Abstract
Rapid and efficient methods for the detection and quantification of infectious viruses are required for public health risk assessment. Current methods to detect infectious viruses are based on mammalian cell culture and rely on the production of visible cytopathic effects (CPE). For hepatitis A virus (HAV), viral replication in cell culture has been reported to be nonlytic and relatively slow. It may take more than 1 week to reach the maximum production and subsequent visualization of CPE. A molecular beacon (MB), H1, specifically targeting a 20-bp 5′ noncoding region of HAV, was designed and synthesized. MB H1 was introduced into fixed and permeabilized fetal rhesus monkey kidney (FRhK-4) cells infected with HAV strain HM-175. Upon hybridizing with the viral mRNA, fluorescent cells were visualized easily under a fluorescence microscope. Discernible fluorescence was detected only in infected cells by using the specific MB H1. A nonspecific MB, which was not complementary to the viral RNA sequence, produced no visible fluorescence signal. This MB-based fluorescence assay enabled the direct counting of fluorescent cells and could achieve a detection limit of 1 PFU at 6 h postinfection, demonstrating a significant improvement in viral quantification over current infectivity assays.
Hepatitis A virus (HAV), a positive single-stranded RNA virus, was previously classified as enterovirus type 72 (14) and has now been classified in the Hepatovirus genus of the Picornaviridae family (15). These single-plus-stranded RNA viruses can directly translate plus-strand RNA genomes into protein using the host ribosomes. The plus strand is transported to the infected cell via specific membrane vesicles, where it is copied into full-length minus strands. These minus strands then serve as templates for the synthesis of plus-strand genomic RNA molecules (1, 2).
HAV is known to cause acute liver infection with a discrete onset of symptoms (e.g., fever, malaise, and nausea), followed in several days by jaundice. Person-to-person transmission through the fecal-oral route is the primary means of HAV transmission. Outbreaks and periodic cases also occur from exposure to fecally contaminated food or water (Centers for Disease Control and Prevention). Most picornaviruses are lytic, causing distinctive cytopathic effects and replicating in an 8-h cycle under one-step growth conditions. The growth of HAV in cell culture systems, however, is much slower and nonlytic and does not produce a detectable cytopathic effect in infected cells (13). Cromeans et al. reported the isolation of a cytopathic HAV variant from the rapidly replicating isolate HM-175 that is lytic for fetal rhesus monkey kidney (FRhK-4) cells (4). However, complete lysis still did not occur until 5 to 6 days postinfection (p.i.).
Conventional methods for detecting infectious HAV rely on viral propagation in cell culture, radioimmunoassay, immunofluorescence, or plaque assay; these methods are difficult to perform, and it may take weeks before the viruses reach sufficiently high amounts in the cell-culture medium to allow detection (4, 5, 9, 10). For public health assessments, this long delay in obtaining infectivity information may affect the process of epidemic control strategies. Improved methods need to be developed for the rapid and reliable detection and quantification of infectious viruses.
Molecular beacons (MBs) were first developed in 1996 (16) and have been applied in numerous in vitro hybridization assays (7, 8, 12). MBs are single-stranded oligonucleotide probes possessing stem-loop structures and are doubly labeled with a fluorophore and a quencher at the 5′ and 3′ ends, respectively. In the presence of a complementary sequence, the MB undergoes a conformational change and the MB fluoresces via fluorescence resonance energy transfer. Because of this property, MBs provide a separation-free detection scheme that produces fluorescence upon binding to the targets.
MBs have been widely used in many areas, such as in real-time monitoring of DNA/RNA amplification during PCR, gene typing, mutation detection, real-time enzymatic cleavage assay (8, 11), and RNA detection in living cells (3, 6). By probing the endogenous RNA with MBs, the dynamic behavior of poliovirus plus-strand RNA in living host cells has been studied (6). The use of MBs as a rapid and sensitive tool to detect and quantify the presence of newly synthesized coxsackievirus RNA has also been demonstrated (17).
In this report, we demonstrate a combined cell culture-MB assay for the detection of infectious HAV with a detection limit of 1 PFU within 6 h p.i. Validation of this MB-based fluorescence assay for viral quantification was demonstrated, with results compatible with those of the conventional plaque assay. Finally, the utility of the assay for the detection of HAV in surface water samples was also demonstrated.
MATERIALS AND METHODS
FRhK-4 cell culture.
