Abstract
A fungal mycelium is typically composed of radially extending hyphal filaments interconnected by bridges created through anastomoses. These bridges facilitate the dissemination of nutrients, water, and signaling molecules throughout the colony. In this study, we used targeted gene deletion and nitrate utilization mutants of the cruciferous pathogen Alternaria brassicicola and two closely related species to investigate hyphal fusion (anastomosis) and its role in the ability of fungi to cause disease. All eight of the A. brassicicola isolates tested, as well as A. mimicula and A. japonica, were capable of self-fusion, with two isolates of A. brassicicola being capable of non-self-fusion. Disruption of the anastomosis gene homolog (Aso1) in A. brassicicola resulted in both the loss of self-anastomosis and pathogenicity on cabbage. This finding, combined with our discovery that a previously described nonpathogenic A. brassicicola mutant defective for a mitogen-activated protein kinase gene (amk1) also lacked the capacity for self-anastomosis, suggests that self-anastomosis is associated with pathogenicity in A. brassicicola.
Fungal colonial growth is often described as long branching filaments; however, in most cases it is better depicted as net-like or an interconnected web. A fungal colony (mycelium) is regarded as an individual and typically establishes itself and grows by hyphal extension and branching as the organism explores the surrounding substrate. The process of vegetative hyphal fusion, or anastomosis, is a fundamental activity for the vast majority of filamentous fungi (9, 10, 15, 22, 26). The capacity of individual hypha within a colony to recognize, grow toward, and anastomose with each other (self-fusion or self-anastomosis) facilitates the interconnectedness between sectors of the organism that enables nutrient distribution (13) and the transduction of chemical signals in response to the environment (21). However, the role of self-anastomosis in phytopathogenic fungi is largely unknown.
Recent studies of Neurospora crassa suggest that prior to self-anastomosis, a complex interplay of extracellular signaling occurs among hyphae, resulting in directed growth (homing) and subsequent fusion (12, 23). Targeted mutagenesis of an N. crassa gene termed so (“soft”) resulted in a loss of preanastomosis homing and, consequently, a failure of adjacent hyphae to fuse (7). In addition to this cellular fusion defect, the so mutant exhibited shortened aerial hyphae, an altered conidiation pattern, and female sterility. Further characterization has revealed that the SO protein localizes to septal plugs within mycelia that are undergoing developmental changes associated with sexual structures or a programmed cell death response to non-self-recognition (8). The precise function of the SO protein is thus far unknown, but it contains a double-tryptophan (WW) domain that is commonly involved in mediating protein-protein interactions by recognizing proline-rich ligands (17). It has been proposed that so encodes part of the biochemical machinery involved in the synthesis and/or perception of an extracellular chemoelicitor leading to hyphal homing (7).
The SO protein hypothetically functions via a phosphorylation cascade resulting in the transcription of genes required for anastomosis. Several mitogen-activated protein kinase (MAPK) mutants have been created in N. crassa that exhibit pleiotropic phenotypes, including the loss of vegetative hyphal fusion (20, 29). MAK-2 (encoding a MAPK) and NRC-1 (encoding a MAPK kinase kinase) are essential for self-anastomosis in N. crassa (20, 23). Orthologs of these two genes are found in Saccharomyces cerevisiae (STE11 and FUS3), where they function in a signal transduction cascade that is activated by a pheromone during the mating process (4, 6). A proposed model of the molecular underpinnings of anastomosis in N. crassa (22) shows the initiation of specialized anastomosis hyphae called conidial anastomosis tubes (CATs [23]) via the MAK-2 MAPK pathway. The initiation of CATs is followed by the production of the SO protein and the homing of adjacent CATs toward one another. Homing is hypothetically mediated by a protein (HAM-2) containing a predicted transmembrane domain (30, 31).
While almost all of what is known about self-anastomosis has been learned from studying N. crassa, non-self-anastomosis has been examined using a wide variety of fungal systems. The prevalence of studies of non-self-anastomosis, defined as the vegetative fusion between genetically distinct fungal individuals, has risen since it was realized that many fungi limit such events through the execution of genetically programmed cell death pathways involving diverse allelic and/or nonallelic loci (9, 15). Although precontact signaling is likely important in non-self encounters (30), they are primarily defined by the sequence of events following fusion. Fungal individuals exhibiting genetic identity with one or more “vegetative compatibility” genes can anastomose successfully to exchange cytoplasm and genetic information, leading to their circumscription as members of the same vegetative compatibility group (VCG). Alternatively, individuals differing at one or more critical vegetative compatibility loci will anastomose, but the resulting fused hyphae will quickly collapse following DNA degradation and cytoplasmic shrinkage (9).
Alternaria brassicicola is a common necrotrophic pathogen of a variety of crucifers. No sexual stage has been identified thus far for A. brassicicola or the vast majority of Alternaria species (11, 28). Here, we initiated a study of anastomosis and vegetative compatibility with A. brassicicola, using a combination of genome mining, targeted gene deletion, and analysis of nitrate utilization (nit) mutants. We evaluated a previously described pathogenicity mutant (amk1) (1) for its ability to self-anastomose and disrupted the homolog of the N. crassa so gene in A. brassicicola. In both instances, mutants were defective in self-anastomosis and pathogenicity on cabbage, strongly suggesting that anastomosis is necessary for the full virulence of this fungal necrotroph.
MATERIALS AND METHODS
Generation and scoring of nit mutants.
The isolates used in this study are listed in Table 1. Cultures were maintained on potato dextrose agar. nit mutants are extremely useful for visualizing anastomosis (e.g., see references 14 and 16) and for assigning isolates to VCGs. Here, nit mutants were generated by placing four mycelial plugs per plate on minimal medium amended with 3% potassium chlorate (KClO3). Plates were incubated for 2 to 3 weeks at 25°C, after which time putative nit mutants were identified as fast-growing, appressed mycelia radiating from slow-growing colonies. nit mutations were identified on minimal medium with a defined nitrogen source as described previously (3). Cultures were grown for 1 to 2 weeks, and putative nit mutants were scored for either appressed or wild-type growth on each medium source. Colonies scored as true nit mutants (nit1, nit3, or nitM) were maintained on minimal medium amended with 3% potassium chlorate. The same procedure was used to generate nit mutations in an aso1 disruption mutant background.
