each epiflora cluster in nine full-scale WWTPs were investigated by using quantitative FISH (Table 5). Some of the WWTPs were sampled three times to obtain an impression of the fluctuation of the biovolume of each cluster over 1 to 3 years. Except for a few occasions, each of the epiflora clusters was present in all of the WWTPs examined. On average, cluster 1 accounted for 2% ± 1% (mean value ± standard deviation), cluster 2 accounted for 1% ± 1%, and cluster 3 accounted for 2% ± 1% of the total bacterial biovolume. The biovolume measured with Epi741 (group level) in each WWTP generally agreed with the biovolume measured with cluster level probes (Table 5).
DISCUSSION
Phylogenetic affiliation of probe-defined epiflora bacteria.
The epiflora group identified in this study consists of at least three probe-defined clusters. Each cluster contains 11 to 16 sequences with 93 to 99% similarity and has less than 82% similarity to the available type stains of the genera in Saprospiraceae and <87% similarity to other Saprospiraceae clones. Therefore, they should be classified as new taxa in the family Saprospiraceae. Moreover, based on 97% similarity or a higher threshold (∼99%) (41) for defining a species, each cluster may consist of several species. The clone library constructed by using probe Bac111 as the primer successfully retrieved the 16S rRNA genes of the epiflora bacteria.
In addition to the three clusters shown in Fig. 1B, the broad probe Epi741 also perfectly matches four other Saprospiraceae clones, AJ318142, AF087054, AF255645, and BQ232435 (>1,200 bp), from the GenBank database which could not be grouped with any of the clusters. However, we found that probe Epi741 also occasionally hybridized with a few filamentous bacteria not hybridizing with any cluster level probe, so for specific and simultaneous detection of the three epiflora clusters, the EpiMix probe consisting of Epi993A, Epi993B, and Epi1004 should be used.
EpiMix or Epi741 targeting clusters 1 to 3 only hybridized with 60 to 90% of the epiflora bacteria hybridizing with the broader probe Bac111, so the detailed identity of the 10 to 30% epiflora bacteria missed by EpiMix or Epi741 is still not clear. Whether their 16S rRNA genes were obtained but not detected by the EpiMix, as many of the clusters associated with Skagen marker clones were not screened, and/or their 16S rRNA genes were not successfully retrieved, is still unknown. However, EpiMix covers a major fraction of the epiflora bacteria in activated sludge, and the morphological and ecophysiological characteristics of the epiflora bacteria hybridizing with EpiMix, Epi741, or Bac111 are very similar. No further efforts, therefore, were made to investigate the detailed phylogeny of the epiflora bacteria missed by EpiMix. The EpiMix-defined epiflora group represents the protein-hydrolyzing epiflora bacteria well, but probe Bac111 should be used to estimate all or most of the epiphytic bacteria belonging to Saprospiraceae.
PCR amplification of the epiflora bacteria containing biomass obtained from micromanipulation followed by clone library construction retrieved only 4 (out of 32) epiflora-related 16S rRNA gene sequences, although a significant number of clones, 180, were sequenced. This is mainly due to the difficulty associated with micromanipulation of filamentous bacteria with epiflora, because they were often partly inside the sludge flocs, so other loosely attached bacteria might also be present.
Ecophysiology of the probe-defined epiflora bacteria.
Each of the epiflora clusters was characterized ecophysiologically in three ways, (i) organic substrate utilization pattern under different electron acceptor conditions, (ii) production of the storage compounds PHA and polyphosphate, and (iii) production of extracellular enzymes. The resolution of our studies did not show any differences, so all three epiflora clusters were very similar with regard to these ecophysiological aspects.
All of the clusters could only utilize amino acids as carbon and energy sources and not the tested short-chain fatty acids, monosaccharides, ethanol, glucosamine, or oleic acid. The utilization occurred under anaerobic (0 to 3 h), aerobic, and anoxic conditions. However, they seemed to be more active under aerobic conditions than under anaerobic conditions because the number of epiflora bacteria taking up amino acids under aerobic conditions (50 to 70%) was higher than under short-term anaerobic conditions (10 to 50%). The lack of activity by some of the epiflora bacteria under both aerobic and anaerobic conditions has also been observed on other probe-defined bacteria in these WWTPs (19, 31). Whether the bacteria were inactive or whether they were species or ecotypes not utilizing amino acids is unknown. Their ability to utilize amino acids under short-term (3 h) anaerobic conditions may indicate a capability to store organic compounds, but the identity of such compounds is not clear, as they did not store PHA.
