Abstract
In eukaryotes, acetyl coenzyme A (acetyl-CoA) produced during peroxisomal fatty acid β-oxidation needs to be transported to mitochondria for further metabolism. Two parallel pathways for acetyl-CoA transport have been identified in Saccharomyces cerevisiae; one is dependent on peroxisomal citrate synthase (Cit), while the other requires peroxisomal and mitochondrial carnitine acetyltransferase (Cat) activities. Here we show that the human fungal pathogen Candida albicans lacks peroxisomal Cit, relying exclusively on Cat activity for transport of acetyl units. Deletion of the CAT2 gene encoding the major Cat enzyme in C. albicans resulted in a strain that had lost both peroxisomal and mitochondrion-associated Cat activities, could not grow on fatty acids or C2 carbon sources (acetate or ethanol), accumulated intracellular acetyl-CoA, and showed greatly reduced fatty acid β-oxidation activity. The cat2 null mutant was, however, not attenuated in virulence in a mouse model of systemic candidiasis. These observations support our previous results showing that peroxisomal fatty acid β-oxidation activity is not essential for C. albicans virulence. Biofilm formation by the cat2 mutant on glucose was slightly reduced compared to that by the wild type, although both strains grew at the same rate on this carbon source. Our data show that C. albicans has diverged considerably from S. cerevisiae with respect to the mechanism of intracellular acetyl-CoA transport and imply that carnitine dependence may be an important trait of this human fungal pathogen.
β-Oxidation of fatty acids is a well-conserved metabolic process that results in the stepwise degradation of fatty acids to acetyl coenzyme A (acetyl-CoA). While this process takes place both in mitochondria and peroxisomes in mammalian cells, oxidation of fatty acids is exclusively peroxisomal in plant and fungal cells (16). To allow further metabolism, the acetyl-CoA produced must be transported from peroxisomes to the mitochondria, where it enters the tricarboxylic acid (TCA) cycle and is oxidized completely to CO2 and H2O. In the yeast Saccharomyces cerevisiae, two pathways for the transport of acetyl-CoA from peroxisomes to mitochondria have been identified; the first pathway depends on peroxisomal citrate synthase (Cit2p), while the second pathway requires carnitine acetyltransferase (Cat2p) (40). In the Cit2p-dependent pathway, the acetyl moiety of peroxisomal acetyl-CoA is linked to oxaloacetate by Cit2p to form citrate and free CoA. Citrate is subsequently transported to the mitochondria, where it enters the TCA cycle. In the Cat2p-dependent pathway, Cat2p catalyzes the formation of acetyl-carnitine from acetyl-CoA and the carrier molecule carnitine. Formation of acetyl-carnitine is necessary to allow the transport of acetyl units over the peroxisomal and mitochondrial membranes. Mitochondrial Cat2p catalyzes the reverse reaction, liberating the acetyl unit from carnitine and coupling it to a molecule of free CoA for further metabolism. Interestingly, in S. cerevisiae, the peroxisomal and mitochondrial forms of Cat2p are encoded by a single gene and the mechanism of differential targeting of the protein has been well established (6). The work of van Roermund et al. (39, 40) has shown that the Cit2p- and Cat2p-dependent pathways of acetyl-CoA export can work in parallel; disruption of either the CIT2 or the CAT2 gene in S. cerevisiae does not lead to a growth defect on fatty acids, while the simultaneous disruption of both genes does. While both pathways function in S. cerevisiae, plants seem to lack the Cat pathway, as disruption of both peroxisomal citrate synthases (CSY2 and CSY3) in Arabidopsis thaliana resulted in a mutant that failed to metabolize triacylglycerol (27).
In S. cerevisiae, acetyl-CoA transport can be studied independently of fatty acid β-oxidation. When yeast cells are growing on acetate or ethanol, acetyl-CoA is formed in the cytosol, which needs to be transported to the mitochondria and fed into the glyoxylate cycle. Transport of acetyl-CoA from the cytosol to mitochondria is probably mediated by one or both of the other carnitine acetyltransferases in S. cerevisiae, Yat1p and Yat2p. Yat1p is localized to the outer mitochondrial membrane (33), while the localization of Yat2p is unknown. The specific functions of either of the Yat proteins are not known; however, a random-mutagenesis screen of an S. cerevisiae cit2Δ strain (which is dependent on the Cat pathway for growth on oleate and ethanol) revealed that all three carnitine acetyltransferases (Cat2p, Yat1p, and Yat2p) are essential for growth on nonfermentable carbon sources such as acetate, ethanol, oleate, and glycerol (35).
The C. albicans genome encodes three putative carnitine acetyltransferases, homologs of S. cerevisiae Cat2p, Yat1p, and Yat2p, which have been named Ctn2p, Ctn1p, and Ctn3p, respectively (28). Mitochondrial and peroxisomal targeting signals are conserved in the C. albicans Cat2p homolog (Ctn2p), and two in-frame start codons are present, as is also the case in S. cerevisiae (6), suggesting that the mechanism of dual localization of Cat2p is conserved between the two yeasts. All three putative carnitine acetyltransferases are up-regulated during phagocytosis by macrophages (28).
To allow growth on fatty acids, ethanol, or acetate, not only must the acetyl-CoA produced be transported to the mitochondria for oxidation in the TCA cycle, but this C2 compound also needs to be converted to C4 units (succinate) that can be used for biosynthetic purposes. The metabolic pathway that allows the net synthesis of C4 units from acetyl-CoA is the glyoxylate cycle (14). The essential role of this metabolic pathway for the utilization of nonfermentable carbon sources has been substantiated through the analysis of fungal mutants lacking one of the key enzymes of the glyoxylate cycle, i.e., isocitrate lyase (Icl1p) or malate synthase (Mls1p). S. cerevisiae icl1 or mls1 mutants are unable to grow on fatty acids, ethanol, or acetate as the sole carbon source (7, 11, 21), and similar phenotypes have been reported for C. albicans mutants lacking Icl1p (20, 26). While the function of the glyoxylate pathway in eukaryotes is well conserved, the subcellular localization of the enzymes may vary from organism to organism. For example, in plants and certain fungi such as C. albicans, Icl1p and Mls1p are exclusively peroxisomal (24; our unpublished data), whereas in S. cerevisiae, Icl1p is cytosolic and Mls1p is only peroxisomal when cells are grown on fatty acids (17, 36).
Interestingly, acetyl-CoA transport and metabolism seem to play an essential role in the virulence of pathogenic fungi. The glyoxylate pathway enzymes Icl1p and Mls1p were shown to be crucial for the virulence of the plant pathogens Magnaporthe grisea (44) and Stagonospora nodorum (34) and the human pathogen C. albicans (20, 26), while carnitine-dependent acetyl unit transport in M. grisea is required for the elaboration of penetration hyphae during plant infection (2, 30).
