Abstract
Rhomboids are serine proteases that cleave their substrates within the transmembrane domain. Toxoplasma gondii contains six rhomboids that are expressed in different life cycle stages and localized to different cellular compartments. Toxoplasma rhomboid protein 1 (TgROM1) has previously been shown to be active in vitro, and the orthologue in Plasmodium falciparum processes the essential microneme protein AMA1 in a heterologous system. We investigated the role of TgROM1 to determine its role during in vitro growth of T. gondii. TgROM1 was localized in the secretory pathway of the parasite, including the Golgi apparatus and micronemes, which contain adhesive proteins involved in invasion of host cells. However, unlike other micronemal proteins, TgROM1 was not released onto the parasite surface during cell invasion, suggesting it does not play a critical role in cell invasion. Suppression of TgROM1 using the tetracycline-regulatable system revealed that ROM1-deficient parasites were outcompeted by wild-type T. gondii. ROM1-deficient parasites showed only modest decrease in invasion but replicated more slowly than wild-type cells. Collectively, these results indicate that ROM1 is required for efficient intracellular growth by T. gondii.
Toxoplasma gondii is the agent of toxoplasmosis, which represents a fatal threat to immunocompromised individuals (22). T. gondii belongs to the phylum Apicomplexa, a group of medically and economically important parasites including Plasmodium falciparum, the agent of malaria, and Cryptosporidium parvum, the cause of cryptosporidiosis. The complex life cycle of T. gondii contains a sexual stage in which oocysts are shed in the environment by cats and asexual stages consisting of dormant, slow-growing forms called bradyzoites and fast-growing forms called tachyzoites, which are extremely efficient at invading a variety of cells and disseminating within the host (13).
T. gondii contains several sets of secretory organelles including micronemes, rhoptries, and dense granules whose sequential secretion is responsible for attachment, penetration, and survival in the infected cell, respectively (8, 14). Attachment to host cells is initiated by secretion of microneme (MIC) proteins, which comprise a family of adhesive molecules (3, 6, 9). MIC proteins are typically found in complexes consisting of a transmembrane protein and a soluble partner(s) (10). MIC protein complexes also form higher-order complexes: for example, MIC2/M2AP is a heterohexamer composed of three molecules of MIC2 associated with three molecules of M2AP (21).
Apicomplexans use a unique form of locomotion called gliding motility, which relies on apical secretion and trafficking of MICs along the surface of the parasite (32). Translocation is mediated by an interaction between the cytoplasmic tail of MIC2 and aldolase, which bridges to the actin-myosin motor of the parasite (20). Upon contact with host cells, MIC2 and its associated binding partner M2AP are released at the tip of the parasite, where they participate in binding to the host cell and then translocated toward the posterior end of the parasite during penetration into the host cell (6, 31). At the posterior end, the complex is released from the surface by proteolysis within the transmembrane domain of MIC2 (7, 30, 40) and this cleavage is required for efficient invasion of host cells (4). MIC6 and TgAMA1 have also been shown to be cleaved within their transmembrane domains (18, 30), and AMA1 provides a vital function in formation of the moving junction (1, 24). Recent data strongly suggest that parasite-derived, rhomboid-like proteases cleave MIC proteins in order to release them from the parasite membrane (5, 12, 36).
Rhomboids are ubiquitous serine proteases that are able to recognize and cleave their substrates within their transmembrane domains (35, 37). Rhomboids have been extensively studied in Drosophila melanogaster, where they are involved in the regulation of development (38). Spitz is an epidermal growth factor-like protein that is a substrate for rhomboids in the fly. Spitz is also used as a universal substrate to characterize heterologous rhomboids in COS cells (36). The genome of T. gondii contains six rhomboid-like genes (TgROMs) (5, 11, 12). Analysis of the predicted TgROM6 sequence suggests that the protein is localized in the mitochondria (11). The in vitro activities of the remaining five rhomboids (TgROM1 to TgROM5) have been characterized by cell biological, molecular, and biochemical methods. TgROM2 and TgROM3 are mainly expressed in the sporozoites, with much lower expression in tachyzoites, as indicated by reverse transcription-PCR (RT-PCR) (5). Expression of a tagged form of TgROM2 in tachyzoites results in accumulation in the trans-Golgi network (12). In contrast, TgROM4 and TgROM5 are normally expressed at the surface of tachyzoites, with TgROM5 being primarily localized at the posterior end of the cell. Like other ROMs, TgROM1 is active against Spitz in a heterologous COS cell assay and this activity critically depends on the conserved transmembrane region of the substrate (5, 12). While TgROM1 is clearly a functional protease, its substrates have not been identified in the parasite. TgROM2 is also able to cleave chimeric proteins composed of the transmembrane domain of MIC2 or MIC12 (12), while only TgROM5 efficiently cleaves full-length MIC2 or chimeric proteins composed of the transmembrane domains of MIC6 or MIC12 (5). Despite demonstration of these in vitro activities, the in vivo roles of these proteases remain uncertain.
In this study, we focused on TgROM1, which is expressed in the tachyzoite stage of T. gondii. To define the role of TgROM1 in vivo, the expression of TgROM1 was conditionally suppressed using the tetracycline-repressible system described previously (25, 26).
MATERIALS AND METHODS
Growth of host cells and Toxoplasma strains.
T. gondii strain tTa (26) and transformants were maintained by growth in monolayers of human foreskin fibroblast (HFFs), propagated in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM glutamine, 20 mM HEPES (pH 7.5), and 20 μg ml−1 gentamicin. Chloramphenicol (20 μg/ml) (Sigma-Aldrich, St. Louis, MO), phleomycin (5 μg/ml) (Invitrogen, San Diego, CA), and anhydrotetracycline (Atc) (1.5 μg/ml) (Clontech, Palo Alto, CA) were added to the media as indicated.
Antibodies.
The HA9 epitope (YPYDVPDYA) was detected using rabbit polyclonal antisera (Zymed, CA), monoclonal antibody (MAb) 16B12 (Covance, CA), or MAb HA-7 (Sigma-Aldrich, St. Louis, MO) for immunofluorescence (IF), Western blotting, and immunoelectron microscopy (immuno-EM) experiments, respectively. MIC2 was detected with MAb 6D10 and rabbit polyclonal anti-C-domain antibodies for IF and immuno-EM experiments, respectively (39). MIC4 was detected with rabbit polyclonal antibodies, generously provided by Dominique Soldati, and MAb 5B1. TgAMA1 was detected with the MAb B3.90, generously provided by Gary Ward (University of Vermont). SAG1 was detected using MAb DG52 directly coupled to Alexa 488 or Alexa 594 (Molecular Probes). RON4 was detected with mouse polyclonal antisera, generously provided by Peter Bradley (University of California). GRA1 was detected with mouse MAb Tg-17-43, a kind gift of Marie France Cesbron-Delauw.
Generation of Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 strains.
The TgROM1 open reading frame (NCBI accession no. AAT84608) with an N-terminal HA9 tag was inserted in the previously described vectors p7TetOS1 and p7TetOS4, downstream of the inducible promoters that consist of seven TetO sequences fused with the upstream regions from the SAG1 and SAG4 genes, respectively (25), to generate the plasmids pS1HA9-ROM1 and pS4HA9-ROM1. Transactivator-expressing parasites, referred to as tTa (26), were cotransfected by electroporation with pS1HA9-ROM1 or pS4HA9-ROM1 and a plasmid containing the ble gene driven by SAG1 flanking sequences, conferring the resistance to phleomycin, as described previously (27). Following two rounds of selection, clones were obtained by limiting dilution on HFF monolayers grown in 96-well plates (27). ROM1/S1HA9-ROM1 or ROM1/S4HA9-ROM1 clones were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis or Western blotting and IF microscopy with anti-HA9 antibodies. The level of expression of HA9-ROM1 was higher when the protein was expressed under the control of the TetOSAG4 promoter compared to the TetOSAG1 promoter, as reported previously (25). ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1 clones were used to generate TgROM1 knockout lines as follows.