TABLE 1.
Isolates included in this study
| Fungal species | Isolate | Origin | Mating typea | Host | Accession numberb |
|---|---|---|---|---|---|
| A. brassicicola | Ab1 | California | MAT1-2 | Brassica oleracea | NA |
| A. brassicicola | Ab2 | California | MAT1-2 | Brassica oleracea | NA |
| A. brassicicola | Ab4 | Lampoc, California | ND | Brassica oleracea var. botrytis | NA |
| A. brassicicola | Ab5 | Chualar, California | MAT1-2 | Brassica oleracea var. botrytis | NA |
| A. brassicicola | Ab6 | Texas | MAT1-2 | Brassica oleracea | NA |
| A. brassicicola | Ab7 | Oregon | MAT1-2 | Brassica oleracea | NA |
| A. brassicicola | 34622 | Unknown | MAT1-1 | Crambe sp. | ATCC 34622 |
| A. brassicicola | 96836 | United States | MAT1-1 | Brassica oleracea | ATCC 96836 |
| A. japonica | Ajap1 | New Zealand | ND | Brassica pekinensis | ATCC 96834 |
| A. mimicula | Amim1 | Georgia | MAT1-1 | Lycopersicon esculentum | ATCC 12251 |
ND, not determined.
NA, not applicable.
Nit complementation testing.
To test for self-anastomosis within strains of A. brassicicola, A. japonica and A. mimicula, nit mutants complementary for each isolate (nit1 versus nit3, nit1 versus nitM, and nit3 versus nitM) were opposed to one another on minimal medium amended with NaNO3 and allowed to grow together for 1 to 2 weeks. Anastomosis events were visualized as the restoration of prototrophic growth in the zone of interacting hyphae. Similarly, non-self-anastomosis was tested between the complementary nit mutants of each isolate. All possible combinations were made with two replicates.
Stability of anastomosis-derived prototrophic growth.
The stability of heterokaryons derived from anastomosis events was evaluated by grinding mycelia and spores from prototrophic zones emerging between complementary self-anastomosing nit mutants of three isolates (Ab4, Ab5, and Ab6) and then collecting single spores or hyphal tips on nitrate medium in a 24-well format. This procedure was repeated, with prototrophic colonies arising in the 24-well plate, and followed for three rounds. Agar blocks from revertants to appressed growth were isolated and cultured on all four medium types with defined nitrogen sources to rescore their nit phenotype.
Cloning of Aso1.
A partial So gene homolog (Aso1) was identified in the A. brassicicola genome data by sequence homology to that of Neurospora crassa. To isolate the complete sequence of Aso1, a combination of genome walking (Clontech, Mountain View, CA) and degenerate PCR was utilized to link together neighboring genomic contigs of isolate Ab96866 containing the entire inferred So gene homolog. Primers were designed 5′ and 3′ of the inferred promoter and the stop codon, respectively, and were also used to amplify the entire Aso1 gene from strain Ab7. The PCR product was purified using a QIAquick PCR purification kit (Qiagen, Inc.) and sequenced using BigDye version 1.1 chemistry (Applied Biosystems, Foster City, CA) and a model 3700 genetic analyzer (Applied Biosystems), using the manufacturer's protocols at the Genome Research Laboratory, North Carolina State University.
Strains and media.
A. brassicicola strains Ab7 and Ab96866 were grown on potato dextrose agar (PDA) (Difco). For protoplast isolation, the fungus was grown on GYB medium (l0 g glucose, 5 g yeast extract per liter). Solid RM (1 M sucrose, 0.1% yeast extract, 0.1% casein amino acids, and 1.5% agar) with 20 μg/ml hygromycin B (Roche, Indianapolis, IN) was used for the selection of transformants. Modified Richard's minimal medium (10 g KNO3, 5 g KH2PO4, 2.5 g MgSO4, 20 g sucrose, 1 g yeast extract, 15 g agar per liter), potato dextrose agar (40 g per liter), and 1% water agar (10 g crude agar per liter) were used for in vitro growth characteristics of wild type and mutants.
Transformation and disruption of Aso1.
Primers were designed to amplify a 466-bp fragment from the 5′ region of the Aso1 open reading frame (ORF) and to incorporate EcoRI and BamHI sites. PCR products amplified from genomic DNA were ligated into pCB1636 (27) after digestion with the appropriate enzymes and were transformed into subcloning efficiency DH5α-competent cells (Invitrogen, Carlsbad, CA). Protoplasts of A. brassicicola strain Ab7 were isolated and transformed using the linear minimal element technique as previously described (1). This technique, developed with A. brassicicola, homologously incorporates a linear piece of DNA with as few as 250 bp of homologous sequence, followed by a selectable marker, into the target loci in over 90% of transformed nuclei. The result is disruption of the recipient gene by the hygromycin resistance gene. Transformants were confirmed by amplifying the hygromycin resistance gene. Disruptants were identified by Southern hybridization; genomic DNA was digested with XhoI and probed with Aso1 DNA from the 5′ end of the gene, outside the disruption cassette, amplified with primers So-seq-F5 and So-seq-R5 (XCXXXX) to identify an XhoI fragment size shift resulting from the incorporation of the hygromycin B resistance gene. Ectopic mutants were confirmed as having a wild-type XhoI fragment, by Southern hybridization, and as PCR positive for the hygromycin B resistance gene.
Complementation with a functional Aso1 gene.