In this study, we investigated the roles of acetyl-CoA transport in fatty acid metabolism and in the virulence of the human fungal pathogen C. albicans. We demonstrate here that the C. albicans genome contains only one citrate synthase (CIT) gene (alleles orf19.4393 and orf19.11871) and show that citrate synthase activity is present in mitochondria but not in peroxisomes, suggesting that for growth on fatty acids and transport of acetyl units from peroxisomes, the fungus is dependent on the Cat pathway. A C. albicans mutant lacking the major Cat protein (Cat2p) is not able to grow on fatty acids or on acetate or ethanol, but virulence of the mutant is not attenuated. While the cat2 deletion strain exhibits no growth deficiency on glucose, the mutant shows a small but significant reduction in its ability to form biofilms in vitro on this carbon source. Our results provide insight into the mechanism of intracellular acetyl-CoA transport in C. albicans and suggest that carnitine dependence may be an important trait of this human fungal pathogen.
MATERIALS AND METHODS
Media and culture conditions.
C. albicans strains were grown at 28°C unless otherwise stated. For routine nonselective culturing of C. albicans strains, YPD (2% Bacto peptone, 1% yeast extract, 2% glucose, 80 μg/ml uridine) was used. C. albicans transformants were selected and grown on minimal solid medium containing 0.67% yeast nitrogen base (YNB) without amino acids (Difco), 2% glucose, 2% agar, 80 μg/ml uridine, and amino acids as needed (20 μg/ml arginine, 20 μg/ml histidine). Plates used for spot assays had the same composition and contained glucose (2%), oleic acid-Tween 80 (0.12%-0.2%), ethanol (2%), or sodium acetate (2% with 0.5% potassium phosphate buffer [pH 6.0]) as a carbon source. For Cat enzyme assays, strains were grown overnight on YPD, YPO (2% Bacto peptone, 1% yeast extract, 0.12%-0.2% oleic acid-Tween 80), YPA (0.5% Bacto peptone, 0.3% yeast extract, 0.5% potassium phosphate buffer [pH 6], 2% sodium acetate), or YPE (0.5% Bacto peptone, 0.3% yeast extract, 0.5% potassium phosphate buffer [pH 6], 2% ethanol). For subcellular fractionation, β-oxidation assay, and immunoelectron microscopy, strains were grown overnight on YPO. In all experiments, strains were pregrown on minimal glucose medium (0.3% glucose, 0.67% YNB) for at least 8 h before being shifted to the medium of choice.
Spot test.
Cells were pregrown on medium containing 0.3% glucose, washed, resuspended to a concentration of about 2.7 × 107 cells/ml, and serially diluted (1:10 dilutions). Four microliters of each dilution was spotted onto agar plates. The pictures were taken after 3 days (glucose) or 5 days (oleate, acetate, and ethanol) of incubation at 28°C. Sensitivity to various stress agents was tested by spotting serial dilutions of cells onto YPD plates containing 25 μg/ml Calcofluor white, 200 μg/ml Congo red, 1.5 M sorbitol, or 0.05% sodium dodecyl sulfate. Plates were incubated at 28°C for 3 days. Hypha formation was tested by spotting dilutions onto YPD plates containing 10% fetal calf serum or 3% glycerol, followed by incubation at 37°C for 7 days.
Strains and plasmids.
The C. albicans strains used in this study (listed in Table 1) are derivatives of SN76 (23). The plasmids and primers used in this study are listed in Tables 2 and 3, respectively. The CAT2 gene (alleles orf19.4591 and orf19.12060) was deleted by using a PCR-based procedure with primers containing 100-bp regions identical to the 5′ and 3′ flanking sequences of the open reading frames (45). A cat2Δ/cat2Δ URA3 strain was created by successive transformation with two disruption cassettes. The first cassette, containing the ARG4 auxotrophic marker, was amplified from plasmid pFA-CaARG4 (8) by using primers CAT2-D-F-FA and CAT2-D-R-FA in combination with extension primers CAT2-F-Ext and CAT2-R-Ext. The same primers were used for construction of the second cassette by amplification on plasmid pFA-CdHIS1 (32), which contains the HIS1 auxotrophic marker from Candida dubliniensis. The cat2Δ/cat2Δ URA3 strain was transformed with XhoI-digested pLUBP (29) to create a prototrophic cat2 null mutant. For complementation of the cat2Δ/cat2Δ strain with the CAT2 gene, a plasmid was constructed by PCR with primers KS128 and JB10 on genomic DNA isolated from SN76. The PCR product, containing the 800-bp promoter region, the full-length CAT2 open reading frame, and the 800-bp 3′ untranslated region, was cloned into pGEM-T and sequenced. The fragment was cloned into pLUBP, resulting in plasmid pKa30. Transformation of the cat2Δ/cat2Δ URA3 strain with XhoI-digested pKa30 resulted in the cat2Δ/Δ + CAT2 strain (the complemented deletion strain). A prototrophic SN76 strain (SN76-P) was constructed by sequential transformation of strain SN76 with NheI-linearized pLUBP (for complementation of the URA3-IRO1 region), a 2.0-kb KpnI/SacI fragment of pRS-ARG4*SpeI (to complement the ARG4 deletion) and a 1.8-kb BamHI/XbaI fragment from pGEM-HIS1 (to complement the HIS1 deletion). Strain SN76-P will be referred to as the wild type. All strains were verified by PCR.
TABLE 1.
Yeast strains used in this study
| Strain | Species | Name | Genotype | Reference |
|---|---|---|---|---|
| BJ1991 | S. cerevisiae | Wild type | MATaleu2 ura3-251 trp1 pbr1-1122 pep4-3 gal2 | 13 |
| SN76 | C. albicans | Wild-type auxotroph | arg4/arg4 his1/his1 ura3::imm434/ura3::imm434iro1::imm434/iro1::imm434 | 23 |
| SN76-P | C. albicans | Wild-type prototroph | arg4Δ/ARG4 his1Δ/HIS1 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 | This study |
| CEM28 | C. albicans | cat2Δ/cat2Δ | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434iro1Δ::imm434/iro1Δ::imm434cat2Δ::CdHIS1/cat2Δ::ARG4 | This study |
| CEM38 | C. albicans | cat2Δ/cat2Δ + URA3 | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 cat2Δ::CdHIS1/cat2Δ::ARG4 | This study |
| CKS58 | C. albicans | cat2Δ/cat2Δ + CAT2 | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434::URA3::CAT2 iro1Δ::imm434/iro1Δ::imm434::IRO1 cat2Δ::CdHIS1/cat2Δ::ARG4 | This study |
| CEM16 | C. albicans | fox2Δ/fox2Δ + URA3 | ura3Δ::imm434/ura3Δ::URA3 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG fox2Δ::ARG4/fox2Δ::HIS1 | 26 |
TABLE 2.
Plasmids used in this study
TABLE 3.
