Abstract
Microsatellites are composed of short tandem direct repeats; deletions or duplications of those repeats through the process of replication slippage result in microsatellite instability relative to other genomic loci. Variation in repeat number occurs so frequently that microsatellites can be used for genotyping and forensic analysis. However, an accurate assessment of the rates of change can be difficult because the presence of many repeats makes it difficult to determine whether changes have occurred through single or multiple events. The current study was undertaken to experimentally assess the rates of replication slippage that occur in vivo in the chloroplast DNA of Chlamydomonas reinhardtii. A reporter construct was created in which a stretch of AAAG repeats was inserted into a functional gene to allow changes to be observed when they occurred at the synthetic microsatellite. Restoration of the reading frame occurred through replication slippage in 15 of every million viable cells. Since only one-third of the potential insertion/deletion events would restore the reading frame, the frequency of change could be deduced to be 4.5 × 10−5. Analysis of the slippage events showed that template slippage was the primary event, resulting in deletions rather than duplications. These findings contrasted with events observed in Escherichia coli during maintenance of the plasmid, where duplications were the rule.
Microsatellite sequences, also called simple sequence repeats (SSRs), are short tandem DNA repeats, 1 to 6 bases long, commonly found in the genomes of eukaryotes and some prokaryotes (4, 6, 12, 40). These repeats, which are present in both the noncoding and coding regions of genomes, are unstable, undergoing additions or deletions of one or more repeat units, leading to variations in the length of the microsatellite (17, 35, 38). In most cases, the insertions or deletions (indels) occur as a consequence of slippage of the template or daughter strand at the replication fork (16, 18). Although a similar indel could theoretically occur through intra- or intermolecular recombination, homologous recombination does not appear to be involved in microsatellite variability, since the variation is independent of homologous recombination factors, such as the RecA protein (18).
Because of their abundance and variability, microsatellite loci have been used extensively as genetic markers in evolutionary and ecological studies of natural populations in eukaryotes (14, 37) and also as highly polymorphic markers for forensics and genotyping of animals (39). Well-saturated microsatellite maps have been developed for nuclear genomes from a number of plants, including rice, maize, barley, Arabidopsis, and soybean (14, 23). In some cases, nuclear microsatellite instability is remarkably high: in chickpea, indels were found at an average rate of 2.9 × 10−3 to 10 × 10−3 per (TAA)n locus per generation (36). An even higher rate of somatic instability was found for mononucleotide microsatellites in reporter genes in Arabidopsis thaliana, with individual leaves having many sectors (2, 3).
Microsatellites and larger tandem repeats have also been identified in all chloroplast genomes from which sequence data are available (reviewed in reference 25), including the unicellular green alga Chlamydomonas reinhardtii (20). In barley, rice, and pine, chloroplast microsatellites have been used to reveal much higher levels of diversity than can be observed through traditional chloroplast restriction fragment length polymorphism analysis (reviewed in reference 25). From their observations of sequence data from Pinus species, Provan et al. (24) concluded that SSR length polymorphisms occur in chloroplast DNA (cpDNA) at frequencies of 3 × 10−5 to 8 × 10−5 per site per year.
The aforementioned studies provide a useful starting point for assessing microsatellite variability, but interpretation of SSR data based solely on an evolutionary context is problematic, because single changes can easily involve multiple repeats. Because the repeats are identical, independent deletions or duplications can result in the same DNA variations. Although such “synapomorphies” occur in parallel, they would be scored as arising from a single event. With a goal of providing an accurate assessment of the rate of chloroplast microsatellite variability, we created an experimental system that would allow us to monitor change in a microsatellite reporter in the cpDNA of the green alga C. reinhardtii. Since such mutational analyses require data from a large number of individuals, the algal system was chosen over a higher plant due to the ease in handling large populations in a limited space. In addition, the easy transformability and selection procedures for Chlamydomonas cpDNA have made it a uniquely accessible system for such a study. To create the microsatellite slippage reporter, a stretch of repeats was introduced into a cpDNA gene essential for photosynthesis (rbcL) so that it created a disruption in the reading frame. Restoration of photosynthetic competence could be achieved only by duplication or deletion events.
MATERIALS AND METHODS
Chlamydomonas strains, media, and growth conditions.