In order to reintroduce a functional Aso1 gene into the aso1 mutant, the wild-type Aso1 allele was amplified from genomic DNA of A. brassicicola strain ATCC 96866, using the primer set So1whF (CCTTTTGTCTCCAACCCAGA) and NRFcSo1whR (TGTGTTTGTTTCCAAGAAAAGACCCATCGCTGGAATAGAAGA). This PCR product was 5,560 bp in length and included the region 1,669 bp upstream of the predicted start codon and 59 bp downstream in relation to the stop codon to ensure isolation of the native promoter and terminator. Concurrently, a 2,076-bp-long nourseothricin-resistant cassette was amplified using the primer set So1whRcpNRF (TCTTCTATTCCAGCGATGGGTCTTTTCTTGGAAACAAACACA ) and pNRR (TCATTCTAGCTTGCGGTCCT ) from the pNR plasmid as template DNA (18). The final construct for transformation was amplified by using two primers, So1whF and pNRR, and the mixture of the two PCR products as template DNA, according to the manufacturer's instructions for an AccuPrime polymerase kit (Invitrogen, Carlsbad, CA). The final products were purified by using a PCR purification kit (Qiagen, CA), and 10 μg of constructs was transformed into the aso1-10 mutant. Putative transformants were selected on PDA plates containing both hygromycin B and nourseothricin antibiotics during transformation and two rounds of single conidium isolation.
Microscopic analysis.
Mycelial blocks from each wild-type strain were inoculated onto water agar plates overlaid with sterile cellophane opposite a different wild type and allowed to grow together. Cellophane with mycelia was excised and placed on a microscope slide with a drop of sterile water or aniline blue and scanned for anastomosis events. A green fluorescent protein (GFP)-labeled mutant of strain ATCC 96866 was opposed to each wild type and analyzed similarly. To evaluate evidence for the presence of CATs, conidia of each wild type (1 × 106 conidia ml−1) were placed on water agar plates and incubated for 4, 8, and 24 h. At each time point, agar blocks were excised and examined for the presence of CATs and anastomosis events.
Preparation of conidia.
Wild-type A. brassicicola and mutant strains were grown on PDA at 28°C in the dark for 2 days and then transferred to fluorescent white light for 8 h, followed by 16 h of darkness for 1 to 2 days to induce sporulation. Sterile water was added to plates, and the resulting conidial suspensions were collected. Conidia were quantified by using a hemocytometer and resuspended in sterile water at 1 × 105 conidia ml−1.
Pathogenicity assays.
A detached leaf assay and 5-week-old plants were used for pathogenicity assays. For the detached leaf assay, fresh 10- to 20-cm leaves were harvested from 8-week-old cabbage plants (cv.'s “Early Green” and “Super Red”) grown in the greenhouse. Two leaves were placed abaxial side up on a wire mesh, which was suspended above a paper towel impregnated with sterile ionized water in a clear polystyrene box (26.67 by 15.24 by 3.81 cm; Althor Products, Bethel, CT) with a hinged lid. One 10-μl drop of conidial suspension (1 × 105 conidia ml−1) of each strain (wild type, Aso1-7E (ectopic), aso1-10a, aso1-11a, and aso1-14a) was placed on each leaf. Incubation boxes were left at room temperature (approximately 25°C), and symptom formation was monitored at 6, 12, and 18 days after inoculation. A total of five boxes were set up for each cultivar.
Phenotypic characterization.
Mycelial plugs were grown on Richard's minimal medium, PDA, and water-agar. Colony growth radii were measured and recorded every other day. Conidia were counted with a hemacytometer, by taking mycelial plugs suspended in water that had been vortexed after 72 h of incubation at 25°C. Tests were performed in triplicates and analyzed by standard t tests.
Nucleotide sequence accession number.
The complete coding sequence of the Aso1 gene from A. brassicicola has been submitted to GenBank and is assigned accession number EF989017.
RESULTS
Generation of nit mutants and complementation testing.
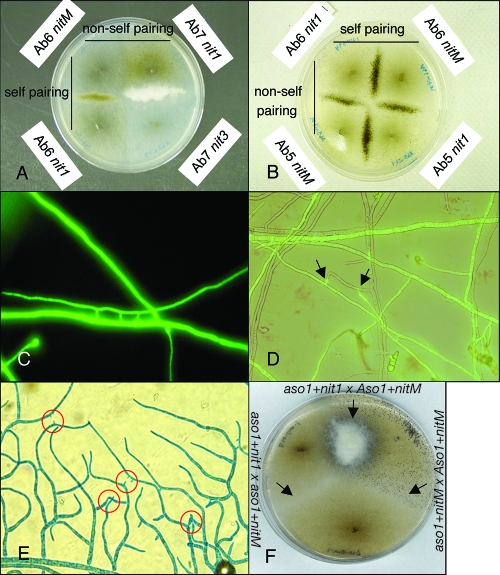
We generated a minimum of 10 nit mutants for each of 10 fungal strains and subsequently characterized them as nit1, nit3, or nitM based upon their growth phenotypes on minimal media containing distinct nitrogen sources. In all cases, complementary nit mutants generated from the same wild-type isolate were able to anastomose and produce prototrophic hyphal growth similar to that of the wild type (Fig. 1A). In contrast, most pairings between complementary nit mutants derived from different wild-type isolates did not result in prototrophic growth, indicating that non-self-anastomosis is restricted. However, one pairing (A. brassicicola strains Ab5 and Ab6) consistently yielded prototrophic hyphae, indicative of non-self-anastomosis (Fig. 1B).
FIG. 1.
Anastomosis phenotypes within and between A. brassicicola strains. (A) Prototrophic growth indicative of anastomosis between complementary nit mutants of the same A. brassicicola isolates (self-pairing). The lack of prototrophic growth between complementary nit mutants of different isolates (non-self-pairing) indicates a failure to anastomose. (B) Anastomosis within and between A. brassicicola strains Ab5 and Ab6. (C) The GFP-labeled strain ATCC 96836 showing “laddering phenotype” of anastomosed hyphae. (D) Rare interisolate fusion between typically incompatible isolates ATCC 96836-GFP and unlabeled Ab4, where the GFP signal is migrating into Ab4 hyphae (arrows). (E) Hyphae of strain Ab5 showing the initiation of anastomosis “pegs” in response to similar structures in adjacent hyphal strands. (F) Pairing of aso1/nit single and/or double mutants on nitrate media. Interacting hyphae involving complementary nit mutants, but both defective in Aso1, show no evidence of prototrophic growth (side arrows), while interacting hyphae of complementary nit mutants do yield prototrophic growth if at least one has a functional Aso1 gene (top arrow).
Instability of anastomosis-derived prototrophy.