Primers used in this study
| Primer | 5′-3′ sequence |
|---|---|
| CaCAT2-D-F-FA | AACTAATCAGAAGAAATAGAGGTCGAAAATAAAGAATAACGACAAGAAAAAAAAAAAGTAATCACATTTGTTCTGATATCATAGAGAAGCTTCGTACGCTGCAGGTC |
| CaCAT2-D-R-FA | AAATTTAAGAACTTTCTATATGTTACTTATACTAATATGAAATAAATAGATATGAATTGAAAAATGAAAAGACTAACCAAATTTCTCTGATATCATCGATGAATTCGAG |
| CaCAT2-Ext-F | TATTCTTTTCAATCCAACTAATCAGAAGAAATAGA |
| CaCAT2-Ext-R | GTGCTAATAATAACTAAATTTAAGAACTTTCTATA |
| CAT2-C-F | ATCAAGTATCCATGACCCCCAC |
| CAT2-C-R | GATGAGAGTAGTGTTGTTGAGG |
| CaA2 | AATGGATCAGTGGCACCGGTG |
| CaA4 | GGGCCCATTGGTTAAGTTCATATGC |
| X2-CdHIS1 | TCTAAACTGTATATCGGCACCGCTC |
| X3-CdHIS1 | GCTGGCGCAACAGATATATTGGTGC |
| JB10 | AGTTGGTACCAGAAGTGTGGGGTGCATAGG |
| KS128 | TGAGGCCTCGTTGATTTAAACTCCGCGATGT |
Transformation.
C. albicans was transformed by using a modified lithium acetate protocol (42). Heat shock was carried out at 44°C for 15 min.
Subcellular fractionation and density gradient analysis of C. albicans.
Cells were inoculated in YNB-2% glucose and grown overnight. On the next morning, the culture was diluted in YNB-0.3% glucose to an optical density at 600 nm (OD600) of 0.15, grown to an OD600 of 1.0 to 1.5, and then inoculated into YPO at an OD600 of 0.01 and grown overnight. On the next morning, cells were collected by centrifugation (10 min at 4,000 × g), washed three times with 25 ml distilled water, and collected again by centrifugation (10 min at 4,000 × g). Cells were resuspended in 5 ml of buffer Z (0.5 M KCl, 5 mM morpholinepropanesulfonic acid [MOPS; pH 7.2], 10 mM Na2SO3) containing 0.25 mg Zymolyase 100T (ICN Biomedicals) per g (wet weight) and incubated for 10 to 30 min at 28°C while shaking at 120 rpm to convert the cells to spheroplasts. Spheroplast formation was monitored microscopically. Spheroplasts were harvested by centrifugation at 2,300 × g for 8 min at 4°C. The pellet was resuspended in 3 ml buffer F (5% Ficoll 400, 0.6 M sorbitol, 5.5 mM MOPS [pH 7.2], 0.5 mM EDTA, 0.1% ethanol) containing 1 mM phenylmethylsulfonyl fluoride and yeast protease inhibitor cocktail and homogenized in a grinding vessel (Potter-Elvehjem) with a tight-fitting pestle by 20 down-and-up strokes while on ice. The homogenate was centrifuged three times for 10 min at 3,000 × g at 4°C to remove cell debris and nuclei, and the low-speed supernatant (H) was separated into an organellar pellet (P) and a cytosolic (S) fraction by centrifugation for 20 min at 20,000 × g at 4°C. The organellar pellet was taken up in 1 ml buffer G (5 mM Tris-HCl [pH 7.5], 3 mM KCl, 0.3 mM EDTA [pH 8], 0.1% ethanol, 0.6 M sorbitol) and loaded onto a 15 to 50% Nycodenz gradient. Gradients were spun in a vertical rotor for 2.5 h at 29,000 × g with gentle acceleration and braking. Gradients were fractionated from the bottom to the top in about 12 fractions. Each fraction was analyzed for the presence of citrate synthase by Western blotting with an antibody raised against S. cerevisiae Cit1p (41), 3-hydroxyacyl-CoA dehydrogenase activity (43) (a peroxisomal marker), fumarase activity (38) (a mitochondrial marker), and carnitine acetyltransferase (Cat) activity (6). Subcellular fractionation and density gradient analysis of S. cerevisiae were performed as described previously (6).
Fatty acid β-oxidation measurements.
The β-oxidation activity in intact cells was measured essentially as described before (39), except that the cells were resuspended at an OD600 of 1. Production of CO2 and incorporation of radiolabel into acid-soluble material were followed over time.
Acyl-CoA measurements.
Acyl-CoA measurements were performed as described by Hammond et al. (10), with some modifications. Approximately 20 mg of freeze-dried material of oleate-grown yeast cells was added to 1.5-ml Eppendorf vials, and the exact weight of the sample was determined with a microbalance. To the sample, 20 μl of internal standard (2H3-acetyl-CoA [100 μM], 2H3-octanoyl-CoA [100 μM], and 2H3-palmitoyl-CoA [100 μM] in 70% acetonitrile) was subsequently added on ice in 50 mM KH2PO4-50% 2-propanol. An equal volume of acetonitrile was added. After 3 min of mixing, the samples were centrifuged at 1,600 × g for 5 min at 4°C and the supernatant was transferred to a glass tube and evaporated under a stream of nitrogen at 40°C. The final residue was taken up in 100 μl methanol-H2O (1:1) and subjected to liquid chromatography-tandem mass spectrometry analysis as described before (10).
Virulence studies.
Virulence assays were performed at the University of Aberdeen by using a murine tail vein injection model. Groups of six BALB/c female mice were challenged intravenously with the C. albicans strains at a dose of 1.5 × 104 CFU/g body weight as previously described (26). Mice were weighed and observed daily. Animals that developed signs of serious illness or lost more than 20% of their initial body weight were humanely terminated and recorded as dying the following day. Survival data were analyzed by log rank statistics, and tissue burden data were analyzed by t test.
Growth and analyses of biofilms.
Biofilms were grown in 96-well plates (Costar; Corning Incorporated, Corning, NY) coated with fetal bovine serum (FBS) as described previously (15), and growth was quantified by crystal violet staining (37). Biofilm thicknesses were measured with a low-load compression tester (LLCT) (25) on biofilms grown on FBS-coated (1.5 by 1.5 cm) polymethylmethacrylate slides in 12-well tissue culture plates. For confocal laser scanning microscopy (CLSM), biofilms were grown in FBS-coated 12-well plates, washed once with 2 ml phosphate-buffered saline (10 mM potassium phosphate, 0.15 M NaCl [pH 7.0]), and incubated with 1 ml phosphate-buffered saline containing BacLight (Molecular Probes, Leiden, The Netherlands), prepared according to the manufacturer's instructions, for 30 min at room temperature in the dark. Confocal laser scanning microscope model LEICA TCS SP2 (Leica Microsystems Heidelberg GmbH, Heidelberg, Germany) equipped with a He-Ne laser and an Ar laser and supplied with the latest version of the Leica Confocal software was used to visualize the submerged biofilms with a 40× water objective. The two components of BacLight were excited at 488 nm, the green signal was recorded with a 510- to 540-nm emission filter setting, and the red signal was recorded with a 600- to 680-nm emission filter setting.