FRhK-4 cells (ATCC CRL-1688; passage level of <40) were grown in 400 ml 1× autoclavable minimal essential medium (Irvine Scientific, Santa Ana, CA) with 4 ml 7.5% sodium bicarbonate, 8 ml 1 M HEPES, 4 ml nonessential amino acids (Gibco BRL, Grand Island, NY), 10 ml A/B-L (1,000 U/ml penicillin, 1,000 U/ml streptomycin, 2 mg/ml kanamycin, 2,000 U/ml nystatin, and 80 mM l-glutamine), and 60 ml fetal bovine serum (Sigma-Aldrich, St. Louise, MO) at 37°C in a 5% CO2 atmosphere. Phosphate-buffered saline (PBS; 0.01 M phosphate, pH 7.4, 0.138 M NaCl, and 2.7 mM KCl) and Tris-buffered saline solution (TBSS; 0.05 M Tris, pH 7.4, 0.28 M NaCl, 10 mM KCl, and 0.82 mM Na2HPO4) were used for washing steps in the plaque assay and MB analysis, respectively.
Virus preparation.
The cytopathic HM-175 strain of HAV (ATCC VR-2089) was inoculated into uninfected FRhK-4 cells (passage numbers 34 to 36) for 7 days at 37°C in a 5% CO2 atmosphere. The cell lysate was purified (−20°C freezing and 37°C thawing by turns) and then extracted with chloroform. The HM-175 virus stock was stored at −80°C.
Plaque assay.
The HM-175 virus stock was thawed, and then a 10-fold serial dilution in PBS was performed. Confluent, 7-day-old FRhK-4 monolayers in 24-well, 15-mm dishes (Costar; Corning Inc., Corning, NY) were infected with 0.5 ml of virus dilutions. After 90 min of adsorption at 37°C in 5% CO2, the solutions were aspirated and the monolayers were washed once with PBS. One milliliter of 100-ml 2% carboxymethylcellulose sodium salt (Sigma-Aldrich, St. Louis, MO) in 100 ml 2× autoclavable minimal essential medium (Irvine Scientific, Santa Ana, CA) with 2 ml 7.5% sodium bicarbonate, 4 ml 1 M HEPES, 2 ml nonessential amino acids, 5 ml A/B-L (1,000 U/ml penicillin, 1,000 U/ml streptomycin, 2 mg/ml kanamycin, 2,000 U/ml nystatin, and 80 mM l-glutamine), and 4 ml fetal bovine serum (Sigma-Aldrich, St. Louis, MO) was added. After 8 days of incubation at 37°C in 5% CO2, the carboxymethylcellulose overlay was removed and the cells were washed once with PBS. One milliliter 0.8% crystal violet-3.7% formaldehyde solution was added, and the plate was returned to the 37°C incubator for overnight incubation before counting. Excess stain was removed by washing with deionized water, and plaques were enumerated.
MB design.
The design of MB H1 was based on an alignment of the sequences of 26 different HAV strains obtained from GenBank. The DNA folding program mfold (www.bioinfo.rpi.edu/applications/mfold/dna/) and IDT SciTools (www.idtdna.com/SciTools/SciTools.aspx) were used to predict the thermodynamic properties and the secondary structures of MBs. MB H1 (5′-6-carboxyfluorescein-CTTGGGCCGCCGCTGTTACCCTATCCCCCAAG-DABCYL-3′ [the stem sequence is underlined]) was designed to be specifically hybridized to a 20-bp region (421 to 480 bp of the genome) of the 5′ untranslated region of HAV genomic RNA. The thermal denaturation profile was determined on an iCycler iQ multicolor real-time PCR detection system (Bio-Rad, Hercules, CA) by monitoring the fluorescence of a 50-μl volume of a solution containing 5 μM MB H1 dissolved in 100 mM Tris-HCl (pH 8.0) containing 1 mM MgCl2 as the temperature was decreased from 90°C to 10°C at a rate of 1°C/min. The fluorescence of 50 μl of a solution of 5 μM MB H1/oligonucleotide hybrids was also monitored for comparison. The probe 5′-6-carboxyfluorescein-CGCTATGCATCCGGTCAGTGGCAGTATAGCG-DABCYL-3′ was used for the nonspecific MB study.
Fixation, permeabilization, and incubation.
FRhK-4 cells were cultured to confluence in eight-well Lab-Tek II CC2 chamber slides (Fisher Scientific, NJ) at 37°C in 5% CO2. After incubation for predetermined time periods, the slides were removed from the 37°C incubator. The medium was aspirated, and the cells were washed twice with TBSS. The cells were fixed with 2% (wt/vol) paraformaldehyde in TBSS for 20 min at room temperature, washed three times with TBSS, and then permeabilized with 0.1% Triton X-100 in TBSS for 5 min at 4°C. Following three washes with TBSS, the cells were incubated with 5 μM MB H1 for 1 h at room temperature in the dark. Later, the slides were washed twice with TBSS and observed under a fluorescence microscope with the chambers containing TBSS.