A knockout construct referred to as plasmid pΔR1 was engineered using the selectable marker cat, which confers resistance to chloramphenicol, controlled by 5′ and 3′ SAG1 flanking sequences. This cat cassette was in turn flanked by 2 kb of sequences upstream of the start and downstream of the stop codons of TgROM1 (sequences retrieved from http://ToxoDB.org). Two tandem yellow fluorescent protein (YFP) genes expressed under the control of the T. gondii α-tubulin (TUBA) promoter (a generous gift of B. Striepen) were inserted downstream into the SacII site of pΔR1, generating the plasmid pΔR1YFP. ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1 parasite strains were transfected by electroporation with 50 μg of linearized pΔR1YFP. After antibiotic selection and fluorescence-activated cell sorting, ∼10% of YFP-negative and chloramphenicol-resistant parasites were determined to be Δrom1 knockouts. Clones were obtained by limiting dilution for the Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 knockdown lines.
IF and deconvolution microscopy.
To detect HA9-ROM1 in intracellular parasites, HFF cell monolayers were infected with parasites and incubated in presence or absence of Atc overnight at 37°C and 5% CO2, prior to being fixed and processed for IF. Permeabilized cells were incubated with anti-HA9, anti-MIC2, or anti-MIC4 antibodies; washed; and incubated with secondary antibodies conjugated to Alexa 494 or Alexa 488 (Molecular Probes, OR). Coverslips were washed and mounted in Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). Fluorescence images were captured using a ×100, 1.4 numerical aperture lens on a Zeiss Axioskop 2 using an Axiocam MRm cooled charge-coupled device camera (Thornwood, New York, NY). Serial Z-stack images were collected at ×100, and images were deconvolved using the nearest-neighbor algorithm in Axiovision and processed using Photoshop 6.0 (Adobe Systems, San Jose, CA).
To localize HA9-ROM1 during host cell invasion, ROM1/S4HA9-ROM1 tachyzoites were allowed to settle by gravity onto 80% confluent cells at 4°C and warmed up for invasion during 5 min at 37°C in Dulbecco's modified Eagle's medium containing 20 mM HEPES, 0.2% sodium bicarbonate, and 3% fetal bovine serum. Invasion was stopped by addition of 2.5% paraformaldehyde. Monolayers were washed, blocked, and incubated with anti-SAG1 or anti-RON4 antibodies to label surface-exposed epitopes. Cells were then permeabilized with 0.5% saponin and incubated with anti-HA9 antibodies, followed by secondary antibodies coupled to Alexa 488 (green) and Alexa 594 (red). Monolayers were washed and mounted in Vectashield containing DAPI. Fluorescence images were captured using a ×63, 1.4 numerical aperture lens on a Zeiss Axioskop 2 using an Axiocam MRm cooled charge-coupled device camera (Thornwood, New York, NY). Serial Z-stacks were collected, and images were deconvolved as described above.
Invasion assay.
Invasion assays were performed as described previously (4). HFF monolayers were infected during a short invasion pulse (5 min or 20 min) with parasites that had been isolated from cultures treated during the previous 144 h with 1.5 μg/ml Atc versus control cultures that were not treated. Extracellular parasites were detected with anti-SAG1 antibodies directly coupled to Alexa 594 (red). Monolayers were then permeabilized with 0.5% saponin, and both intracellular and extracellular parasites were labeled with anti-SAG1 antibodies directly coupled to Alexa 488 (green). Monolayers were washed and mounted in Vectashield containing DAPI. The percentage of intracellular parasites was determined as the inverse of the ratio of red extracellular parasites versus the entire green population. More than 100 parasites were counted for each condition in four independent experiments, and the values reported are means ± standard errors.
EM.
Parasites were fixed in 4% paraformaldehyde-0.01% glutaraldehyde in 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] for 1 h at 4°C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose-20% polyvinylpyrrolidone in PIPES at 4°C. Samples were frozen in liquid nitrogen and sectioned with a cryo-ultramicrotome. Sections were probed with mouse or rabbit anti-HA9 and rabbit anti C-terminal domain of MIC2 followed by 18- and 12-nm colloidal gold-conjugated antispecies antibodies, respectively, stained with uranyl acetate/methylcellulose, and analyzed by transmission EM. Parallel controls omitting the primary antibody were consistently negative at the concentration of colloidal gold-conjugated secondary antibodies used in these studies. The distribution of immunogold particles was determined from samples that were immunostained for TgROM1 and MIC2 using primary rabbit antibodies followed by secondary antibodies conjugated to gold. Representative images of cross sections through the Golgi region and apical end were chosen for study. The cumulative length of membranes surrounding micronemes and Golgi apparatus, as identified by morphological criteria, were measured using Volocity 4.0 (Improvision, Lexington, MA), and the density of gold in particles per μm of membrane was determined.
RNA extraction and RT-PCR.
Total RNAs were obtained by treatment of parasites with TRIzol (Invitrogen, CA) for 5 min at room temperature, extraction with 20% chloroform, and precipitation with 50% isopropanol. RNAs were resuspended in water to a final concentration of 1 mg/ml. Aliquots of 1 μg of total RNA were used in separate, parallel reactions to reverse transcribe TgROM1 and ACT1 genes using SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Segments consisting of 800 and 400 bp of the cDNAs encoding ACT1 and ROM1, respectively, were then amplified for 30 cycles using Taq DNA polymerase (Sigma-Aldrich, MO) using the primer pairs actF-actR (for ACT1) and F6-R6 (for ROM1), respectively (see Table S1 in the supplemental material). RT-PCR products were loaded onto a 0.7% agarose gel stained with ethidium bromide.
Real-time qPCR.
Parasites were cultivated in presence or absence of 1.5 μg/ml Atc for 48 h. Three independent cultures were performed for each experimental condition. Total RNAs were extracted as described above. One microgram of total mRNA was used to reverse transcribe TgROM1 and TgACT1 with SuperScript III reverse transcriptase according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) using primers R7 and TgActinRTR, respectively (see Table S1 in the supplemental material). Quantitative PCR (qPCR) was performed using a SmartCycler (Cepheid, Sunnyvale, CA) with a reaction mixture volume of 25 μl containing SYBR green qPCR premix (Clontech, Palo Alto, CA), 200 nM of each primer, and 2 μl of reverse-transcribed cDNA. Following cDNA synthesis, primer pairs F8-R8 and TgActinRTR-TgActinRTF were used to amplify TgROM1 and TgACT1, respectively (see Table S1 in the supplemental material). The reaction conditions for PCR were 95°C for 1 min and 62°C for 30 s for 45 cycles. Data analysis was conducted using SmartCycler software (Cepheid). The relative TgROM1 expression levels were calculated as previously described (15) as the change (fold) using the formula 2−ΔΔCT, where ΔCT = threshold cycle (CT) of actin − CT of TgROM1 and ΔΔCT = ΔCT of parasites cultivated in absence of Atc − ΔCT of parasites cultivated in the presence of Atc.
Competition assay.
ROM1/S1HA9-ROM1 and Δrom1/S1HA9-ROM1 or ROM1/S4HA9-ROM1 and Δrom1/S4HA9-ROM1 strains were cocultivated (1:1) in the presence of Atc for 16 days (8 passages). After each round of egress, extracellular parasites were purified and proteins were digested with 10 mg/ml of proteinase K (Sigma-Aldrich, St. Louis, MO), as previously described (23). Samples containing crude extracts of genomic DNA were kept at −20°C until further processing. For PCR analysis, sets of primers specific for the endogenous TgROM1 gene (primers F1 and R1) or the cat gene (primers F5 and R5) were used to analyze the ratio of intermediate clones that were diploid (i.e., ROM1/S1HA9-ROM1 or ROM1/S4HA9-ROM1) versus knockout strains containing only the regulatable copy of TgROM1 (i.e., Δrom1/S1HA9-ROM1 or Δrom1/S4HA9-ROM1), respectively. Total genomic DNA corresponding to 103 parasites was used as a template. PCR products obtained after 30 cycles were loaded onto a 1% agarose gel and stained with ethidium bromide, and the intensity of each band was quantified using a PhosphorImager FLA-5000 (Fuji Medical Systems, Stamford, CT). The ratios of values obtained for the Δrom1/S1HA9-ROM1 versus ROM1/S1HA9-ROM1 or Δrom1/S4HA9-ROM1 versus ROM1/S4HA9-ROM1 strains were plotted, and the curve of best fit was added using linear regression analysis in Excel (Microsoft).