RESULTS
C. albicans contains one citrate synthase that is localized to mitochondria.
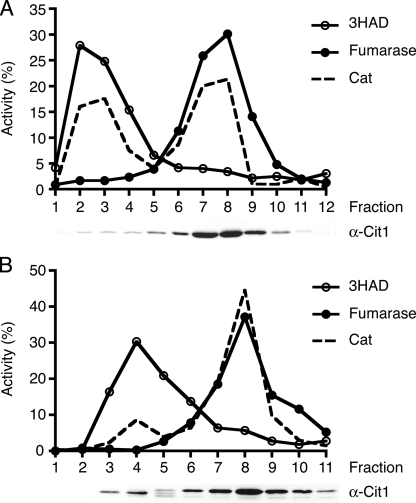
The S. cerevisiae genome encodes three citrate synthases, of which two are mitochondrial (Cit1p and Cit3p) and one is peroxisomal (Cit2p) (9, 12, 18, 31). We performed BLASTp searches with the S. cerevisiae Cit proteins and found that the C. albicans genome (http://www.candidagenome.org) contains only one gene predicted to encode citrate synthase. The putative C. albicans citrate synthase (orf19.4393) shows 77% sequence identity with ScCit1p and 72% and 45% identity with ScCit3p and ScCit2p, respectively, and has a predicted mitochondrial targeting signal in its N-terminal region (4). To determine the subcellular localization of the C. albicans citrate synthase, we performed subcellular fractionation, followed by density gradient analysis, on cells grown in rich medium containing oleic acid (Fig. 1A). Analysis of the C. albicans gradient fractions by immunoblotting with an antibody directed against S. cerevisiae Cit1p revealed a single band with an apparent molecular mass of about 45 kDa, corresponding to the predicted molecular mass of the C. albicans Cit1p protein that cofractionated with the mitochondrial marker enzyme fumarase. Very little signal was detected in the peroxisomal peak fractions, suggesting that the single CIT gene in C. albicans encodes a mitochondrion-targeted protein.
FIG. 1.
Subcellular localization of citrate synthase and carnitine acetyltransferase in S. cerevisiae and C. albicans. Nycodenz density gradient analysis of the organellar pellet fraction obtained from wild-type C. albicans (A) and wild-type S. cerevisiae (B) grown on rich oleic acid medium. Enzyme assays were performed on the gradient fractions to determine the distribution of the peroxisomal marker enzyme 3-hydroxyacyl-CoA dehydrogenase (3HAD), the mitochondrial marker enzyme fumarase, and Cat. Activities are given as percentages of the total activity present in the whole gradient. Relative Cat activities were determined for the peroxisomal and mitochondrial peak fractions of both gradients, 4,088.8 nmol/min/mg for the C. albicans peroxisomal peak fraction (3), 445.6 nmol/min/mg for the mitochondrial peak fraction (8), 82.2 nmol/min/mg for the S. cerevisiae peroxisomal peak fraction (4), and 122.6 nmol/min/mg for the mitochondrial peak fraction (8). The localization of citrate synthase in the gradients was determined by immunoblotting with an antibody directed against S. cerevisiae citrate synthase 1 (Cit1p).
As a control, oleate-grown S. cerevisiae cells were analyzed in a similar way. Figure 1B shows that the anti-Cit1 antibody recognizes at least two citrate synthase proteins in S. cerevisiae, a band running at about 45 kDa that cofractionates with the mitochondrial marker and most likely represents Cit1p and/or Cit3p and a band enriched in the peroxisomal fractions that runs slightly faster, presumably corresponding to peroxisomal Cit2p. Because of the almost identical predicted molecular masses of Cit1p and Cit3p, it is very possible that both proteins run at the same position in the gel and are observed as a single band or, alternatively, that either of the two proteins is not expressed under our experimental conditions. Notwithstanding the above, these data confirm previous results of Lewin et al. (18), who showed the presence of a peroxisomal citrate synthase (Cit2p) in S. cerevisiae and imply that peroxisomes in C. albicans lack this protein.
Carnitine acetyltransferase is abundantly present in peroxisomes and mitochondria of C. albicans.
The absence of peroxisomal citrate synthase in C. albicans predicts that for export of acetyl units, the organism depends on the carnitine acetyltransferase pathway. We measured Cat activity in the gradient fractions of both C. albicans and S. cerevisiae (Fig. 1A and B). Cat activity in the C. albicans gradient colocalized with the peroxisomal and mitochondrial peaks, and the activities in the two peaks are comparable. In concordance with Elgersma et al. (6), we also found a bimodal distribution of the Cat activity in S. cerevisiae, with about five times more activity in the mitochondrial peak fraction than in the peroxisomal peak fraction.
CAT2 encodes the major carnitine acetyltransferase in C. albicans.
In S. cerevisiae, the CAT2 gene encodes both the peroxisomal and mitochondrial forms of Cat2p and contributes about 95% of the total Cat activity in oleate-grown cells, while the remaining activity is ascribed to Yat1p and Yat2p (6, 35). To determine the role of Cat2p in C. albicans, we constructed a CAT2 (orf19.4591 and orf19.12060) deletion strain by a PCR-based gene disruption procedure (45) (see Materials and Methods). To generate the complemented strain (cat2Δ/cat2Δ + CAT2), the CAT2 gene, together with the 800-bp upstream promoter region, was cloned into the pLUBP vector (29) and transformed into the cat2Δ/cat2Δ strain. URA3 prototrophy was restored to the cat2Δ/cat2Δ strain by introducing the empty pLUBP vector.
Total Cat activity was determined in lysates of the wild-type, cat2Δ/cat2Δ, and complemented strains grown on rich medium containing glucose, oleate-Tween 80, ethanol, or acetate (Table 4). The Cat activity of the wild-type and complemented strains was about twofold higher on oleate, acetate, and ethanol compared to that on glucose. The Cat activity of the cat2Δ/cat2Δ strain was just above the background levels under the growth conditions tested, in spite of the fact that the C. albicans genome encodes two other putative carnitine acetyltransferases, potential homologs of S. cerevisiae Yat1p and Yat2p (28, 33, 35). We tried to detect residual Cat activity in the cat2Δ/cat2Δ strain by varying the conditions of the assay (different pH, addition of a nonionic detergent) by varying the method of protein lysate preparation and by using purified peroxisomal and mitochondrial fractions instead of total protein lysates (data not shown). We were, however, not able to detect significant amounts of Cat activity in the cat2 null mutant by any of these methods, suggesting that the expression of the YAT genes is very low under the conditions tested. Taken together, these results show that the C. albicans CAT2 gene, like the S. cerevisiae CAT2 gene (6), encodes both the major peroxisomal and mitochondrial forms of Cat.
TABLE 4.
Cat activities in lysates of C. albicans cells grown on various carbon sources
| Strain | Mean Cat activitya ± SD
|
|||
|---|---|---|---|---|
| Glucose | Oleate | Acetate | Ethanol | |
| Wild type | 653.4 ± 18.2 | 1,189.5 ± 403.3 | 1,273.8 ± 3.0 | 1,486.2 ± 39.9 |
| cat2Δ/cat2Δ | 1.1 ± 0.3 | 0.1 ± 0.1 | 3.4 ± 5.5 | 0.9 ± 1.8 |
| cat2Δ/cat2Δ + CAT2 | 517.3 ± 59.3 | 1,403.9 ± 115.4 | 939.5 ± 11.9 | 679.9 ± 21.4 |
Nanomoles of [14C-]acetylcarnitine formed per minute per milligram of protein.