To discern whether homokaryotic polyploids derived from self-pairings (nit1 × nitM) were stable, we tracked the breakdown of prototrophy for three crosses. We defined stability as the maintenance of prototrophic growth, and reversion to the sparse, appressed growth habit typical of nit mutants as the evidence for genomic reduction (instability). For each self-complement tested, 96 single spores or hyphal tips were isolated from the prototrophic growth zone and plated on nitrate media. Of these, one prototrophic colony emerged for two of the isolates, and none for the third. The procedure was repeated, with a single prototrophic colony arising for one of the two remaining isolates. After the third round of isolation with the remaining prototrophic colony, no prototrophic growth was observed. Mycelial blocks were isolated from 20 colonies, with the appressed phenotype arising from the single spore or hyphal tip isolation and rescored on the defined nitrogen sources. Of these, fifteen had the nit1 phenotype and five had the nitM phenotype, showing the reassortment of nuclei and the reestablishment of a haploid state.
Microscopic visualization of hyphal interactions.
To investigate whether the lack of complementation and prototrophic growth observed in our nit mutant studies was due to a simple failure to anastomose or was potentially due to a postfusion cell death response, we examined each wild-type strain grown opposed to a GFP-labeled isolate (Fig. 1C) of strain ATCC 96866. Fusion was observed between hyphae of the same isolate but very rarely between hyphae of the wild-type and the GFP strain. A rare exception is shown in Fig. 1D; not only did the hyphae fuse, but the GFP appears to have been translocated into the wild-type hyphae of Ab4 (Fig. 1D, arrows). With the exception of these events, non-self-hyphae appeared to intermingle and even grow over one another without any evidence of recognition or adverse effects. We observed that most self-fusion events were initiated by branching of one hypha, followed by elicitation of a similar branch initiation in the adjacent hyphae (Fig. 1E). Homing oriented the branches to grow toward each other, eventually leading to fusion. This response was completely lacking in non-self-hyphal interactions. Homing was also not observed between germinated conidia, and we were unable to find evidence for the formation of CATs that have been described for other fungal species (23). Self-anastomosis events in Alternaria brassicicola appear to be confined to the interior regions of mature colonies.
Molecular characterization of Aso1.
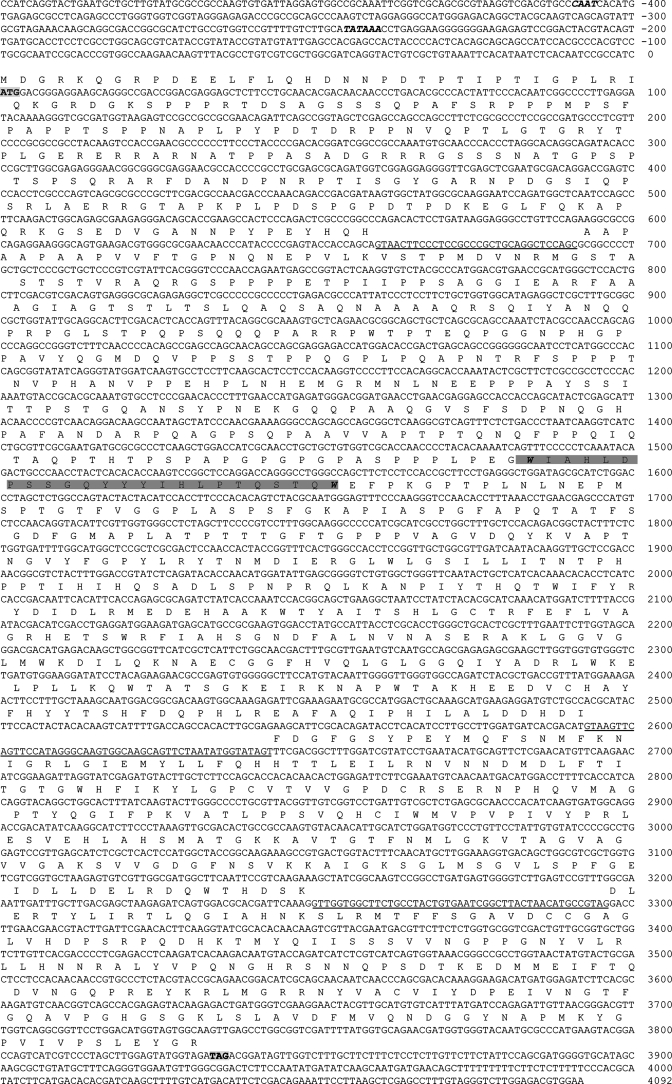
An initial homology search of the A. brassicicola genome, using the so gene sequence from N. crassa, revealed that a partial ORF that was truncated at the 5′ end due to nonoverlapping supercontigs in the genome assembly. To obtain the remaining portion of the A. brassicicola so homolog (Aso1), we created degenerate primers based upon the aligned so homologs from other filamentous fungi and amplified a fragment of genomic DNA that we then used to probe an A. brassicicola genomic bacterial artificial chromosome library. Two clones were retrieved and sequenced to yield the entire Aso1 ORF and 5′-untranslated region. Based upon the inferred promoter regions and start and stop codons, the entire ORF is 3,836 bp in length and contains four predicted exons, with three introns predicted to be 31 bp, 50 bp, and 47 bp in length (Fig. 2). The Aso1 gene has two tryptophan residues at positions 1583 and 1649. The alignment of the inferred amino acid sequence of Aso1 to homologs identified in 10 additional filamentous fungi revealed striking differences in homology between the 5′ and 3′ ends of the gene. The amino end of the protein shares roughly 45% identity with homologs from other fungi to which it was compared, while the carboxyl end shares, on average, greater than 85% identity (data not shown). The single identifiable domain in the ORF (WW; double tryptophan), located just within the amino end, is highly conserved among filamentous fungi (three nonsilent substitutions).
FIG. 2.
Nucleotide and amino acid sequence of the inferred Aso1 ORF. The predicted translational start and stop codons are in bold and shaded in gray, respectively. The inferred CAAT and TATA promoter elements are in bold italics. Introns are underlined. The double-tryptophan domain is shaded in dark gray, bounded with two tryptophan residues (bold italic).
Generation of Aso1 mutants.