Growth assays.
Parasite strains were cultured in the presence of 1.5 μg/ml of Atc for three serial passages (2 days each) in T25 flasks containing HFF monolayers and complete culture medium. Purified parasites were then used to inoculate 96-well plates seeded with confluent HFF monolayers and cultured in the presence of 1.5 μg/ml Atc for 72 h. Monolayers were washed in phosphate-buffered saline, fixed with methanol, and stained with 0.1% crystal violet. Parasite growth was determined by the loss of monolayer integrity as monitored by absorbance when monitored at 570 nm using the EL800multiwell plate reader (Bio-Tek Instruments, VT). Parasite lines were tested in four separate experiments using quadruplicate wells for each sample.
Growth was also monitored using an intracellular replication assay. Freshly egressed parasites were used to challenge monolayers of HFF cells grown on coverslips. Following culture for 24 h, monolayers were fixed, permeabilized, and stained with fluorescently conjugated MAb DG52 to the surface protein SAG1. Monolayers were mounted in Vectashield containing DAPI and examined by epifluorescence microscopy. The average number of parasites per vacuole was determined by counting 100 or more cells from each of three coverslips in two or more experiments.
Statistics.
Statistical calculations were performed in Excel using Student's t test under the assumption of equal variance and using a two-tailed test.
RESULTS
TgROM1 localized to the Golgi apparatus and micronemes.
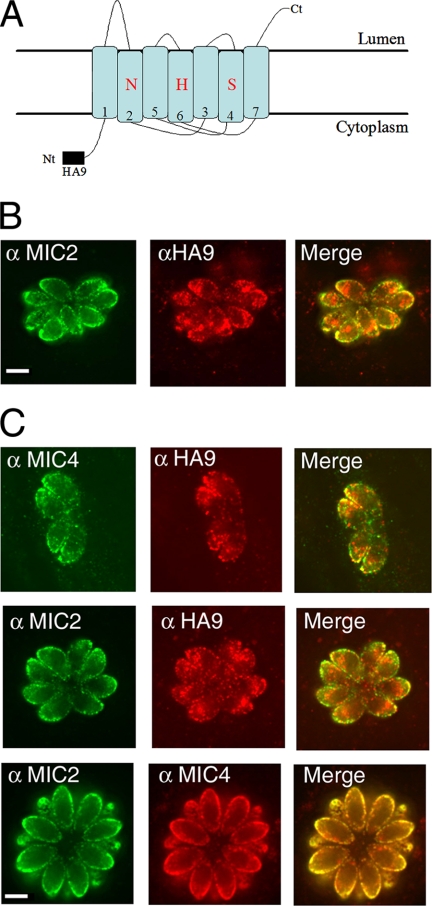
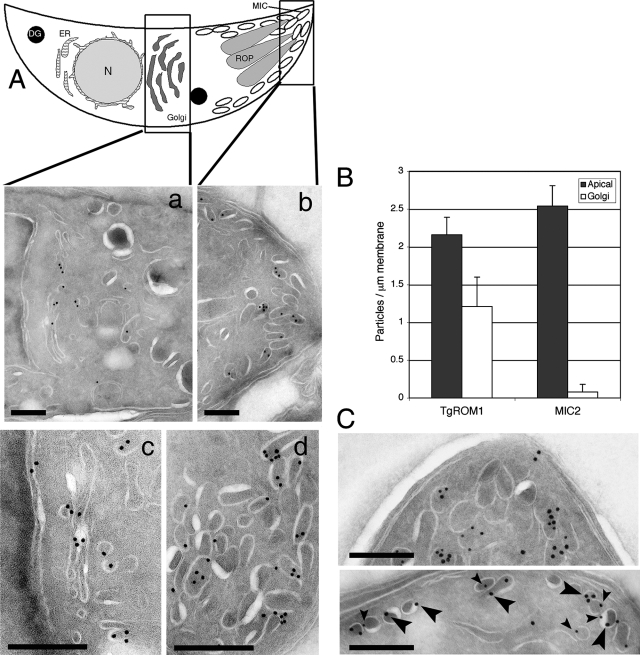
Epitope-tagged TgROM1 was previously reported to be associated with the apical end of T. gondii, where it partially colocalized with the microneme protein MIC2 (5, 12). We investigated the nature of the apical compartments harboring TgROM1 using deconvolution IF microscopy and cryoimmuno-EM. Rhomboids are poorly immunogenic due to the fact that the vast majority of the protein is embedded in the membrane; hence, specific antibodies against TgROM1 are not available. Therefore, we used the previously described construct encoding an amino-terminal, HA9-epitope-tagged TgROM1 under control of the endogenous promoter (1 kb upstream of the ATG) (5). TgROM1 is predicted to be a transmembrane protein with the N terminus extending into the cytosol along with the epitope tag added here and the C terminus in the lumen of the secretory system (Fig. 1A). Epitope-tagged TgROM1 was detected in intracellular parasites using anti-HA9 antibodies for indirect IF microscopy followed by deconvolution (Fig. 1B). HA9-ROM1 was partially colocalized with MIC2 at the apical margin of the cell and was also found in other perinuclear structures in the parasite (Fig. 1B). The distribution of HA9-ROM1 was also examined under the control of the pTetOSAG4 tetracycline-regulatable promoter, described previously (25). A similar pattern of partial colocalization with MIC2 and MIC4 was observed, with additional staining of HA9-ROM1 being found throughout the apical end of the parasite (Fig. 1C). In contrast, MIC2 and MIC4 were precisely colocalized in the micronemes at the apical margin of the cell (Fig. 1C). Immuno-EM revealed that HA9-ROM1 was associated with membranes of the micronemes and was also abundant in the Golgi stacks and occasionally observed in small vesicles anterior to the Golgi apparatus (Fig. 2A). Quantitative analysis of the distribution of HA9 labeling revealed that the abundant staining of the Golgi apparatus was characteristic of TgROM1 and not the microneme protein MIC2 (Fig. 2B). Double labeling with antibodies against MIC2 and the HA9 epitope showed considerable overlap in the micronemes, although some secretory vesicles were only labeled with one antibody (Fig. 2C). Collectively, these results reveal that TgROM1 is localized within the Golgi apparatus and micronemes of the parasite, in a pattern that partially overlaps with resident microneme proteins.
FIG. 1.
Subcellular localization of HA9-tagged TgROM1 in intracellular parasites. (A) Model of the tagged construct of TgROM-1HA9. The protein is predicted to adopt a transmembrane topology with the N terminus (Nt) in the cytosol along with the epitope tag. The C terminus (Ct) is predicted to extend into the lumen. Catalytic triad residues (N, asparagine; H, histidine; and S, serine), are shown in transmembrane domains 2, 6, and 4). (B) IF localization of HA9-ROM1 driven by the endogenous promoter in transiently transfected parasites. HA9-ROM1 was partially colocalized with MIC2 at the tip of intracellular parasites but also extended throughout the apical half of the parasite. At 16 h posttransfection, cells were fixed, permeabilized, and incubated with anti-HA9 (α HA9) or anti-MIC2 (α MIC2) antibodies and revealed using secondary antibodies coupled to Alexa 594 (red), or Alexa 488 (green). IF images were processed by deconvolution microscopy, and a single Z-slice is shown in each example. Scale bar, 5 μm. (C) Transgenic parasites expressing HA9-TgROM1 under control of the TetOSAG4-regulatable promoter showed partial colocalization with MIC2 and MIC4, again extending throughout the apical end of the parasite. In contrast, MIC2 and MIC4 show almost perfect overlap at the apical border of the parasite. Cells were fixed; permeabilized; incubated with anti-HA9, anti-MIC2, or anti-MIC4 (α MIC4) antibodies; and revealed using secondary antibodies coupled to Alexa 594 (red) or Alexa 488 (green). IF images were processed by deconvolution microscopy, and a single Z-slice is shown in each example. Scale bar, 5 μm.