The C. albicans cat2 null mutant is unable to utilize fatty acids, ethanol, or acetate.
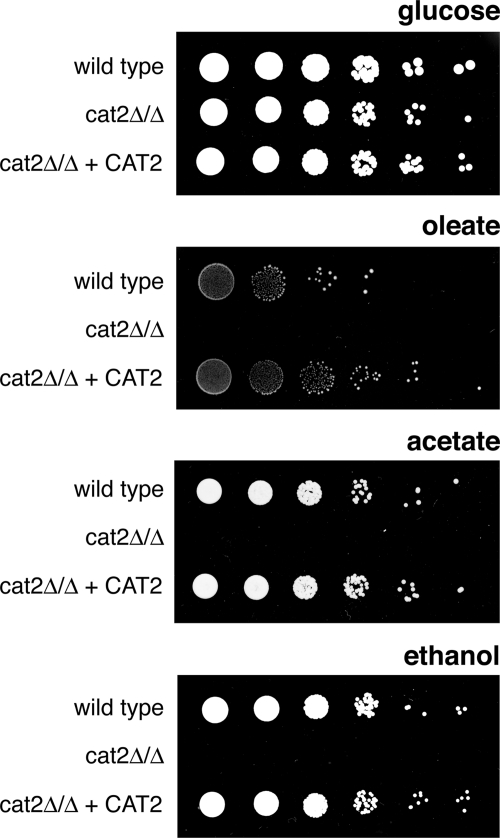
To determine whether, in the absence of a peroxisomal citrate synthase, peroxisomal Cat2p plays an essential role in the export of the acetyl-CoA produced during fatty acid β-oxidation, we tested the cat2Δ/cat2Δ strain for the ability to utilize oleate as the sole carbon source. Serial dilutions of the wild-type, cat2Δ/cat2Δ, and complemented strains were spotted onto plates containing YNB and oleate or glucose (as a control) as the carbon source. All of the strains grew well on glucose; however, the cat2Δ/cat2Δ strain was unable to grow on oleate (Fig. 2), a phenotype that was confirmed by carrying out growth assays in liquid media (data not shown). These results support the hypothesis that C. albicans is dependent on the activity of Cat2p to export acetyl units produced during fatty acid β-oxidation and confirm that C. albicans lacks a peroxisomal citrate synthase. As the cat2Δ/cat2Δ strain lacks both the peroxisomal and mitochondrial forms of Cat2p, it is unclear what each form contributes to the phenotype on oleate. Remarkably, the cat2Δ/cat2Δ strain is also not able to grow on the C2 carbon source acetate or ethanol (Fig. 2), a condition under which acetyl-CoA is produced in the cytosol. Together, these results show that both the peroxisomal and mitochondrial forms of Cat2p are essential for growth on nonfermentable carbon sources.
FIG. 2.
The cat2 null mutant is unable to grow on nonfermentable carbon sources. Glucose-grown cultures (about 2.7 × 107 cells/ml) of the indicated strains were serially diluted (1:10), and 4 μl of each dilution was spotted on a minimal plate containing glucose, oleate, acetate, or ethanol as the sole carbon source. Pictures were taken after 3 to 7 days of incubation at 28°C.
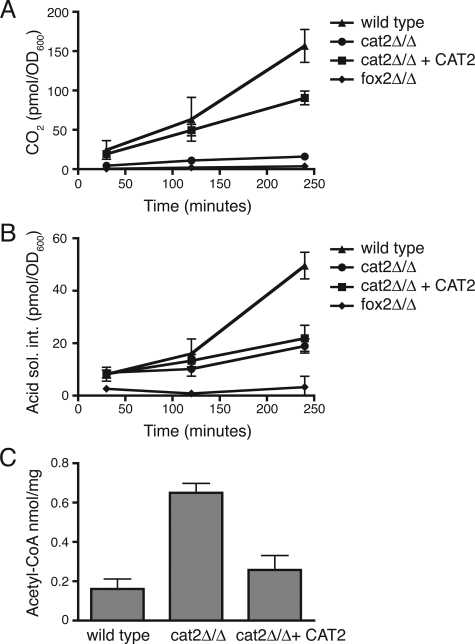
The cat2 null mutant shows reduced β-oxidation activity and elevated levels of acetyl-CoA.
To study the oleic acid metabolism of the cat2 null mutant in more detail, we determined its total β-oxidation activity by incubating intact cells with 1-14C-labeled oleic acid and measuring the labeled CO2 and acid-soluble counts produced (representing carbon metabolism intermediates) (39). The C. albicans fox2Δ/fox2Δ strain, lacking the second enzyme of the β-oxidation pathway, served as a negative control, as this mutant has virtually no fatty acid β-oxidation activity (26). The fox2Δ/fox2Δ and cat2Δ/cat2Δ strains showed greatly reduced production of CO2 compared to the wild-type and CAT2 complemented strains (Fig. 3A). The incorporation of radiolabel into acid-soluble material was very low in the fox2Δ/fox2Δ strain, but significant amounts were found in the cat2Δ/cat2Δ strain, suggesting that the labeled fatty acid can still be converted to acetyl-CoA and/or other carbon metabolism intermediates in this strain (Fig. 3B). We determined total acetyl-CoA levels by mass spectrometry analysis and found them to be three- to fourfold higher in the cat2Δ/cat2Δ strain than in the wild-type or complemented strain (Fig. 3C). The accumulation of acetyl-CoA in the cat2 mutant may account for the relatively large amount of acid-soluble material produced in this strain compared to the fox2 null strain (Fig. 3B). Together, these data show that acetyl-CoA transport from peroxisomes to mitochondria is defective in cells lacking Cat2p.
FIG. 3.
The cat2 mutant shows greatly reduced β-oxidation activity and accumulates acetyl-CoA. Oleate-grown cells of the indicated strains were incubated with 1-14C-labeled oleic acid, and release of CO2 (A) and conversion into acid-soluble intermediates (B) were measured over a period of 4 h. Total acetyl-CoA levels in the three strains were determined as described in Materials and Methods (C). Error bars represent standard deviations of three or four measurements from two independent experiments.
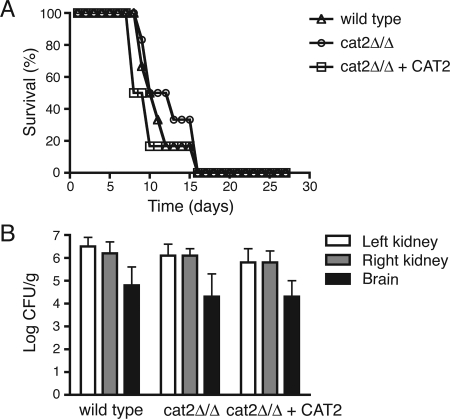
The cat2 null mutant is not attenuated in virulence in the mouse model of systemic candidiasis.