Protoplasts of A. brassicicola strain Ab7 were isolated and transformed as described previously (1). A total of 12 putative transformants (designated aso1-1 through aso1-12) were obtained and purified further by transferring a single spore to a hygromycin B-containing medium and renaming the products aso1-1a through aso1-12a. PCR indicated gene disruption for all transformants except for Aso1-7E (data not shown), and this was confirmed by Southern hybridization (Fig. 3). To confirm that Aso1-7E indeed resulted from an ectopic integration of the disruption cassette, the presence of the hygromycin B resistance gene was confirmed by PCR (data not shown), and the presence of the wild-type Aso1 allele was shown by PCR (data not shown) and Southern hybridization (Fig. 3).
FIG. 3.
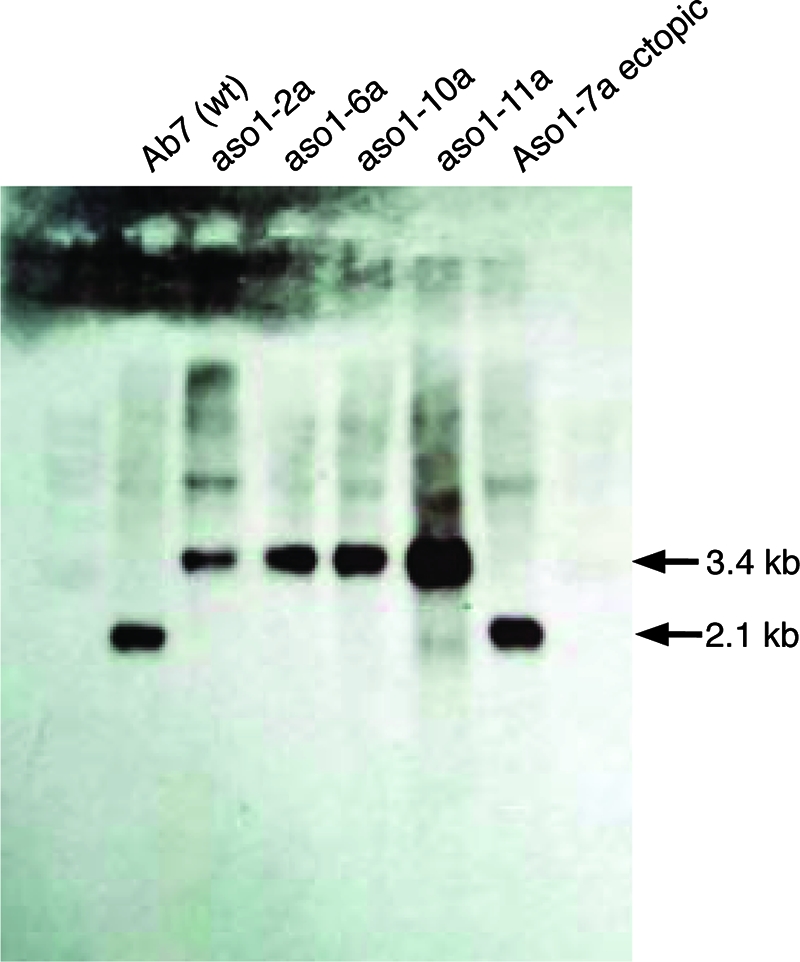
Autoradiograph of a DNA gel blot of XhoI-genomic digests of A. brassicicola wild-type Ab7 and aso1 mutants (aso1-2a, aso1-6a, aso1-10a, and aso1-11a) and the ectopic transformant Aso1-7E probed with the Aso1 gene fragment labeled with an ECL direct nucleic acid labeling and detection system (Amersham Biosciences, Piscataway, NJ).
Phenotypic evaluation of aso1 mutants.
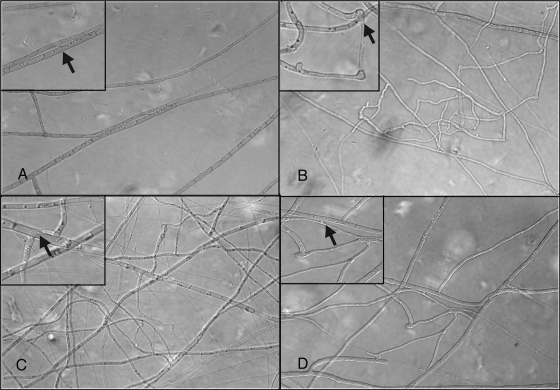
In contrast to observations with the wild-type isolates or the ectopic transformant Aso1-7E, homing and self-fusion were not observed for the colony periphery or interior of all aso1 mutants (Fig. 4). Despite this lack of anastomosis, growth rates, pigmentation, and sporulation of the mutants were not significantly different from ectopic or wild-type strains (data not shown). We generated nit1 and nitM mutants in the aso1 mutant background (aso1 nit1 and aso1 nitM). When the complementary aso1 nit double mutants were opposed on nitrate media, no prototrophic growth was observed. Interestingly, when either double mutant was paired with the complementary nit mutant of the wild-type strain, prototrophic growth was observed in some cases (Fig. 1F), suggesting that only one isolate with a functional Aso1 allele is required for anastomosis.
FIG. 4.
Anastomosis phenotype of A. brassicicola aso1 mutants. (A) Wild type; (B) aso1-1a; (C) aso1-9a; (D) aso1-7E (ectopic). Arrows indicate fusion events (panels A and D) or instances of close contact without fusion (panels B and C).
Pathogenicity of the aso1 mutant.
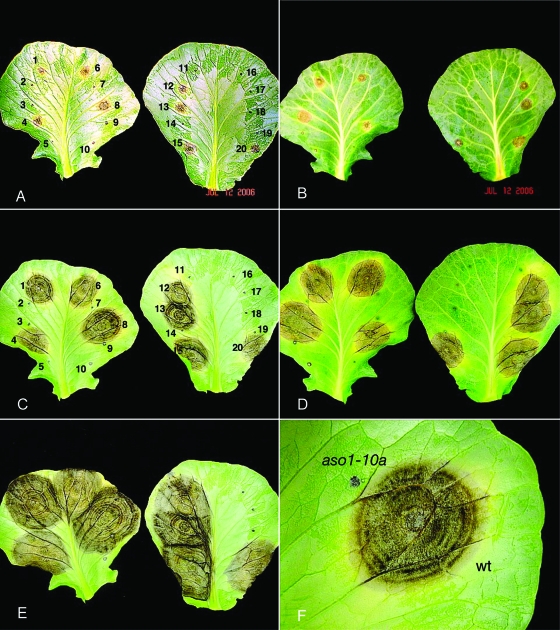
Spreading necrotic lesions typical of A. brassicicola were observed on cabbage leaves inoculated with the wild-type strain and the ectopic transformant Aso1-7E (Fig. 5). Necrosis was evident on the abaxial (underside) inoculated side and the adaxial side, and it spread rapidly from the initial site of infection. In contrast, all transformants with disrupted aso1 genes failed to spread from the site of initial infection (Fig. 5).