FIG. 2.
Cryoimmuno-EM localization of TgROM1 in intracellular parasites. (A) Immuno-EM revealed that TgROM1 was localized in micronemes and the Golgi apparatus. Schematic representation of T. gondii showing dense granules (DG), rhoptries (ROP), micronemes (MIC), endoplasmic reticulum (ER), Golgi apparatus (Golgi), and nucleus (N). Cryoimmuno-EM was performed on extracellular parasites expressing HA9-ROM1 and detected with anti-HA9 antibody followed by secondary antibodies conjugated to 18-nm colloidal gold particles. Two separate images show the middle (a) and the apical end (b) of the parasite, respectively. Two other images taken at higher magnification show the Golgi apparatus (c) and the micronemes (d). Scale bars, 200 nm. (B) Quantitative distribution of immunogold from representative images of the Golgi apparatus and apical regions as defined in panel A. The distribution of TgROM1 was determined from 10 representative negatives stained as described above. The distribution of MIC2 was determined by staining with rabbit anti-C-domain followed by secondary antibodies conjugated to gold. Values shown are means ± standard deviations. (C) Double labeling with HA9-ROM1 (18-nm gold; large arrows) and MIC2 (12-nm gold; small arrows) revealed costaining of some compartments, while others contained only one of the markers. Scale bars, 200 nm.
TgROM1 was not detectable on the parasite surface or at the moving junction during cell invasion.
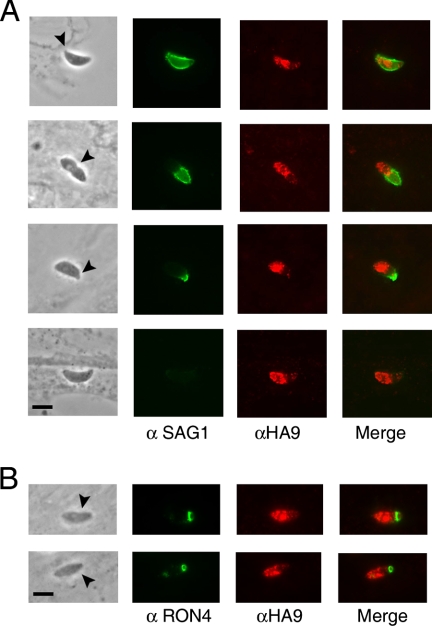
Micronemal proteins such as MIC2 are released at the apical end of the parasite and then translocated posteriorly during invasion (20). In order to test whether TgROM1 follows a similar path, pulse-infected monolayers were first stained with antibodies to the parasite surface protein SAG1, to reveal exposed epitopes. This procedure readily identifies parasites in the process of invading since only the posterior portion of the parasite body, which is outside the moving junction, is accessible to antibodies. Monolayers were then permeabilized and incubated with antibodies to reveal the HA9 epitope, which is predicted to be on the cytoplasmic side of the parasite membrane. Parasites that were apically attached or partially invading monolayers of host cells were examined to determine if HA9-ROM1 was exposed at the surface or if it translocated toward the posterior end of the parasite. TgROM1 was only detected at the apical end of the parasite and was not found at the junction or along the surface of the parasite (Fig. 3A). These observations are consistent with the localization in micronemes and secretory compartments. We also examined the distribution of TgROM1 with respect to the junctional marker RON4, which is released from the neck of the rhoptries and which defines the moving junction (1, 24). While RON4 demarked a clear ring at the junction, this did not coincide with staining for TgROM1 (Fig. 3B). Collectively these findings indicate that despite being found in micronemes, TgROM1 is not released in appreciable amounts onto the parasite surface during host cell invasion.
FIG. 3.
Localization of TgROM1 during cell invasion. (A) TgROM1 was found in apical compartments but was not detected at the surface of the parasite during invasion of host cell monolayers. Following a short invasion pulse of 5 min, monolayers were fixed and stained with antibodies to SAG1 (αSAG1) to reveal externally exposed epitopes. Monolayers were then permeabilized and incubated with anti-HA9 (αHA9) antibodies, followed by secondary antibodies coupled to Alexa 488 (green indicates SAG1) and Alexa 594 (red indicates HA9), respectively. IF images were processed by deconvolution microscopy. Arrowheads mark the site of apical attachment or the moving junction. Scale bars, 5 μm. (B) TgROM1 was not associated with the moving junction. Following pulse invasion, monolayers were fixed in 2.5% formaldehyde without detergent and incubated with anti-RON4 (αRON4) antibodies. Monolayers were then permeabilized and incubated with anti-HA9 antibodies followed by secondary antibodies coupled to Alexa 488 (green indicates RON4) and Alexa 594 (red indicates HA9). Scale bars, 5 μm. Arrowheads indicate the moving junction.
Generation of TgROM1 knockdown parasites.
To investigate the role of TgROM1 in T. gondii, we attempted to delete the endogenous gene by double homologous crossover using the cat selectable marker flanked by upstream and downstream sequences from TgROM1 as diagrammed in Fig. 4A. Despite repeated attempts, this approach did not yield viable knockouts (data not shown). This result suggested that the expression of TgROM1 was critical for the survival of T. gondii. Consequently, the role of TgROM1 was investigated using the tetracycline (Tet)-repressible system that allows analysis of essential genes in T. gondii (26). We used two regulatable promoters that are known to have different strengths, to drive the expression of HA9-ROM1, as reported previously (26).
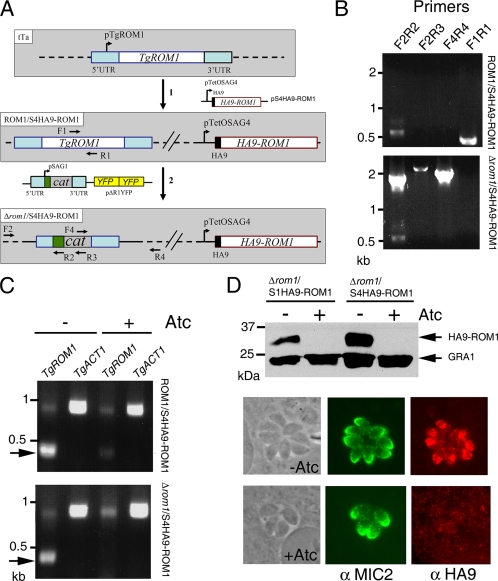
FIG. 4.