Previously, it was shown that carnitine acetyltransferase plays an essential role during plant infection by the rice blast fungus M. grisea (2). We used the mouse model of systemic infection to test the role of Cat2p in the virulence of C. albicans. BALB/c mice were injected intravenously with cells of the wild-type, cat2Δ/cat2Δ, or complemented strain at a challenge dose of 1.5 × 104 CFU/g body weight. No significant differences were found between the strains with respect to survival times (as calculated by log rank test) or tissue burdens (t test) (Fig. 4A and B). These results indicate that carnitine acetyltransferase does not play an essential role in C. albicans infection in the mouse model.
FIG. 4.
Virulence of the cat2 mutant is not attenuated in a mouse model of systemic candidiasis. Survival of BALB/c mice injected with each of the three different strains at 1.5 × 104 CFU/g body weight is shown (A). Tissue burdens were determined for the left and right kidneys and the brain (B).
The cat2 null mutant shows a small but significant defect in biofilm formation.
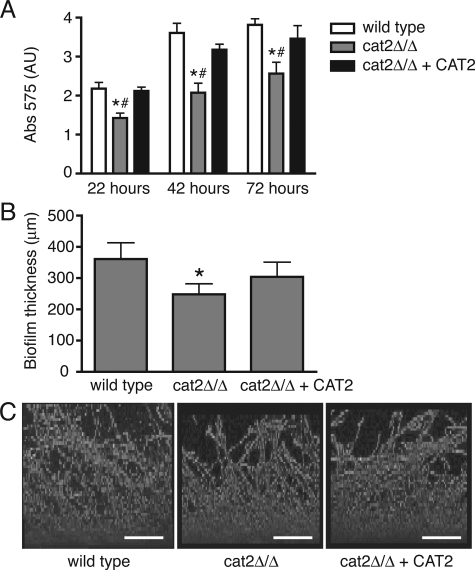
We tested whether the absence of the metabolic enzyme Cat2p affected biofilm formation over a period of 72 h and observed that significantly less biofilm was formed by the cat2 null mutant compared to the wild-type and complemented strains as quantified by crystal violet staining (Fig. 5A). Direct measurement of biofilm thickness with an LLCT (25) revealed that the cat2 null mutant formed 30% thinner biofilms compared to the wild-type strain (350 and 250 μm thick, respectively) and that biofilm formation was partially restored in the complemented strain (300 μm thick), which is in line with the gene dosage effect (Fig. 5B). CLSM analysis revealed no difference among the three strains with respect to biofilm architecture or morphology and confirmed the reduced ability of the cat2 null mutant to form biofilm (Fig. 5C).
FIG. 5.
Biofilm formation is reduced in the cat2 mutant. (A) Biofilm formation as measured by crystal violet staining after 22, 42, and 72 h. Two independent experiments were performed, with similar results. Shown are the results of one experiment (mean plus standard deviation). For each time point and each strain, five individual biofilms were measured. A significant reduction in biofilm formation by the cat2 mutant was observed compared to the wild-type (*, P < 0.005) and complemented strains (#, P < 0.05), as tested by the Student t test. Abs 575, absorbance at 575 nm; AU, arbitrary units. (B) Quantitative assessment of biofilm thickness determined after 72 h by LLCT. Error bars represent standard deviations of six to eight independent measurements on two individual biofilms for each strain. The asterisk represents a significant difference between the cat2 null mutant and the wild-type or complemented strain (P < 0.005, Student t test). (C) CLSM analysis of 72-h biofilms formed by the three strains indicated. Bars represent 100 μm.
DISCUSSION
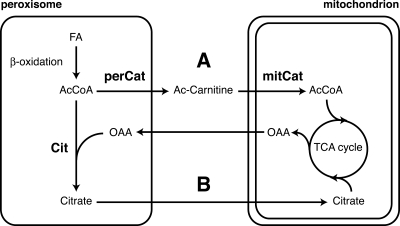
Two possible pathways for the transport of acetyl units from peroxisomes to mitochondria have been described in plants and yeasts, of which one is reliant on peroxisomal citrate synthase and the other is dependent on peroxisomal carnitine acetyltransferase. Here we show that the human fungal pathogen C. albicans lacks a peroxisomal citrate synthase and exclusively relies on Cat activity for the export of acetyl units from peroxisomes (Fig. 1). A C. albicans cat2 null mutant that lacks the major Cat enzyme is unable to grow on a fatty acid as the sole carbon source, shows greatly reduced β-oxidation activity, and accumulates acetyl-CoA (Fig. 2 and 3). In S. cerevisiae, it was shown that the Cit and Cat routes work in parallel since deletion of either peroxisomal citrate synthase (cit2Δ) or the major Cat protein (cat2Δ) does not affect growth on oleate but deletion of both does (39). Current evidence suggests that plant peroxisomes lack carnitine acetyltransferase and use the citrate synthase pathway to shuttle acetyl units to mitochondria (27). Thus, whereas S. cerevisiae can employ both pathways, A. thaliana and C. albicans are each dependent on a single pathway, Cit in A. thaliana and Cat in C. albicans (Fig. 6). A BLAST search of the sequenced fungal genomes (http://www.broad.mit.edu/annotation/fgi/ and http://cbi.labri.fr/Genolevures/BLAST.php) revealed that at least three other closely related fungal species (Candida tropicalis, Debaryomyces hansenii, and Candida lusitaniae) contain a single CIT gene predicted to encode a mitochondrion-targeted citrate synthase, suggesting that they, like C. albicans, are dependent on peroxisomal Cat activity for growth on fatty acids. The fact that C. albicans grows very well on mineral oleate medium lacking carnitine (Fig. 2 and our unpublished observations), together with the observed strict requirement for Cat activity under these growth conditions, implies that C. albicans possesses a functional carnitine biosynthesis pathway. Experiments to provide evidence of this are under way in our laboratory.
FIG. 6.
Schematic representation of two possible pathways for the transport of acetyl units from peroxisomes to mitochondria. Peroxisomal β-oxidation of fatty acids (FA) generates acetyl-CoA (AcCoA) as the end product. In pathway A, the acetyl (Ac) moiety of acetyl-CoA is linked to carnitine by carnitine acetyltransferase (perCat) and transported to the mitochondrion, where mitochondrial Cat (mitCat) catalyzes the reverse reaction, resulting in the generation of mitochondrial acetyl-CoA entering the TCA cycle. In pathway B, acetyl groups are linked to oxaloacetate (OAA) by peroxisomal citrate synthase (Cit) and transported as citrate to the mitochondrial TCA cycle. For simplicity, the other enzymes of the glyoxylate cycle (isocitrate lyase, malate synthase, and aconitase) are not depicted. S. cerevisiae was shown to possess both the Cat- and Cit-dependent pathways (40), and A. thaliana only employs the Cit-dependent pathway (27), while C. albicans is solely dependent on the Cat pathway for export of acetyl units from peroxisomes (this report). (Figure adapted from reference 27 with permission of the publisher.)