FIG. 5.
Pathogenicity assay with the A. brassicicola wild type, aso1 mutants, and ectopic transformant. (A, C, and E) Inoculated, abaxial surface at 6, 12 and 18 days postinfection; (B and D) adaxial (uninoculated) surface of the same cabbage leaves at 6 and 12 days postinfection; (F) wild type (wt) and aso1 lesions, at a magnification of ×2. Panel A and C isolate numbers are identified as follows: wild type, 1, 8, 13, and 15; aso1-10a, 2, 7, 14, and 17; aso1-11a, 5, 9, 11, and 19; aso1-14a, 3, 10, 16, and 18; aso1-7E (ectopic), 4, 6, 12, and 20.
Complementation of aso1.
Double mutants were selected on regeneration medium containing 20 μg hygromycin and 200 μg nourseothricin. Complementation of the aso1-10a mutant strain with the full-length Aso1 ORF from strain Ab96866 fully restored the capacity for self-anastomosis (Fig. 6) and pathogenicity on cabbage (not shown).
FIG. 6.
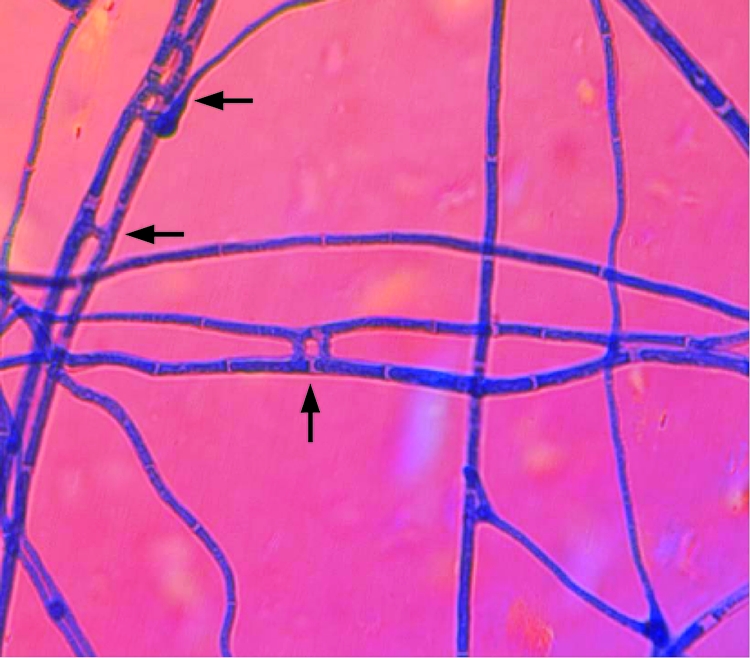
Anastomosis phenotype of complemented strain aso1-10. Arrows indicate self-fusion events.
DISCUSSION
We report here the first investigation of the phenomena of anastomosis and vegetative compatibility in the fungal pathogen Alternaria brassicicola and provide evidence for a correlation between self-anastomosis and pathogenicity. The disruption of the so homolog Aso1 in A. brassicicola results in severely restricted infectivity of the pathogen, which fails to spread from the site of inoculation. The lack of lesion development was not due to a failure to penetrate and initially colonize, as small lesions were evident that appeared on both the inoculated side and the underside of the leaf, indicating that some colonization had occurred. Following this initial growth, the wild-type and ectopic isolates continued to ramify through the leaf, while the growth of the aso1 mutants failed to progress. The only other detectable phenotype of this mutant was the inability to successfully home toward and fuse with adjacent hyphae from the same colony. As discussed further below, this phenotype is significantly different from that of the pleiotropic effects of the so mutation in N. crassa.
The failure of the aso1 mutants to necrotize enough host tissue to sustain a progressive infection may very well result from an inability to translocate sufficient nutrition or signaling molecules throughout the fungal colony. In contrast to biotrophic infections, necrotrophs must secrete compounds that cause host cell death in advance of colonization. In several Alternaria species, particularly the A. alternata sp. group, a diverse array of host-selective toxins facilitates this tissue death through various mechanisms that typically mimic components of, or actively disrupt, normal plant programmed cell death pathways. Although there have been reports of nonhost-specific toxic secondary metabolites, as well as the production of a host-selective protein toxin by A. brassicicola (19), the precise nature of these compounds remains obscure. Regardless, the lack of anastomosis observed with the aso1 mutants may undermine the synthesis and secretion of the toxin(s) and/or its translocation through the mycelium, resulting in a failure to necrotize sufficient amounts of host tissues to support infection.
A second line of evidence that anastomosis and pathogenicity are intimately linked in A. brassicicola is shown by our analysis of the amk1 mutant. AMK1 is a homolog of the pheromone response MAP kinases Fus3p and Kss1p in Saccharomyces cerevisiae and MAK-2 in N. crassa. Disruption of MAK-2 in N. crassa has been shown to result in a pleiotropic phenotype, including derepressed conidiation, shortened aerial hyphae, female sterility, autonomous ascospore lethality, and a lack of vegetative hyphal fusion (20). Previous work has shown that the amk1 mutant of A. brassicicola lacks pathogenicity on cruciferous hosts, does not conidiate, and has altered hyphal characteristics (1). Our work shows that amk1 is also similar to mak-2 in that the ability for self-anastomosis is lost (data not shown). Intriguingly, results obtained from reverse transcription-PCR show that Aso-1 is expressed constitutively in culture and in vivo, even in the amk1 mutant background (data not shown). This suggests that Aso1 is not directly dependent upon Amk1 and that the homing response mediated by the former is distinct from the initiation of hyphal anastomosis mediated by the latter.