All of the clusters were able to hydrolyze proteins but not the other macromolecules tested. They could not store polyphosphate or PHA and were most likely not able to denitrify. Therefore, they most probably grow in activated sludge by hydrolyzing proteins and using the hydrolysates (small peptides or amino acids) as energy and carbon sources. The exact types of proteases they excrete and the amino acids they take up are, however, not known because the BODIPY-labeled casein and bovine serum albumin conjugates respond to different proteases (45). Similarly, the labeled amino acid mixture they took up consists of 14 different amino acids. Some of the amino acids in the mixture were tested individually (aspartic acid, leucine, glutamic acid, and glycine), but none of them were utilized, implying either that these amino acids could not be utilized or that they could only be utilized in the presence of other amino acids.
Colonization to filaments and distribution in full-scale WWTPs.
Interestingly, the epiphytic bacteria mainly colonized filamentous bacteria belonging to the phylum Chloroflexi, candidate phylum TM7, and filaments related to the genus Aquaspirillum but not several other filamentous bacteria present in the sludge samples, such as “Candidatus Microthrix parvicella” and other unidentified bacteria. All of these filamentous bacteria are common or abundant in most activated-sludge WWTPs with nutrient removal (21, 37, 42, 43). Moreover, no obvious preference for the colonization of a specific filamentous bacterial group was found in any of the epiflora clusters, with the exception of cluster 1, which did not colonize TM7 filaments. Therefore, attachment may rely on differences in surface structure or other, unknown, factors.
The advantage of being attached to the filamentous bacteria is still speculative. There might be a sort of symbiotic relationship between the epiflora bacteria and the filamentous bacteria. The attachment not only prevents the epiflora bacteria from being washed out from the WWTP with the effluent but most importantly facilitates the adsorption of macromolecules from the wastewater as the filaments often protrude into bulk water, where most of the organic matter exists in the form of colloids and small particles (29). In return, the epiflora bacteria may, as protein-hydrolyzing organisms, provide substrates (amino acids) to their hosts.
This study showed that the EpiMix-defined epiflora group is an important component of the microbial community of activated sludge. It constituted up to 10% of the bacterial biovolume in the nine WWTPs examined. The group was more abundant in EBPR WWTPs (6%, mean value of 10 samples from five WWTPs) than in N removal WWTPs with chemical precipitation of phosphorus (4%, mean value of 8 samples from four WWTPs). However, whether this is due to an ecological selection of the epiflora clusters (or the filamentous bacteria they colonize) by the anaerobic period in the EBPR systems (the main difference between EBPR and N removal WWTPs) or if there are any other reasons has to be investigated further.
Taxonomic consideration: proposal of “Candidatus genus Epiflobacter.”
The epiflora group that has been characterized phylogenetically and ecophysiologically in this study consists of three closely related clusters with 90 to 95% similarity. Each cluster contains different species. However, they are all rods attached to mainly three probe-defined filamentous bacteria and have similar ecophysiology, all being specialized in protein hydrolysis. Therefore, we propose to classify them into a new genus, “Candidatus Epiflobacter,” on the basis of the requirement of the International Committee on Systematic Bacteriology (30).
“Candidatus Epiflobacter” spp. are uncultured bacteria belonging to the family Saprospiraceae in the phylum Bacteroidetes and are defined by probe EpiMix (a mixture of probes Epi1004, Epi993A, and Epi993B). They are gram-negative rods (1.3 to 1.9 μm in length and 0.3 to 0.4 μm in width) that attach to a range of filamentous bacteria in activated sludge. They are strict aerobes and specialists in protein hydrolysis that are capable of utilizing only (or primarily) amino acids as energy and carbon sources. They do not form PHA or polyphosphate granules.
Acknowledgments
This study was supported by Danish Technical Research Council grant 26-04-0115 (Identification and Characterization of Uncultured Bacteria Involved in Hydrolysis and Fermentation in Nutrient Removal Plants).
C. Kragelund, J. L. Nielsen, and A. Schramm are acknowledged for helpful suggestions and discussions.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björnsson, L., P. Hugenholtz, G. W. Tyson, and L. L. Blackall. 2002. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148:2309-2318. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal-RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Eikelboom, D. H., and H. J. J. van Buijsen. 1983. Microscopic sludge investigation manual, 2nd ed. TNO Research Institute of Environmental Hygiene, Delft, The Netherlands.