Fluorescence microscopy and image processing.
The optical setup for fluorescence imaging consisted of a BX51 fluorescence microscope, a 100W Osram mercury burner, filter wheels for excitation filters, emission filters, diachronic mirrors, a 10×/0.30 objective lens and a QImaging Retiga EXI monochrome camera (Olympus America, Inc., San Diego, CA). Both phase-contrast and fluorescent images were taken and analyzed using Image-Pro Plus (Media Cybernetics, Inc., Bethesda, MD).
Enumeration of fluorescent cells.
A magnification of ×100 (10× objective and 10× eyepiece) was used for observing fluorescent cells. For each 0.7-cm2 glass chamber well, approximately 117 fields could be visualized at a magnification of ×100. To calculate the value for infected cells in each chamber well, both phase-contrast (to count the total number of cells) and fluorescent (to count the number of infected cells) images of 30 randomly chosen areas were taken at a magnification of ×100. The number of fluorescent cells within the chosen area was recorded by Image-Pro Plus software.
RESULTS AND DISCUSSION
Visualization of infected FRhK-4 cells using MBs.
MB H1 was designed to target a 20-bp common sequence of the 5′ untranslated region of 26 different HAV strains. In our previous study (17), fixation and permeabilization were used to introduce MBs into BGMK cells. To test whether the same procedure would work with FRhK-4 cells, MB H1/oligonucleotide hybrids were first introduced into uninfected cells. After 1 h of incubation, 100% of cells were brightly fluorescent, indicating that the same procedure works well even with FRhK-4 cells (Fig. 1, panels III). Next, the ability to detect HAV viral RNA was tested by introducing MB H1 into highly infected cells (105 PFU for 24 h) and uninfected cells. As expected, 100% of the highly infected cells were brightly fluorescent (Fig. 1, panels IV), while only background fluorescence was observed for the uninfected cells (Fig. 1, panels I). These results confirm that the MBs specifically recognize the HAV RNA. In addition, the introduction of a nonspecific MB that is not complementary to the HAV RNA produced a negligible amount of fluorescence (Fig. 1, panels II), demonstrating the specificity of the reported method.
FIG. 1.
Visualization of uninfected or infected FRhK-4 cells by introducing MBs. Cells were fixed and permeabilized before 1 h of incubation with 5 μM MB. (Panels I) Uninfected cells with MB H1. (Panels II) Highly infected (105 PFU) FRhK-4 cells at 24 h p.i. with a nonspecific MB. (Panels III) Uninfected cells with MB H1/oligonucleotide hybrids. (Panels IV) Highly infected (105 PFU) FRhK-4 cells at 24 h p.i. with MB H1. Scale bars, 100 μm.
Visualization of HAV replication.
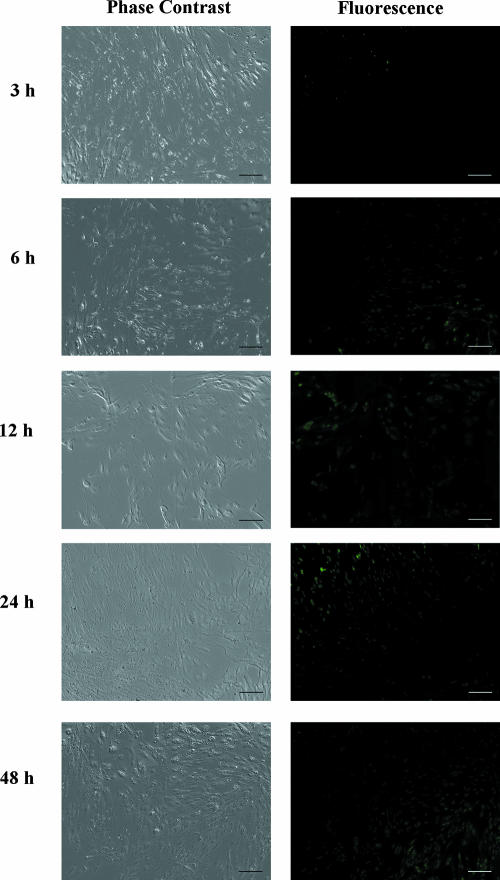
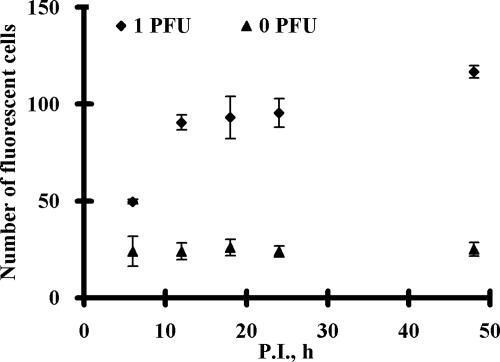
To investigate whether this approach can be used to rapidly detect low doses of infectious HAV, cultures infected with 1 PFU of HAV were analyzed from 3 to 48 h p.i. in order to investigate the minimum time required to consistently detect a positive fluorescent signal. As shown in Fig. 2, fluorescent cells can be visualized even within 6 h p.i. and the number of fluorescent cells increased with increasing infection times. The average number of fluorescent cells at different p.i. time points was calculated by direct counting using a fluorescence microscope at ×100 magnification to