Construction of TgROM1 knockdowns. (A) Schematic representation of the strategy used to obtain knockdowns in T. gondii. The construction of the Δrom1/S4HA9-ROM1 strain is shown as an example. The tTa strain was transfected with plasmid pS4HA9-ROM1 containing HA9-tagged TgROM1 expressed under the control of the TetOSAG4 promoter generating the ROM1/S4HA9-ROM1 strain (step 1). Recombinant ROM1/S4HA9-ROM1 parasites contained both the endogenous and the tagged copy of TgROM1. ROM1/S4HA9-ROM1 clones were then transfected with plasmid pΔR1YFP designed to delete TgROM1 by replacing the endogenous copy with the chloramphenicol resistance marker (cat) surrounded by 5′ and 3′ untranscribed regions and flanking regions of TgROM1 and driven by the SAG1 promoter (step 2). Parasites were subsequently cloned by limiting dilution to obtain the Δrom1/S4HA9-ROM1 strain. The chromosome of each strain is shown as a dashed line. (B) PCR analysis demonstrating the disruption of TgROM1. PCR was performed using genomic DNAs from ROM1/S4HA9-ROM1 and Δrom1/S4HA9-ROM1 strains as templates and primers described in panel A. Primer pairs F2-R2, F2-R3, and F4-R4 amplified fragments of the expected sizes only in the Δrom1/S4HA9-ROM1 strain. Primer pair F1-R1 amplified a fragment of the expected size only in the ROM1/S4HA9-ROM1 strain. The smaller band seen in the first lane in each case is a nonspecific product. PCR products were resolved on a 0.7% agarose gel stained with ethidium bromide. (C) TgROM1 transcripts were not detected in the Δrom1/S4HA9-ROM1 strain cultivated in the presence of Atc. ROM1/S4HA9-ROM1 and Δrom1/S4HA9-ROM1 strains were cultivated in the absence (−) or presence (+) of Atc. Total RNAs were extracted, and RT-PCR was performed using primers specific for TgROM. The single-copy ACT1 gene was amplified in parallel as a control (TgACT1). RT-PCR products were resolved on a 0.7% agarose gel stained with ethidium bromide. An arrow indicates the TgROM1 amplification product. The band seen at 0.9 kbp is a nonspecific amplification product. (D) Repression of HA9-TgROM1 by culture in Atc. Clones were incubated in the absence (−) or presence (+) of Atc for 16 h for IF and 48 h for Western blotting. Western blotting was performed on parasite lysates using anti-HA9 antibodies or MAb Tg17-43 against GRA1 as a loading control. The IF assay was performed using antibodies against MIC2 (α MIC2) and against the HA9 epitope (α HA9) to detect HA9-ROM1 in intracellular parasites. Scale bar, 2 μm.
The parental tTa strain of T. gondii, harboring a transactivator that binds to the Tet operator, was transfected with the vector pS4HA9-ROM1 to obtain parasites whose genomes contained both endogenous and tagged copies of TgROM1 (referred to as ROM1/S4HA9-ROM1) (Fig. 4A). In a second step, a ROM1/S4HA9-ROM1 clone was transfected with the knockout vector pΔR1YFP to generate knockouts that lacked the endogenous gene but contained a regulatable HA9-tagged copy of TgROM1 (referred to as Δrom1/S4HA9-ROM1) (Fig. 4A). A Δrom1/S4HA9-ROM1 clone was obtained by limiting dilution and characterized at the genomic level. A similar strategy was used to obtain a Δrom1/S1HA9-ROM1 clone in which HA9-ROM1 was expressed under the control of the pTetOSAG1 promoter (25). The phenotypes reported below were similar for clones of both knockdowns (i.e., Δrom1/S4HA9-ROM1 and Δrom1/S1HA9-ROM1).
PCR was performed with primers designed to verify the integration of the cat gene in place of TgROM1 at the correct locus (Fig. 4A and B and see Table S1 in the supplemental material). Primer pairs F2-R2, F2-R3, and F4-R4 can only amplify products if the cat gene replaced TgROM1 following homologous recombination. This pattern was observed when PCR was performed with genomic DNA from Δrom1/S4HA9-ROM1 but not with that from ROM1/S4HA9-ROM1. In contrast, primers F1 and R1 hybridize within two separate introns of the genomic copy of TgROM1. Since the plasmid pS4HA9-ROM1 was generated from HA9-ROM1 cDNA, primer set F1-R1 can only amplify the endogenous TgROM1 gene. A product for the endogenous TgROM1 gene was observed when PCR was performed with genomic DNA from ROM1/S4HA9-ROM1 but not with that from Δrom1/S4HA9-ROM1. Taken together, these data indicate that the endogenous TgROM1 gene has been deleted in the Δrom1/S4HA9-ROM1 clone. Similar results confirmed that the TgROM1 gene was disrupted in the Δrom1/S1HA9-ROM1 clone (data not shown).
We determined the capacity of the nontoxic tetratcycline analogue, Atc, to down regulate the expression of HA9-ROM1 in the transgenic ROM1/S4HA9-ROM1 and Δrom1/S4HA9-ROM1 parasite clones following treatment for 48 h (Fig. 4C). The expression of TgROM1 was determined by RT-PCR, using primers that detect expression of both TgROM1 and HA9-ROM1 compared to actin (TgACT1) as an internal control. mRNA encoding TgROM1 was detected in absence or presence of Atc in the ROM1/S4HA9-ROM1 clone, as expected, since it contains the endogenous copy as well as a regulatable copy (Fig. 4C). In the absence of Atc, the signal corresponding to the TgROM1 transcript was more intense than that observed in presence of Atc, confirming that HA9-ROM1 was down regulated in the presence of Atc. In the Δrom1/S4HA9-ROM1 clone, mRNA encoding TgROM1 was detected only in the absence of Atc. These data indicate that Atc efficiently down regulates the expression of HA9-ROM1.
Suppression of HA9-ROM1 was confirmed by immunofluorescence and Western blotting using anti-HA9 antibodies to probe the transgenic Δrom1/S4HA9-ROM1 and Δrom1/S1HA9-ROM1 clones (Fig. 4D). The expression level of HA9-ROM1 was approximately twofold higher in the S4HA9-ROM1 clone versus the S1HA9-ROM1 clone, and addition of Atc for 48 h reduced the expression of HA9-ROM1 to undetectable levels in both clones. In intracellular parasites examined by indirect IF labeling, HA9-ROM1 partially colocalized at the apical end of the parasite with MIC2 in the absence of Atc. In contrast, HA9-ROM1 was not detectable by IF in parasites incubated with Atc for 16 h (Fig. 4D). These results indicate a tight down regulation of the expression of HA9-ROM1 by Atc to a level that is not readily detectable.
In order to more precisely define the degree of suppression of TgROM1 expression over time, mRNA levels were determined using qPCR. The expression of TgROM1 in the Δrom1/S4HA9-ROM1 and Δrom1/S1HA9-ROM1 clones, which express only the regulated copy of the gene, was compared to the tTA line, which expresses only the endogenous copy of the gene. Primers designed to amplify TgROM1 from both backgrounds were normalized for actin message (see Materials and Methods) to determine the degree of repression in the knockdown clones. When parasites were grown in Atc for 48 h, TgROM1 mRNAs were repressed by 97% in the Δrom1/S4HA9-ROM1 clone and 89% in the Δrom1/S1HA9-ROM1 clone, respectively (Table 1). However, when clones were gown for 96 h in Atc, the degree of suppression was greater and reached ∼98 to 99% in both clones (Table 1).
TABLE 1.
Repression of Tet-regulatable HA9-ROM1 expression in the presence of Atca
| Strain | Result atb:
|
|||
|---|---|---|---|---|
| 48 h
|
96 h
|
|||
| 2−ΔΔCT | Repression (%) | 2−ΔΔCT | Repression (%) | |
| tTa | ||||
| Δrom1/S1HA9-ROM1 | 0.103 | 89.7 | 0.021 | 97.9 |
| Δrom1/S4HA9-ROM1 | 0.033 | 96.7 | 0.015 | 98.5 |
Primers detect both wild-type TgROM1 and the HA9-ROM1 copies as described in Materials and Methods.
ΔΔCT = ΔCT of tTa − ΔCT of Δrom1 clones. The data are an average of three separate replicates. Repression is relative to the wild-type level of TgROM1.
TgROM1 was essential for growth of T. gondii.
The failure to obtain a direct knockout suggested there is an advantage to the parasite to retain the ROM1 gene. Initial attempts to define a phenotype in TgROM1 knockdown clones failed to identify any growth or invasion defect when the clones were cultured for 48 h in Atc and then tested in the presence or absence of Atc (data not shown). The results of qPCR experiments (Table 1) suggest that shutdown achieved in this time frame only results in partial suppression, leaving a residual level of ∼10% of wild-type levels in the Δrom1/S1HA9-ROM1 clone and ∼5% in the Δrom1/S4HA9-ROM1 clone. This low level of expression was evidently sufficient to mask the phenotype of ROM1 in vitro.