A failure to create interconnected webs of self-anastomosed hyphae may prohibit a fungus from producing structures of increasing complexity and organization, such as asexual and sexual fruiting bodies (5), rhizomorphs, and sclerotia. The dense tissues that characterize such structures are almost certainly composed of fused hyphae that may serve to impart structural integrity and resistance to environmental degradation. It is interesting to note that naturally occurring self-incompatible “mutants” have been described for several species of Fusarium (15). Although the mutated gene(s) was never identified, sexual crosses between self-anastomosing and nonanastomosing strains of Fusarium moniliforme yielded meiotic progeny segregating 1:1 for the nonanastomosing phenotype, suggesting that the trait was under the control of a single nuclear gene. Intriguingly, sexual crosses between two nonanastomosing strains could not be made because all such strains examined were sterile females. We propose that it may be the so homolog in Fusarium that has been disrupted in such instances and that the mutants have been rendered incapable of producing the fused hyphae necessary to form sexual, maternally derived fruiting bodies. Recently, the so homolog in Fusarium oxysporum (termed Fso1) was disrupted, and the mutant was slightly less virulent on tomato (24). However, like A. brassicicola, the teleomorphic stage of F. oxysporum has not been defined, and thus, the effects of eliminating anastomosis on the production of higher order structures (fruiting bodies) cannot be evaluated. We are currently pursuing gene disruption in other fungi that are known to have a sexual stage to evaluate this hypothesis.
Historically, the research of fungal anastomosis systems has focused on a very specific type of interisolate postfusion reaction, where incompatible unions result in cell death, a result that can be easily assayed macroscopically as a barrage zone. Although the number of Alternaria isolates we studied is limited, we have shown that non-self-anastomosis between these isolates is rare and does not involve a programmed cell death-like response to restrict such encounters. Our set of eight A. brassicicola isolates can be grouped into seven distinct anastomosis groups, with Ab5 and Ab6 belonging to the same compatibility group. While it would be premature to reject the notion of an incompatible response in A. brassicicola until additional isolates are investigated, we propose the possibility that pheromones or other external signals may regulate such events at the prefusion stage, with “incompatible” strains or species simply failing to recognize each other. In support of this, we have observed that subapical branching within a colony appears to stimulate similar branch initiation in adjacent hyphae as a prerequisite for anastomosis (Fig. 1C). This elicitation is completely lacking in non-self-encounters. The role of the Aso1 gene in non-self-interactions is unknown, although the disruption of so in Neurospora eliminates interisolate anastomoses between individuals of the same VCG (30). We suggest that these two temporally and genetically distinct recognition systems can operate in tandem, as in Neurospora; or, perhaps only the prefusion system mediated by Aso1 is active in Alternaria. If this is the case, the selective forces that result in the emergence of one or both of these recognition systems may differ substantially.
The A. brassicicola system also differs from that of N. crassa in that self-anastomosis does not occur between conidia (via CATs) or young germlings. All germ tubes emerging from conidia are relatively uniform in width and show no evidence of chemotropism toward other conidia or nearby germ tubes. Read and Roca (22) have stated that CATs are extremely prevalent among fungi and postulate two functions for CAT fusion: (i) sharing of heterogeneously distributed nutrients and water, leading to improved chances for colony establishment and (ii) gene transfer and nonmeiotic recombination in non-self-anastomoses. In support of the second notion, they find that the incompatible response typically resulting from heterokaryon formation is suppressed in very early stages of colony establishment. We propose that the early formation of hyphal networks may be unnecessary in A. brassicicola as it is a facultative saprophyte with an impressive arsenal of degredative enzymes at its disposal and it may be able to obtain nutrition from a wide variety of substrates easily and immediately after germination. Indeed, Alternaria spp. have been cultured from an astoundingly diverse array of substrates such as leather, sewage, stone monuments, optical instruments, and even jet fuel (25). Furthermore, we note that the mutant was fully capable of normal growth and development on various medium types and was able to initially penetrate host tissue. This is what we would expect if the cellulolytic and degredative capabilities of the aso1 mutant were not affected by its failure to anastomose. Once inside the plant, however, a progressing infection would require efficient toxin biosynthesis and secretion, and thus, the defective phenotype becomes readily apparent. As to the second potential benefit of CAT fusion proposed by Read and Roca (22), the authors imply that the strong selection imposed by an incompatibility response may have in turn selected for a brief relaxation of the response (during CAT formation and fusion) to allow a period of potential gene flow between otherwise incompatible genotypes. We have found no evidence of an incompatible response in A. brassicicola. Without such selection imposed by a vegetative incompatibility system, there may be no requirement to form early networks between germ tubes and/or CATs. In support of this hypothesis, a second genus of ascomycetous fungi, the Epichloë endophytes of grasses, also lack a vegetative incompatibility system (2) and do not form CATs (K. D. Craven, personal observation).
Elucidating the underlying mechanisms and regulation of vegetative anastomosis, as well as appreciating the potential diversity of mechanisms that facilitate such events, will greatly advance how we understand the biology, lifestyles, and evolution of these fungi, as well as those that are less experimentally tractable. This understanding may also enhance efforts to advance the quantity and quality of food crops worldwide. Homologs of Aso1 have only been identified in filamentous ascomycetes, albeit with various degrees of relatedness and distinct lifestyles. This suggests that this gene may play a more fundamental role in fungal biology. Interestingly, the 3′ portion of these Aso1 homologs shares a remarkable level of conservation at the amino acid level, while the 5′ end is quite variable (data not shown). Further dissection of this gene and the elucidation of functional domains beyond the highly conserved double-tryptophan domain will be illuminating.
Acknowledgments
We thank Barry Pryor (University of Arizona) for providing the A. brassicicola isolates. We thank Dennis Knudson and Susan Brown (Colorado State University) for providing an A. brassicicola bacterial artificial chromosome library. We thank Mauricio La Rota (Virginia Bioinformatics Institute) for bioinformatics analysis and Julie Macialac, Robyn Hicks, and Mark Leyer (North Carolina State University) for laboratory assistance. We also thank Rao Uppalati and Marilyn Roosinck (Noble Foundation) for critically reviewing the manuscript.