provide a better quantitative assessment. A significantly higher number of fluorescent cells (∼50) was detected for the infected cultures at 6 h p.i. than for the uninfected cultures (0 PFU), where roughly 20 fluorescent cells were detected independently of infection time (Fig. 3). The number of fluorescent cells increased rapidly from 50 to 90 from 6 to 12 h p.i. and remained relatively constant thereafter. This result is indicative of the fairly slow replication process for HAV, suggesting that a complete lytic cycle may take more than 48 h, consistent with previous reports (4, 5, 9, 10). Unlike the long lytic cycle, less than 6 h was required to detect the synthesis of HAV RNA, enabling the rapid detection of infectious HAV.
FIG. 2.
Visualization of FRhK-4 cells infected with 1 PFU at various p.i. time points. Cells infected with 1 PFU were fixed and permeabilized before 1 h of incubation with 5 μM MB H1. Scale bars, 100 μm.
FIG. 3.
The numbers of fluorescent cells at different p.i. time points. FRhK-4 cells infected with 0 or 1 PFU for 6, 12, 18, 24, and 48 h p.i. were fixed and permeabilized before 1 h of incubation with 5 μM MB H1. The number of fluorescent cells was counted using a fluorescence microscope at ×100 magnification. Error bars represent the standard deviations for three replicates.
Quantification of infectious HAV dosages.
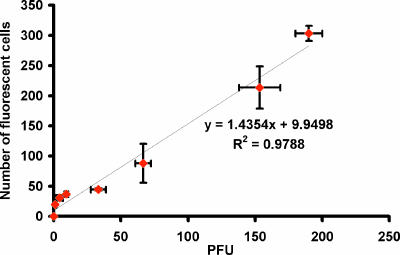
To investigate whether this 6-h infection window could be used to quantify infectious HAV, cells were infected with 1 to 200 PFU HAV and the average number of fluorescent cells was recorded. A linear correlation was obtained by plotting the number of fluorescent cells versus the number of PFU (Fig. 4), indicating that this MB-based fluorescence assay can be used as a quantification tool for detecting infectious HAV. The number of PFU can be calculated easily by using the correlation obtained after direct counting of fluorescent cells. More importantly, the method can be used to provide the rapid quantification of infectious HAV doses within 6 h p.i. compared to that with the 8-day incubation period for the conventional plaque assay.
FIG. 4.
The correlation between the number of fluorescent cells and the number of PFU at 6 h p.i. The fluorescent images from 30 different fields within a chosen chamber well were captured under a fluorescence microscope at ×100 magnification. The numbers of fluorescent cells were recorded and averaged. The 8-day plaque assay was performed to confirm the average infectious doses. Error bars represent the standard deviations for three replicates.
Detection of HAV in environmental water samples and comparison with plaque assay.
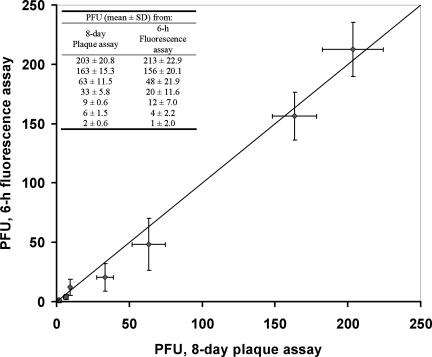
Using the 6-h infection window, the utility of the MB-based fluorescence assay for environmental samples was tested by using runoff water samples collected from Davis, CA, spiked with HAV. FRhK-4 cells were incubated with water samples spiked with different amounts of HAV, and the resulting infectious doses were determined based on the calibration curve shown in Fig. 4. In parallel, the conventional 8-day plaque assay was carried out to determine the infectious doses. The infectious dosages obtained from the two assays were remarkably similar (Fig. 5, table inset), with a maximum of 25% difference, validating the utility of the assay for viral quantification in real water samples.