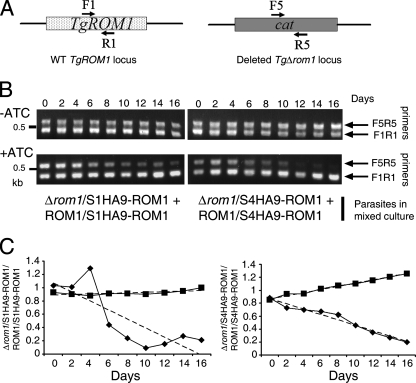
To test the role of ROM1 in long-term growth, we designed a competition assay in which the Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 parasite clones were cocultured in the presence of Atc with their respective pseudodiploid strains (i.e., ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1, respectively). Addition of Atc during prolonged growth shuts down the expression of HA9-ROM1 in all strains. While the ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1 strains can still express TgROM1, the Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 clones have had this gene deleted and thus do not express detectable levels of TgROM1. Multiplex PCR analysis was performed after each round of host cell egress (serially passaged on a 2-day cycle). Primer pair F1-R1 was used to detect the wild-type copy of TgROM1 and thus to selectively track the ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1 strains. Primer pair F5-R5 was used to detect the cat gene in the Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 clones (Fig. 5A). The intensity of PCR products obtained with primers F5 and R5 was initially the same for up to 4 to 6 days and then steadily decreased over time (Fig. 5B). In contrast, the intensity of PCR products obtained with primers F1 and R1 increased over time. Quantification of the ratio of the PCR bands over the 16-day period confirmed these observations (Fig. 5C). These results indicate that the TgROM1 knockdown clones (i.e., Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1) were outcompeted by wild-type parasites, (i.e., ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1) under conditions in which expression of HA9-ROM1 is repressed. In order to control for possible inherent growth differences in the backgrounds of these clones, they were also tested in a competition assay in the absence of Atc. Transgenic clones harboring HA9-ROM1 in addition to the endogenous TgROM1 gene grew at the same rate as the Δrom1 clones that only contained the tagged copy (Fig. 5B and C). These results indicate that the defect in Δrom1 clones in the presence of Atc is specific to the absence of ROM1 and not an artifact of the transgenic lines used.
FIG. 5.
Competition assay demonstrating that TgROM1 was required for efficient growth of T. gondii. (A) Schematic representation of primers used for detection of the wild type (WT) and deleted ΔTgrom1 loci. Primers F1 and R1 selectively amplify the TgROM1 gene. Primers F5 and R5 selectively amplify the cat gene. (B) Δrom1 clones were outcompeted by strains containing an endogenous copy of TgROM1 in a mixed culture over time when the regulatable copy of HA9-ROM1 was repressed by continuous culture in Atc. In contrast, Δrom1 clones showed no growth defect when the regulatable copy was not repressed with Atc. Parasites were added at 1:1 at the starting time and cocultured in the absence (−) or presence (+) of Atc. Every 2 days, extracellular parasites were purified and genomic DNA was extracted for multiplex PCR analysis. ROM1/S1HA9-ROM1 and ROM1/S4HA9-ROM1 strains were tracked using primer pair F1-R1, and Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 strains were tracked using primer pair F5-R5. PCR products were resolved on a 1% agarose gels stained with ethidium bromide. (C) The intensity of the bands seen in panel B was quantified using a PhosphorImager. The ratios of the values obtained for the knockdowns in the Δrom1 background versus the nondeleted strains are shown for each passage in the absence (solid squares) or presence (solid diamonds) of Atc. The lines of best fit (dashed lines) were determined by linear regression.
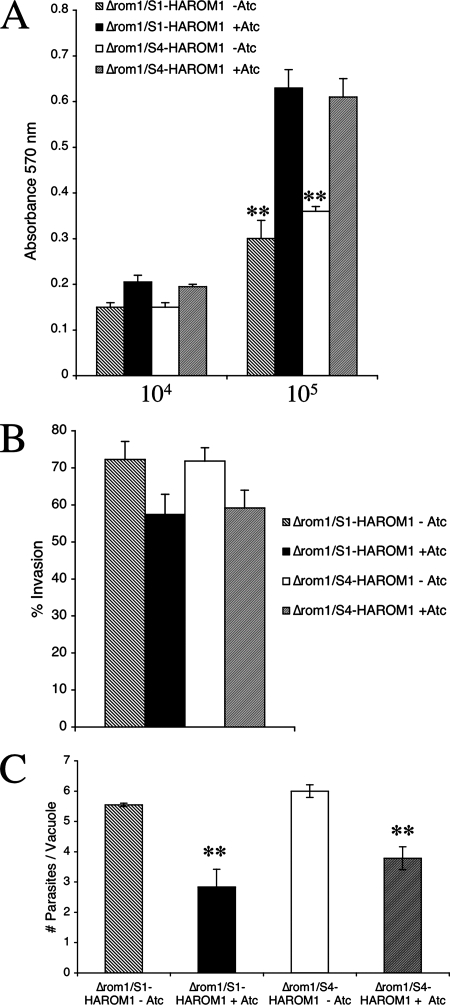
The gradual loss of TgROM1 knockdown clones when grown in the presence of wild-type parasites could reflect competition for host cell receptors, nutrients, or other factors that are necessary for growth. To test whether growth of the TgROM1 knockdowns was also reduced when they were cultured alone, we employed a monolayer lysis assay following extended passage in Atc to efficiently shut down expression of TgROM1 (see Materials and Methods). When parasites were inoculated at a multiplicity of infection of 1:1 (104 parasites in Fig. 6A) with the host cell monolayers, no defect in the extent of lysis was observed. In contrast, when inoculated at a multiplicity of infection of 1:10 (105 parasites in Fig. 6A), there was a 50% reduction in the extent of lysis by TgROM1 knockdown clones (i.e., Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1) in the presence of Atc compared to growth in the absence of Atc (Fig. 6A). This result indicates that the knockdowns have a growth defect that is dependent on the inoculation dose but that does not depend on competition with wild-type parasites.
FIG. 6.
Decreased growth of TgROM1 knockdown clones was associated with slower growth and less-efficient invasion. (A) Monolayer lysis assay revealed a dose-dependent defect in the TgROM knockdown clones following suppression of ROM1. Monolayers of HFF cells were inoculated with Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 parasites in the presence (+) or absence (−) of Atc. After culture for 3 days, monolayers were rinsed, fixed, and stained with crystal violet and the absorbance was read at 570 nm. Decreased absorbance (staining of host cells) was a result of lysis due to parasite growth, as confirmed by microscopic examination. The x axis shows the challenge dose of parasites. Values are means ± standard deviations (n = 4 wells per sample), representative of four experiments. **, P < 0.001 by Student's t test. (B) Invasion assays revealed a modest reduction in invasion by TgROM1 knockdown clones following suppression of ROM1. Parasites were grown for several passages in the presence or absence of Atc, harvested from freshly egressed cultures, and used to challenge monolayers of HFF cells cultured on glass coverslips. Following a 5-min pulse invasion, parasites were classified as intracellular versus extracellular based on accessibility to staining of the surface antigen SAG1. Repression of ROM1 led to an ∼15% reduction in invasion in both the Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 strains. Values are means ± standard errors of the mean (n = 3 experiments). (C) Intracellular replication was more severely decreased in TgROM1 knockdown clones following suppression of TgROM1. Parasites were grown for several passages in the absence or presence of Atc, harvested from freshly egressed cultures, and used to challenge monolayers of HFF cells cultured on glass coverslips. Following 24 h of incubation, parasites were fixed and stained for the surface protein SAG and with DAPI to reveal nuclei, and the average number of parasites per vacuole was determined by counting under epifluorescence microscopy. Values are means ± standard deviations (n = 3 coverslips per group), representative of duplicate experiments. **, P < 0.001 by Student's t test.