We have shown that the cat2Δ/cat2Δ strain not only is unable to grow on fatty acids but also does not grow on ethanol or acetate (Fig. 2). Under the latter conditions, the acetyl-CoA is produced in the cytosol. Since the peroxisomal and mitochondrial membranes are impermeable to acetyl-CoA, it is conceivable that the acetyl units must be linked to carnitine to allow their transport over the peroxisomal and mitochondrial membranes and enter the glyoxylate cycle and TCA cycle, respectively. The absence of the mitochondrial and peroxisomal Cat enzymes in the cat2 null mutant prevents the reformation of acetyl-CoA inside the organelles and thus further metabolism. Which enzyme(s) is involved in linking the acetyl units to carnitine in the cytosol is unclear. This function may be carried out by either of the two other predicted CAT genes, CTN1 and CTN3, which are potential homologs of S. cerevisiae YAT1 and YAT2, respectively (28). In S. cerevisiae, both YAT genes are expressed on ethanol and Yat1p was shown to be associated with the outer mitochondrial membrane, suggesting that Yat1p and/or Yat2p may be involved in the generation of acetyl-carnitine in the cytosol (33, 35). The subcellular localization of Ctn1p and Ctn3p in C. albicans has not been addressed experimentally, but the absence of a clear mitochondrial or peroxisomal targeting signal 1 (http://ihg.gsf.de/ihg/mitoprot.html and http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp) suggests that they may function in the cytosol. However, no significant amounts of Cat activity could be detected in the cat2 null mutant grown on glucose, acetate, ethanol, or oleate (Table 4), despite our efforts to measure this activity under a variety of assay conditions. It remains possible, however, that Ctn1p and Ctn3p are not active in our assay or that they are expressed under very specific conditions. Indeed, Prigneau et al. (28) have shown by Northern blot analysis that CTN1, CTN3, and CAT2 transcripts are induced during macrophage infection but enzyme activities were not determined. How the cytosolic acetyl-CoA produced enters the peroxisomal and mitochondrial compartments remains therefore unresolved.
Previously, we and others investigated the role of fatty acid metabolism in the virulence of C. albicans (19, 26). Piekarska et al. showed that a C. albicans fox2Δ/fox2Δ strain lacking the second enzyme of the β-oxidation pathway shows attenuated virulence in mice but that the virulence defect of this strain is probably caused by a dysfunctional glyoxylate cycle (26). The phenotype of the cat2 null mutant is similar to that of the fox2Δ/fox2Δ strain in that it exhibits greatly reduced β-oxidation activity and an inability to grow on oleate, acetate, and ethanol. However, the glyoxylate cycle in the oleate-grown cat2Δ/cat2Δ strain may still be partially functional, as can be inferred from the conversion of fatty acids into acid-soluble material representing carbon metabolism intermediates (Fig. 3). The wild-type virulence of the cat2 strain in the mouse model corroborates previous data obtained with the fox2Δ/fox2Δ strain and shows independently that fatty acid β-oxidation is not required for the survival of C. albicans in infected mice (Fig. 4). In line with this, Barelle et al. recently reported that the dominant metabolic mode of C. albicans single cells in vivo is glycolytic rather that gluconeogenic (1).
Carnitine acetyltransferase does play an essential role in the virulence of the plant-pathogenic fungus M. grisea, as shown by the inability of a pth2 mutant lacking the major Cat enzyme to form penetration hyphae and cause plant infection (2, 30). Interestingly, the pth2 mutant is also not able to grow on fatty acids (olive oil) or acetate, indicating that M. grisea, like C. albicans, is dependent on the Cat pathway for growth on these carbon sources. By using green fluorescent protein (GFP)-tagged versions of Pth2p, a unique peroxisomal localization was found for the protein (2). Since the Pth2p ortholog Cat2p in S. cerevisiae and C. albicans has dual localizations and contains targeting signals in both the N-terminal (mitochondrial) and C-terminal (peroxisomal) parts of the protein, it is very well possible that GFP tagging of Pth2 has disturbed its mitochondrial targeting, causing it to localize only to peroxisomes. However, the Pth2-GFP construct did fully complement the pth2 mutant phenotype, suggesting that peroxisome-targeted Pth2p is functional. Determination of the subcellular distribution of the endogenous untagged protein is required to resolve this issue.
Cell-cell adherence, the ability to form hyphae, and production of an extracellular matrix are required for robust biofilm formation by C. albicans (3, 5). Because a C. albicans ctn3 null mutant showed reduced hypha formation (28) and an M. grisea pth2 mutant showed an increased sensitivity to cell wall stress (2, 30), we determined whether our cat2 null mutant exhibited similar phenotypes. No noticeable differences in sensitivity to cell wall-perturbing compounds (Calcofluor white, Congo red, sorbitol, and sodium dodecyl sulfate) or the ability to form hyphae were found between the wild type and the null mutant (data not shown). However, a small but significant reduction in the ability to form biofilms in vitro was observed in the cat2 deletion mutant (Fig. 5), possibly because the mutant is unable to metabolize nonfermentable carbon sources, which may cause a growth disadvantage in the later, mature stages of biofilm formation when glucose is exhausted. Further experiments are required to address this issue.
In conclusion, our studies revealed that C. albicans exclusively relies on Cat activity for the transport of acetyl units among peroxisomes, cytosol, and mitochondria during growth on nonfermentable carbon sources. While the lack of Cat activity does not affect the virulence of C. albicans in mice, maximum biofilm formation does require a functional Cat pathway. Following the observations of Mukherjee and colleagues on the role of alcohol dehydrogenase (22), Cat2p is now the second example of a metabolic enzyme affecting biofilm formation in C. albicans.
ADDENDUM
While the manuscript was under review, a paper reporting similar data was published by the group of Lorenz (46).
Acknowledgments
We thank Rick Rachubinski (University of Alberta, Edmonton, Alberta, Canada) for advice on C. albicans subcellular fractionation and density gradient analysis and Rob Benne for valuable comments and suggestions. The antibody directed against S. cerevisiae Cit1p was a kind gift from Véronique Berteaux-Lecellier (Université Paris-Sud, Paris, France).