FIG. 5.
Comparison of infectious HAV doses determined by the conventional 8-day plaque assay and the MB-based fluorescence assay. FRhK-4 cells infected with unknown viral doses for 6 h were fixed and permeabilized before 1 h of incubation with 5 μM MB H1. The number of fluorescent cells from 30 different fields within a chosen chamber well was recorded, and the PFU value was calculated based on the calibration equation from Fig. 4. In parallel, the infectious viral doses were determined independently by using the 8-day plaque assay. Error bars represent the standard deviations (SD) for three replicate experiments. The inset shows the infectivity data obtained from two assays.
In conclusion, the reported fluorescence assay combines a 32-fold reduction in detection time with a quantitative capability similar to that for the 8-day conventional plaque assay. The reported MB-based fluorescence assay is particularly attractive for environmental viral quantification, which focuses particularly on the lower infectious doses.
Acknowledgments
This work was funded by a grant from USEPA (R833008).
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Andino, R., N. Boddeker, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore, D. 1971. Expression of animal virus genomes. Bacteriol. Rev. 35:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratu, D. P., B.-J. Cha, M. M. Mhlanga, F. R. Kramer, and S. Tyagi. 2003. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA 100:13308-13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cromeans, T., M. D. Sobsey, and H. A. Fields. 1987. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J. Med. Virol. 22:45-56. [DOI] [PubMed] [Google Scholar]
- 5.Cromeans, T., H. A. Fields, and M. D. Sobsey. 1989. Replication kinetics and cytopathic effect of hepatitis A virus. J. Gen. Virol. 70:2051-2062. [DOI] [PubMed] [Google Scholar]
- 6.Cui, Z. Q., Z. P. Zhang, X. E. Zhang, J. K. Wen, Y. F. Zhou, and W. H. Xie. 2005. Visualizing the dynamic behavior of poliovirus plus-strand RNA in living host cells. Nucleic Acids Res. 33:3245-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake, T. J., and W. Tan. 2004. Molecular beacon DNA probes and their bioanalytical applications. Appl. Spectrosc. 58:269-280. [DOI] [PubMed] [Google Scholar]
- 8.Fang, X., J. J. Li, J. Perlette, W. Tan, and K. Wang. 2000. Molecular beacons: novel fluorescent probes. Anal. Chem. 72:747A-753A. [DOI] [PubMed] [Google Scholar]
- 9.Flehmig, B. 1980. Hepatitis A virus in cell culture. I. Propagation of different hepatitis A virus isolates in a fetal rhesus monkey kidney cell line (FRhK-4). Med. Microbiol. Immunol. 168:239-248. [DOI] [PubMed] [Google Scholar]
- 10.Flehmig, B. 1981. Hepatitis A virus in cell culture. II. Growth characteristics of hepatitis A virus in FRhK-4/R cells. Med. Microbiol. Immunol. 170:73-81. [DOI] [PubMed] [Google Scholar]
- 11.Galil, K. H. A. E., M. A. E. Sokkary, S. M. Kheira, A. M. Salazar, M. V. Yates, W. Chen, and A. Mulchandani. 2004. Combined immunomagnetic separation-molecular beacon-reverse transcription-PCR assay for detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 70:4371-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel, G., A. Kumar, A. K. Puniya, W. Chen, and K. Singh. 2005. Molecular beacon: a multitask probe. J. Appl. Microbiol. 99:435-442. [DOI] [PubMed] [Google Scholar]
- 13.Koch, F., and G. Koch. 1985. The molecular biology of poliovirus. Springer-Verlag, New York, NY.
- 14.Melnick, J. L. 1982. Classification of hepatitis A virus as enterovirus type 72 and of hepatitis B virus as hepadnavirus type 1. Intervirology 18:105-106. [DOI] [PubMed] [Google Scholar]
- 15.Minor, P. D., F. Brown, E. Domingo, E. Hoey, A. King, N. Knowles, S. Lemon, A. Palmenberg, R. R. Rueckert, G. Stanway, E. Wimmer, and M. Yin-Murphy. 1995. Family Picornaviridae. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy. Sixth report of the International Committee for the Taxonomy of Viruses. Springer-Verlag, New York, NY.
- 16.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 17.Wang, A., A. M. Salazar, M. V. Yates, A. Mulchandani, and W. Chen. 2005. Visualization and detection of infectious coxsackievirus replication using a combined cell culture-molecular beacon assay. Appl. Environ. Microbiol. 71:8397-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]