- 9.Eikelboom, D. H. 1975. Filamentous organisms observed in activated sludge. Water Res. 9:365-388. [Google Scholar]
- 10.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Ginige, M. P., J. Keller, and L. L. Blackall. 2005. Investigation of an acetate-fed denitrifying microbial community by stable isotope probing, full-cycle rRNA analysis, and fluorescent in situ hybridization-microautoradiography. Appl. Environ. Microbiol. 71:8683-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 13.Hosoya, S., V. Arunpairojana, C. Suwannachart, A. Kanjana-Opas, and A. Yokota. 2006. Aureispira marina gen. nov., sp. nov., a gliding, arachidonic acid-containing bacterium isolated from the southern coastline of Thailand. Int. J. Syst. Evol. Microbiol. 56:2931-2935. [DOI] [PubMed] [Google Scholar]
- 14.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima, H., Y. Koizumi, and M. Fukui. 2006. Community structure of bacteria associated with sheaths of freshwater and brackish Thioploca species. Microb. Ecol. 52:765-773. [DOI] [PubMed] [Google Scholar]
- 17.Kong, Y. H., Y. Xia, J. L. Nielsen, and P. H. Nielsen. 2007. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology 153:4061-4073. [DOI] [PubMed] [Google Scholar]
- 18.Kong, Y. H., J. L. Nielsen, and P. H. Nielsen. 2005. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 71:4076-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, Y. H., J. L. Nielsen, and P. H. Nielsen. 2004. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 70:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, Y. H., Y. Xia, J. L. Nielsen, and P. H. Nielsen. 2006. Ecophysiology of a group of uncultured gammaproteobacterial glycogen-accumulating organisms in full-scale enhanced biological phosphorus removal wastewater treatment plants. Environ. Microbiol. 8:479-489. [DOI] [PubMed] [Google Scholar]
- 21.Kragelund, C., C. Levantesi, A. Borger, K. Thelen, D. Eikelboom, V. Tandoi, Y. H. Kong, J. van der Waarde, J. Krooneman, S. Rossetti, T. R. Thomsen, and P. H. Nielsen. 2007. Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Microbiol. Ecol. 59:671-682. [DOI] [PubMed] [Google Scholar]
- 22.Kragelund, C., J. L. Nielsen, T. R. Thomsen, and P. H. Nielsen. 2005. Ecophysiology of the filamentous alphaproteobacterium Meganema perideroedes in activated sludge. FEMS Microbiol. Ecol. 54:111-122. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991.16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Modern microbial methods: nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 25.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgenroth, E., R. Kommedal, and P. Harremoës. 2002. Processes and modeling of hydrolysis of particulate organic matter in aerobic wastewater treatment—a review. Water Sci. Technol. 45:25-40. [PubMed] [Google Scholar]
- 30.Murray, R. G. E., and E. Stackebrandt. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described prokaryotes. Int. J. Syst. Bacteriol. 45:186-187. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 32.Ouellette, B. F. F. 1998. The GenBank sequence database. Bioinformatics 39:16-45. [DOI] [PubMed] [Google Scholar]
- 33.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramey, B. E., M. Koutsoudis, S. B. von Bodman, and C. Fuqua. 2004. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 7:602-609. [DOI] [PubMed] [Google Scholar]
- 35.Reichenbach, H. 1991. The order Cytophagales, p. 3631-3675. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer, (ed.), The prokaryotes, 2nd ed. Springer Verlag, New York, NY.
- 36.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossetti, S., M. C. Tomei, P. H. Nielsen, and V. Tandoi. 2005. “Microthrix parvicella,” a filamentous bacterium causing bulking and foaming in activated sludge systems: a review of current knowledge. FEMS Microbiol. Rev. 29:49-64. [DOI] [PubMed] [Google Scholar]
- 38.Schauer, M., and M. W. Hahn. 2005. Diversity and phylogenetic affiliations of morphologically conspicuous large filamentous bacteria occurring in the pelagic zones of a broad spectrum of freshwater habitats. Appl. Environ. Microbiol. 71:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 40.Seviour, E. M., C. Williams, B. Degrey, J. A. Soddell, R. J. Seviour, and K. C. Lindrea. 1994. Studies on filamentous bacteria from Australian activated sludge plants. Water Res. 28:2335-2342. [Google Scholar]
- 41.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 42.Thomsen, T. R., B. V. Kjellerup, J. L. Nielsen, P. Hugenholtz, and P. H. Nielsen. 2002. In situ studies of the phylogeny and physiology of filamentous bacteria with attached growth. Environ. Microbiol. 4:383-391. [DOI] [PubMed] [Google Scholar]
- 43.Thomsen, T. R., J. L. Nielsen, N. B. Ramsing, and P. H. Nielsen. 2004. Micromanipulation and further identification of FISH-labelled microcolonies of a dominant denitrifying bacterium in activated sludge. Environ. Microbiol. 6:470-479. [DOI] [PubMed] [Google Scholar]
- 44.van Veen, W. L., D. van der Kooij, E. C. Geuze, and A. W. van der Vlies. 1973. Investigations on sheathed bacterium Haliscomenobacter hydrossis gen. nov., sp. nov., isolated from activated sludge. Antonie van Leeuwenhoek J. Microbiol. 39:207-216. [DOI] [PubMed] [Google Scholar]
- 45.Xia, Y., Y. H. Kong, and P. H. Nielsen. 2007. In situ detection of protein-hydrolysing microorganisms in activated sludge. FEMS Microbiol. Ecol. 60:156-165. [DOI] [PubMed] [Google Scholar]