To further define the defect in TgROM1 knockdown clones, we compared the invasion efficiencies of TgROM1 knockdown clones (i.e., Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1) that were grown in Atc for several passages to shut down expression completely. Freshly egressed parasites were then used to challenge HFF monolayers in a short-term pulse (i.e., 5 min), and parasites were scored for invasion based on staining with a surface antibody to SAG1 both before and after detergent permeabilization. Suppression of ROM1 expression by treatment with Atc resulted in only a slight reduction in the percentage of parasites that invaded compared to parasite lines grown in the absence of Atc (Fig. 6B).
Analysis of the intracellular growth of the TgROM1 knockdowns revealed a much stronger phenotype (Fig. 6C). The Δrom1/S1HA9-ROM1 and Δrom1/S4HA9-ROM1 knockdown clones were grown in Atc for several passages to shut down expression. Freshly egressed parasites were then used to challenge HFF monolayers grown on coverslips, and parasites were allowed to replicate for 24 h. The average number of parasites per vacuole was reduced by almost half when the TgROM1 knockdowns where grown in the presence of Atc (Fig. 6C). Collectively, these results suggest that suppression of TgROM1 has only a modest effect in invasion but impairs intracellular replication, resulting in slower growth.
DISCUSSION
We analyzed the role of the rhomboid protease TgROM1 in invasion of host cells and intracellular replication by T. gondii. TgROM1 was localized to the Golgi apparatus and micronemes, although it was not detected on the parasite surface during invasion, suggesting it fulfills its role in an intracellular compartment. Using the tetracycline-repressible system, we have demonstrated that the rhomboid protease TgROM1 plays an important role in parasite growth in vitro. This requirement was attributed to a decrease in the rate of replication in the absence of detectable ROM1 expression. Complete repression of ROM1 required extended treatment with Atc to achieve levels of shutdown of >98% and to appreciate phenotypes. Repression of TgROM1 led to a significant growth disadvantage over time versus wild-type parasites. The Tet-repressible strategy, combined with in vitro competition assays described here, allows the evaluation of genes that have subtle yet important phenotypes in the survival of parasites in vitro.
Deconvolution IF and immuno-EM were used to demonstrate that TgROM1 is localized in micronemes as well as in the Golgi apparatus of T. gondii. Such prominent Golgi apparatus staining is not seen when other ROMs are tagged in this manner (12), suggesting this result is not due to the influence of the HA9 tag, although this remains a formal possibility. The expression level of the transgenes was also closely matched to endogenous levels (within two- to threefold), indicating this pattern is unlikely to be due to overexpression. TgROM1 was largely localized to the Golgi apparatus, and only limited staining of post-Golgi secretory vesicles was observed, other than mature micronemes. This pattern differs from the post-Golgi localization of pro-MIC2, which occupies a compartment thought to be important for maturation of microneme proteins (17). Within the apical microneme organelles, the pattern of ROM1 staining only partially overlapped that of MIC2. Whether this indicates heterogeneity of secretory organelles or inefficient labeling is uncertain; however, it might represent a functional separation of the protease from potential substrates.
Transmembrane microneme proteins are released from the micronemes and transiently occupy the cell surface before being shed by proteolysis, which often occurs by intramembrane cleavage by a rhomboid protease. The Tet-repressible system has previously been used to knock down the expression of two transmembrane microneme proteins, MIC2 and TgAMA1 (19, 28). Both proteins play crucial roles in the invasion of host cells by parasites, although they act at different steps. MIC2 is required for attachment and hence invasion, as well as helical gliding motility of tachyzoites (19). TgAMA1 is required for the tight apical interaction with the host cell that leads to rhoptry secretion and invasion into the parasitophorous vacuole (28). As with other transmembrane MIC proteins, TgAMA1 is processed primarily within its transmembrane domain, as shown by matrix-assisted laser desorption ionization-time of flight analysis of peptides generated from secreted TgAMA1 (18). Previous studies have failed to demonstrate a role for TgROM1 in processing MIC proteins in vitro (5, 12), and results described here suggest that ROM1 does not traffic to the cell surface and thus may not be involved in processing micronemal proteins. In contrast, in vitro assays have demonstrated that TgROM5 is able to efficiently process TgAMA1 (S. Urban, personal communication) and MIC2 (5), suggesting it is responsible for processing these key surface proteins involved in invasion by T. gondii.
In contrast to known transmembrane MICs, TgROM1 was not detected at the surface of the parasite upon invasion of host cells, although we cannot rule out the possibility that low levels of the protein might not be detected. Overall, our findings are most consistent with TgROM1 performing its function within the secretory system, rather than on the cell surface. During intracellular growth, the parasite undergoes an unusual form of cell division termed endodyogeny (34). During this process, maternal organelles are largely resorbed before forming de novo again in the daughter cells. While these processes are not well understood, it is possible that ROM1 plays a role in recycling protein components within the secretory pathway during cell division. Despite suggesting a role for proteolytic processing during cell division, our studies do not identify the precise targets of ROM1 within the cell. TgROM2 has also been localized to the Golgi apparatus (12), raising the possibility that it performs an overlapping or redundant function.
Plasmodium spp. express orthologues of ROM1, although several features of their localization and potential activities differ from the results described here for T. gondii. While P. falciparum ROM1 (PfROM1) is capable of cleaving PfAMA1 in a heterologous cellular assay (2), this is thought to be a rare event in the parasite, based on mass spectrometric data (18). PfROM1 has been associated with the micronemes (29) and more recently with a novel apical organelle called the mononeme (33). Our results with TgROM1 do not reveal a similar organelle in T. gondii, although TgROM1 was a prominent component in the Golgi apparatus. Collectively, these differences indicate that ROM1 may function differently between these parasites rather than underlying a function(s) common to the phylum.
While reverse genetics tools allow dissection of the role of specific genes, the normal definition of “essential” genes implies that they have a strong phenotype in a defined assay. TgROM1 is not essential for invasion, yet absence of this protein led to a decrease in replication efficiency. This effect was somewhat modest in that it could be overcome by a high inoculum. However, the deficiency was cumulative and led to a significant loss in a competitive assay against wild-type parasites over time. In this regard, TgROM1 and similar proteins are essential for fitness, since mutants lacking such genes would rapidly be lost from the population. This may also explain why parasites contain multiple ROM genes (5), which may encode isoforms with either different substrate specificity or with the potential to enhance the effectiveness of substrate processing.
Similarly to the growth defect observed with TgROM1 knockdowns, a subtle growth phenotype was also obtained with a knockout of PP2C-hn in T. gondii (16). The PP2C-hn gene encodes a protein phosphatase 2C that is secreted from the rhoptries and translocated to the nucleus of host cells upon invasion. Δpp2c-hn knockout parasites display a growth defect that is only revealed by competition assay with a wild-type strain. Taken together, it appears that two categories of genes are important for efficient survival of T. gondii: those whose expression is absolutely critical for events like motility and cell invasion, such as MIC2 or TgAMA1, and those whose contribution is less dramatic yet still important for long-term survival, such as TgROM1 and PP2C-hn. Given the complex strategies parasites have evolved for optimizing survival, it is likely that a large proportion of genes will have such subtle phenotypes, which are nonetheless essential in a larger context.
Supplementary Material
Acknowledgments
We thank Dominique Soldati (University of Geneva, Geneva, Switzerland) for the generous gift of rabbit anti-MIC4 polyclonal antibodies, the tTa strain, and the p7TetOS1 and p7TetOS4 plasmids; Gary Ward (University of Vermont) for the MAb B3.90 against TgAMA1; Marie France Cesbron-Delauw (Universite Joseph Fourier, Grenoble, France) for the MAb to GRA1; Peter Bradley (University of California, Los Angeles) for the mouse anti-RON4 polyclonal antibodies; and Boris Striepen (University of Georgia, Athens) for the TUBA-YFPYFP plasmid.