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant number 2004-35600-15030. The project was also supported by National Science Foundation award DBI-0443991.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Cho, Y., R. Cramer, K. Kwang-Hyung, J. Davis, T. Mitchell, P. Figuli, B. Pryor, E. Lemasters, and C. B. Lawrence. 2007. The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of hydrolytic enzyme genes in the fungus Alternaria brassicicola. Fungal Genet. Biol. 44543-553. [DOI] [PubMed] [Google Scholar]
- 2.Chung, K.-R., and C. L. Schardl. 1997. Vegetative compatibility between and within Epichloë species. Mycologia 89558-565. [Google Scholar]
- 3.Cove, D. J. 1976. Chlorate toxicity in Aspergillus nidulans: studies of mutants altered in nitrate assimilation. Heredity 36191-203. [DOI] [PubMed] [Google Scholar]
- 4.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3573-581. [DOI] [PubMed] [Google Scholar]
- 5.Engh, I., C. Würtz, K. Witzel-Schlömp, H. Y. Zhang, B. Hoff, M. Nowrousian, H. Rottensteiner, and U. Kück. 2007. The WW domain protein PRO40 is required for fungal fertility and associates with woronin bodies. Eukaryot. Cell 6831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farley, F. W., B. Satterberg, E. J. Goldsmith, and E. A. Elion. 1999. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics 1511425-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleissner, A., S. Sarker, D. J. Jacobson, M. G. Roca, N. D. Read, and N. L. Glass. 2005. The so locus is required for vegetative cell fusion and post-fertilization events in Neurospora crassa. Eukaryot. Cell 4920-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleissner, A. 2007. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 684-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass, N. L., C. Rasmussen, M. G. Roca, and N. D. Read. 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12135-141. [DOI] [PubMed] [Google Scholar]
- 10.Glass, N. L., and A. Fleissner. 2006. Re-wiring the network: understanding the mechanism and function of anastomosis in filamentous ascomycete fungi. p. 123-139. In U. Kües and R. Fischer (ed.), The mycota. 1. Growth, differentiation and sexuality. Springer-Verlag, Munich, Germany.
- 11.Hawksworth, D. L., P. M. Kirk, B. C. Sutton, and D. N. Pegler. 1995. Ainsworth and Bisby's dictionary of the fungi, 8th ed. CAB International, Wallingford, Oxfordshire, United Kingdom.
- 12.Hickey, P. C., D. J. Jacobson, N. D. Read, and N. L. Glass. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37109-119. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, H., G. P. Boswell, C. M. Scrimgeour, F. A. Davidson, G. M. Gadd, and K. Ritz. 2004. Translocation of carbon by Rhizoctonia solani in nutritionally-heterogenous microcosm. Mycol. Res. 108453-462. [DOI] [PubMed] [Google Scholar]
- 14.Jo, Y.-K., S. W. Chang, J. Rees, and G. Jung. 2008. Reassessment of vegetative compatibility of Sclerotinia homeocarpa using nitrate-nonutilization mutants. Phytopathology 98108-114. [DOI] [PubMed] [Google Scholar]
- 15.Leslie, J. F. 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31127-150. [DOI] [PubMed] [Google Scholar]
- 16.Liu, W., and L. Sundheim. 1996. Nitrate nonutilizing mutants and vegetative compatibility groups in Fusarium poae. Fungal Genet. Biol. 2012-17. [DOI] [PubMed] [Google Scholar]
- 17.Macias, M. J., S. Wiesner, and M. Sudol. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 51330-37. [DOI] [PubMed] [Google Scholar]
- 18.Malonek, S., M. C. Rojas, P. Hedden, P. Gaskin, P. Hopkins, and B. Tudzynski. 2004. The NADPH-cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem. 27925075-25084. [DOI] [PubMed] [Google Scholar]
- 19.Otani, H., A. Kohnobe, M. Kodama, and K. Kohmoto. 1998. Production of a host-specific toxin by germinating spores of Alternaria brassicicola. Physiol. Mol. Plant Pathol. 52285-295. [Google Scholar]
- 20.Pandey, A., M. G. Roca, N. D. Read, and N. L. Glass. 2004. Role of a MAP kinase during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayner, A. D. 1996. Interconnectedness and individualism in fungal mycelia. p. 193-232. In B. C. Sutton (ed.), A century of mycology. University of Cambridge Press, Cambridge, United Kingdom.
- 22.Read, N. D., and M. G. Roca. 2006. Vegetative hyphal fusion in filamentous fungi. p. 87-98. In F. Baluska, D. Volkmann, and P. W. Barlow (ed.), Cell-cell channels. Landes Bioscience, Austin, TX.
- 23.Roca, M. G., J. Arlt, C. E. Jeffree, and N. D. Read. 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosales, R. C. P., and A. Di Pietro. 2008. Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum. Eukaryot. Cell 7172-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotem, J. 1994. The genus Alternaria: biology, epidemiology and pathogenicity. APS Press, St. Paul, MN.
- 26.Saupe, S. J. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64489-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweigard, J., F. Chumley, A. Carroll, L. Farrall, and B. Valent. 1995. A series of vectors for fungal transformation. Fungal Genet. Newsl. 4452-53. [Google Scholar]
- 28.Thomma, B. P. H. J. 2003. Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4225-236. [DOI] [PubMed] [Google Scholar]
- 29.Wei, H. J., N. Requena, and R. Fischer. 2003. The MAPKK kinase SteC regulates conidiophore morphology and is essential for heterokaryon formation and sexual development in the homothallic fungus Aspergillus nidulans. Mol. Microbiol. 471577-1588. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, J. F., and J. A. Dempsey. 1999. A hyphal fusion mutant in Neurospora crassa. Fungal Genet. Newsl. 46:31. [Google Scholar]
- 31.Xiang, Q., C. Rasmussen, and N. L. Glass. 2002. The ham-2 locus, encoding a putative transmembrane domain, is required for hyphal fusion in Neurospora crassa. Genetics 160169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]