This work was supported by a grant from the Academic Medical Center.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhambra, G. K., Z.-Y. Wang, D. M. Soanes, G. E. Wakley, and N. J. Talbot. 2006. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 6146-60. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship, J. R., and A. P. Mitchell. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9588-594. [DOI] [PubMed] [Google Scholar]
- 4.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241779-786. [DOI] [PubMed] [Google Scholar]
- 5.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 1130-36. [DOI] [PubMed] [Google Scholar]
- 6.Elgersma, Y., C. W. van Roermund, R. J. Wanders, and H. F. Tabak. 1995. Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 143472-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández, E., F. Moreno, and R. Rodicio. 1992. The ICL1 gene from Saccharomyces cerevisiae. Eur. J. Biochem. 204983-990. [DOI] [PubMed] [Google Scholar]
- 8.Gola, S., R. Martin, A. Walther, A. Dunkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 201339-1347. [DOI] [PubMed] [Google Scholar]
- 9.Graybill, E. R., M. F. Rouhier, C. E. Kirby, and J. W. Hawes. 2007. Functional comparison of citrate synthase isoforms from S. cerevisiae. Arch. Biochem. Biophys. 46526-37. [DOI] [PubMed] [Google Scholar]
- 10.Hammond, L. E., S. Neschen, A. J. Romanelli, G. W. Cline, O. R. Ilkayeva, G. I. Shulman, D. M. Muoio, and R. A. Coleman. 2005. Mitochondrial glycerol-3-phosphate acyltransferase-1 is essential in liver for the metabolism of excess acyl-CoAs. J. Biol. Chem. 28025629-25636. [DOI] [PubMed] [Google Scholar]
- 11.Hartig, A., M. M. Simon, T. Schuster, J. R. Daugherty, H. S. Yoo, and T. G. Cooper. 1992. Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 205677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia, Y. K., A. M. Becam, and C. J. Herbert. 1997. The CIT3 gene of Saccharomyces cerevisiae encodes a second mitochondrial isoform of citrate synthase. Mol. Microbiol. 2453-59. [DOI] [PubMed] [Google Scholar]
- 13.Jones, E. W. 1977. Proteinase mutants of Saccharomyces cerevisiae. Genetics 8523-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg, H. L. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 991-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krom, B. P., J. B. Cohen, G. E. McElhaney Feser, and R. L. Cihlar. 2007. Optimized candidal biofilm microtiter assay. J. Microbiol. Methods 68421-423. [DOI] [PubMed] [Google Scholar]
- 16.Kunau, W.-H., V. Dommes, and H. Schulz. 1995. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog. Lipid Res. 34267-342. [DOI] [PubMed] [Google Scholar]
- 17.Kunze, M., F. Kragler, M. Binder, A. Hartig, and A. Gurvitz. 2002. Targeting of malate synthase 1 to the peroxisomes of Saccharomyces cerevisiae cells depends on growth on oleic acid medium. Eur. J. Biochem. 269915-922. [DOI] [PubMed] [Google Scholar]
- 18.Lewin, A. S., V. Hines, and G. M. Small. 1990. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol. Cell. Biol. 101399-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 31076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 41283-86. [DOI] [PubMed] [Google Scholar]
- 21.McCammon, M. T. 1996. Mutants of Saccharomyces cerevisiae with defects in acetate metabolism: isolation and characterization of Acn− mutants. Genetics 14457-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee, P. K., S. Mohamed, J. Chandra, D. Kuhn, S. Liu, O. S. Antar, R. Munyon, A. P. Mitchell, D. Andes, M. R. Chance, M. Rouabhia, and M. A. Ghannoum. 2006. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect. Immun. 743804-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen, L. J., W. F. Ettinger, B. Damsz, K. Matsudaira, M. A. Webb, and J. J. Harada. 1993. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paramonova, E., E. D. de Jong, B. P. Krom, H. C. van der Mei, H. J. Busscher, and P. K. Sharma. 2007. Low-load compression testing: a novel way of measuring biofilm thickness. Appl. Environ. Microbiol. 737023-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piekarska, K., E. Mol, M. van den Berg, G. Hardy, J. van den Burg, C. van Roermund, D. MacCallum, F. Odds, and B. Distel. 2006. Peroxisomal fatty acid β-oxidation is not essential for virulence of Candida albicans. Eukaryot. Cell 51847-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pracharoenwattana, I., J. E. Cornah, and S. M. Smith. 2005. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 172037-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prigneau, O., A. Porta, and B. Maresca. 2004. Candida albicans CTN gene family is induced during macrophage infection: homology, disruption and phenotypic analysis of CTN3 gene. Fungal Genet. Biol. 41783-793. [DOI] [PubMed] [Google Scholar]
- 29.Ramón, A. M., and W. A. Fonzi. 2003. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell 2718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos-Pamplona, M., and N. I. Naqvi. 2006. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 6161-75. [DOI] [PubMed] [Google Scholar]
- 31.Rosenkrantz, M., T. Alam, K. S. Kim, B. J. Clark, P. A. Srere, and L. P. Guarente. 1986. Mitochondrial and nonmitochondrial citrate synthases in Saccharomyces cerevisiae are encoded by distinct homologous genes. Mol. Cell. Biol. 64509-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaub, Y., A. Dunkler, A. Walther, and J. Wendland. 2006. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J. Basic Microbiol. 46416-429. [DOI] [PubMed] [Google Scholar]
- 33.Schmalix, W., and W. Bandlow. 1993. The ethanol-inducible YAT1 gene from yeast encodes a presumptive mitochondrial outer carnitine acetyltransferase. J. Biol. Chem. 26827428-27439. [PubMed] [Google Scholar]
- 34.Solomon, P. S., R. C. Lee, T. J. Wilson, and R. P. Oliver. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 531065-1073. [DOI] [PubMed] [Google Scholar]
- 35.Swiegers, J. H., N. Dippenaar, I. S. Pretorius, and F. F. Bauer. 2001. Carnitine-dependent metabolic activities in Saccharomyces cerevisiae: three carnitine acetyltransferases are essential in a carnitine-dependent strain. Yeast 18585-595. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, K. M., C. P. Kaplan, X. Gao, and A. Baker. 1996. Localization and targeting of isocitrate lyases in Saccharomyces cerevisiae. Biochem. J. 319255-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Merode, A. E., D. C. Pothoven, H. C. van der Mei, H. J. Busscher, and B. P. Krom. 2007. Surface charge influences enterococcal prevalence in mixed-species biofilms. J. Appl. Microbiol. 1021254-1260. [DOI] [PubMed] [Google Scholar]
- 38.van Roermund, C. W. T., R. Drissen, M. van Den Berg, L. Ijlst, E. H. Hettema, H. F. Tabak, H. R. Waterham, and R. J. A. Wanders. 2001. Identification of a peroxisomal ATP carrier required for medium-chain fatty acid β-oxidation and normal peroxisome proliferation in Saccharomyces cerevisiae. Mol. Cell. Biol. 214321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Roermund, C. W., Y. Elgersma, N. Singh, R. J. Wanders, and H. F. Tabak. 1995. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 143480-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Roermund, C. W., E. H. Hettema, M. van den Berg, H. F. Tabak, and R. J. Wanders. 1999. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 185843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vélot, C., S. Lebreton, I. Morgunov, K. C. Usher, and P. A. Srere. 1999. Metabolic effects of mislocalized mitochondrial and peroxisomal citrate synthases in yeast Saccharomyces cerevisiae. Biochemistry 3816195-16204. [DOI] [PubMed] [Google Scholar]
- 42.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42339-343. [DOI] [PubMed] [Google Scholar]
- 43.Wanders, R. J., L. Ijlst, A. H. van Gennip, C. Jakobs, J. P. de Jager, L. Dorland, F. J. van Sprang, and M. Duran. 1990. Long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: identification of a new inborn error of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 13311-314. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Z. Y., C. R. Thornton, M. J. Kershaw, L. Debao, and N. J. Talbot. 2003. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 471601-1612. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, H., and M. C. Lorenz. 2008. Carnitine acetyltransferases are required for growth on non-fermentable carbon sources but not for pathogenesis in Candida albicans. Microbiology 154500-509. [DOI] [PubMed] [Google Scholar]