This work was supported in part by National Institutes of Health grant AI 34036.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alexander, D. L., J. Mital, G. E. Ward, P. J. Bradley, and J. C. Boothroyd. 2005. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, R. P., R. Wijetilaka, and S. Urban. 2006. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2922-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brecht, S., V. B. Carruthers, D. J. Ferguson, O. K. Giddings, G. Wang, U. Jaekle, J. M. Harper, L. D. Sibley, and D. Soldati. 2001. The Toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem. 2764119-4127. [DOI] [PubMed] [Google Scholar]
- 4.Brossier, F., T. J. Jewett, J. L. Lovett, and L. D. Sibley. 2003. C-terminal processing of the Toxoplasma protein MIC2 is essential for invasion into host cells. J. Biol. Chem. 2786229-6234. [DOI] [PubMed] [Google Scholar]
- 5.Brossier, F., T. J. Jewett, L. D. Sibley, and S. Urban. 2005. A spatially-localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc. Natl. Acad. Sci. USA 1024146-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carruthers, V. B., O. K. Giddings, and L. D. Sibley. 1999. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell. Microbiol. 1225-236. [DOI] [PubMed] [Google Scholar]
- 7.Carruthers, V. B., G. D. Sherman, and L. D. Sibley. 2000. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J. Biol. Chem. 27514346-14353. [DOI] [PubMed] [Google Scholar]
- 8.Carruthers, V. B., and L. D. Sibley. 1997. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur. J. Cell Biol. 73114-123. [PubMed] [Google Scholar]
- 9.Cérede, O., J. F. Dubremetz, D. Bout, and M. Lebrun. 2002. The Toxoplasma gondii protein MIC3 requires pro-peptide cleavage and dimerization to function as an adhesin. EMBO J. 212526-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowse, T., and D. Soldati. 2004. Host cell invasion by the apicomplexans: the significance of microneme protein proteolysis. Curr. Opin. Microbiol. 7388-396. [DOI] [PubMed] [Google Scholar]
- 11.Dowse, T., and D. Soldati. 2005. Rhomboid-like proteins in Apicomplexa: phylogeny and nomenclature. Trends Parasitol. 21254-258. [DOI] [PubMed] [Google Scholar]
- 12.Dowse, T. J., J. C. Pascall, K. D. Brown, and D. Soldati. 2005. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. Int. J. Parasitol. 35747-756. [DOI] [PubMed] [Google Scholar]
- 13.Dubey, J. P. 1998. Advances in the life cycle of Toxoplasma gondii. Int. J. Parasitol. 281019-1024. [DOI] [PubMed] [Google Scholar]
- 14.Dubremetz, J. F., A. Achbarou, D. Bermudes, and K. A. Joiner. 1993. Kinetics and pattern of organelle exocytosis during Toxoplasma gondii/host-cell interaction. Parasitol. Res. 79402-408. [DOI] [PubMed] [Google Scholar]
- 15.Fux, B., J. Nawas, A. Khan, D. B. Gill, C. Su, and L. D. Sibley. 2007. Toxoplasma gondii strains defective in oral transmission are also defective in developmental stage differentiation. Infect. Immun. 752580-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert, L. K., S. Ravindran, J. M. Turetzky, J. C. Boothroyd, and P. J. Bradley. 2007. Toxoplasma gondii targets a protein phosphatase 2C to the nuclei of infected host cells. Eukaryot. Cell 673-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper, J. M., M. H. Huynh, I. Coppens, F. Parussini, S. N. Moreno, and V. B. Carruthers. 2006. A cleavable propeptide influences Toxoplasma infection by facilitating the trafficking and secretion of the TgMIC2-M2AP invasion complex. Mol. Biol. Cell 174551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell, S. A., F. Hackett, A. M. Jongco, C. Withers-Martinez, K. Kim, V. B. Carruthers, and M. J. Blackman. 2005. Distinct mechanisms govern proteolytic shedding of a key invasion protein in apicomplexan pathogens. Mol. Microbiol. 571342-1356. [DOI] [PubMed] [Google Scholar]
- 19.Huynh, M. H., and V. B. Carruthers. 2006. Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jewett, T. J., and L. D. Sibley. 2003. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell 11885-894. [DOI] [PubMed] [Google Scholar]
- 21.Jewett, T. J., and L. D. Sibley. 2004. The Toxoplasma proteins MIC2 and M2AP for a hexameric complex necessary for intracellular survival. J. Biol. Chem. 2799362-9369. [DOI] [PubMed] [Google Scholar]
- 22.Joynson, D. H., and T. J. Wreghitt. 2001. Toxoplasmosis: a comprehensive clinical guide. Cambridge University Press, Cambridge, United Kingdom.
- 23.Khan, A., C. Su, M. German, G. A. Storch, D. Clifford, and L. D. Sibley. 2005. Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains. J. Clin. Microbiol. 435881-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebrun, M., A. Michelin, H. El Hajj, J. Poncet, P. J. Bradley, H. J. Vial, and J. F. Dubremetz. 2005. The rhoptry neck protein RON4 relocalizes at the moving junction during Toxoplasma gondii invasion. Cell. Microbiol. 71823-1833. [DOI] [PubMed] [Google Scholar]
- 25.Meissner, M., S. Brecht, H. Bujard, and D. Soldati. 2001. Modulation of myosin A expression by a newly established tetracycline repressor based inducible system in Toxoplasma gondii. Nucleic Acids Res. 29E115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meissner, M., D. Schluter, and D. Soldati. 2002. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298837-840. [DOI] [PubMed] [Google Scholar]
- 27.Messina, M., I. R. Niesman, C. Mercier, and L. D. Sibley. 1995. Stable DNA transformation of Toxoplasma gondii using phleomycin selection. Gene 165213-217. [DOI] [PubMed] [Google Scholar]
- 28.Mital, J., M. Meissner, D. Soldati, and G. E. Ward. 2005. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol. Biol. Cell 164341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Donnell, R. A., F. Hackett, S. A. Howell, M. Treeck, N. Struck, Z. Krnajski, C. Withers-Martinez, T. W. Gilberger, and M. J. Blackman. 2006. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 1741023-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opitz, C., M. Di Cristina, M. Reiss, T. Ruppert, A. Crisanti, and D. Soldati. 2002. Intramembrane cleavage of the microneme proteins at the surface of apicomplexan parasite Toxoplasma gondii. EMBO J. 211577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabenau, K. E., A. Sohrabi, A. Tripathy, C. Reitter, J. W. Ajioka, F. M. Tomley, and V. B. Carruthers. 2001. TgM2AP participates in Toxoplasma gondii invasion of host cells and is tightly associated with the adhesive protein TgMIC2. Mol. Microbiol. 41537-547. [DOI] [PubMed] [Google Scholar]
- 32.Sibley, L. D. 2004. Invasion strategies of intracellular parasites. Science 304248-253. [DOI] [PubMed] [Google Scholar]
- 33.Singh, S. B., M. Plassmeyer, D. Gaur, and L. H. Miller. 2007. Mononeme: a new secretory organelle in Plasmodium falciparum merozoites identified by localization of rhomboid-1 protease. Proc. Natl. Acad. Sci. USA 10420043-20048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Striepen, B., C. N. Jordan, S. Reiff, and G. G. van Dooren. 2007. Building the perfect parasite: cell division in Apicomplexa. PLoS Pathog. 3691-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urban, S. 2006. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 203054-3068. [DOI] [PubMed] [Google Scholar]
- 36.Urban, S., and M. Freeman. 2003. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 111425-1434. [DOI] [PubMed] [Google Scholar]
- 37.Urban, S., J. R. Lee, and M. Freeman. 2001. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107173-182. [DOI] [PubMed] [Google Scholar]
- 38.Urban, S., J. R. Lee, and M. Freeman. 2002. A family of rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 214277-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan, K. L., V. B. Carruthers, L. D. Sibley, and J. W. Ajioka. 1997. Molecular characterisation of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Mol. Biochem. Parasitol. 84203-214. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, X. W., M. J. Blackman, S. A. Howell, and V. B. Carruthers. 2004. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol. Cell. Proteomics 3565-576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.