The C. reinhardtii strain used for biolistic transformation and all replication slippage experiments was the wild-type CC-125 (mating type plus [mt+]). For crosses, the two strains CC-67 (mt−; cpDNA with erythromycin resistance) and CC-3455 (mt+; cpDNA with a dominant-negative mutation of the Escherichia coli recA) were used as parents at two different stages in the experiments. Strain 18-7G, which carries an rbcL mutation (34), was used for growth comparisons. All of the strains were obtained from the Chlamydomonas Culture Collection when it was located at Duke University. C. reinhardtii was grown at room temperature (22 to 25°C) in Tris-acetate-phosphate (TAP) medium (10), either on solid plates or in liquid medium. The liquid cultures were grown on a rotary shaker at a speed of ∼200 rpm under 245-mol per m2 per s photosynthetic photon flux (245-PPF) continuous “high” light or at about 10 PPF for “low-light” conditions. Growth curves were plotted from hemocytometer counts (13) of aliquots removed from 50-ml cultures grown in TAP medium lacking acetate (TMP). When testing for photosynthetic competence on plates, either TMP or HS (10) medium was used, with incubation under aquarium lights at ∼40 PPF. Crosses were performed as previously described (28). Zygospores were induced to germinate, and meiotic progeny were separated as described previously (13).
Construction of the microsatellite reporter.
The plasmids for chloroplast transformation, pIF24 and pOF28, were constructed by using a 3.9-kb BamHI-EcoRI subclone of p67 (9) named p698, which contains the wild-type rbcL gene, in the pUC8 vector. Plasmid p547, which contains the aadA gene with 5′ and 3′ control regions from psbA (kindly provided by Heriberto Cerutti), was used as the source of an aadA chloroplast expression cassette, which was inserted into the MfeI cut site of p698, giving plasmid p699 (Fig. 1). A 1.2-kb EcoRV-EcoRI fragment at the right end of p699 was subcloned in pBlueScript+ for inserting the slippage substrates at the BspMI cut site in the sixth codon position of rbcL to avoid other BspMI sites present in the larger clone. The engineered EcoRV-EcoRI fragment was then used to replace the equivalent portion of p699. The pIF24 construct contained six 4-bp repeats, giving a 24-bp in-frame insertion in the rbcL gene, while pOF28 contained seven repeats, resulting in a 28-bp insertion at the same site. These constructs were transformed into the dam mutant and dcm mutant GM2163 bacterial strain. The oligonucleotide pairs used to generate the 24-bp (IF24) and 28-bp (OF28) insertions were as follows:
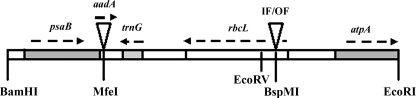
FIG. 1.
Diagram of the cpDNA segment in the transforming plasmids pIF24 and pOF28. The shaded boxes represent the chloroplast-borne genes of C. reinhardtii, and the arrows indicate the directions of their transcription. Two triangles represent the insertions in the transforming plasmid construct: the aadA cassette is the selectable marker that was inserted between two chloroplast genes; the IF24 (24-bp) and OF28 (28-bp) SSRs were inserted at the sixth codon position of the rbcL gene in plasmids pIF24 and pOF28, respectively. The enzymes used for cloning are indicated. The size of the reporter construct was 8.1 kb, including the pUC8 vector.
IF24-F, 5′-AAAC AAAG AAAG AAAG AAAG AAAG-3′
IF24-R, 3′-TTTC TTTC TTTC TTTC TTTC TTTG-5′
OF28-F, 5′-AAAC AAAG AAAG AAAG AAAG AAAG AAAG-3′
OF28-R, 3′-TTTC TTTC TTTC TTTC TTTC TTTC TTTG-5′
Small-scale plasmid isolations followed the alkaline lysis procedure of Sambrook et al. (26). Larger-scale preparations used a kit produced by Qiagen (Valencia, CA).
The biolistic method (5, 15) was used for chloroplast transformation, with CC-125 as the recipient strain and the plasmids pIF24 and pOF28 as the transforming DNAs. After the biolistic procedure, the cells were resuspended in TAP and immediately transferred to TAP agar supplemented with 100 μg/ml spectinomycin (Sigma Chemical Co., St. Louis, MO) for direct selection of the transformants.
Assay for changes in the microsatellite reporter in cpDNA of C. reinhardtii.
The cells with the out-of-frame microsatellite (OF28) were grown in liquid TAP culture to late log phase (2 × 106 to 4 × 106 cells/ml) on a rotary shaker for about 7 days at room temperature in low light. Cell counts were determined with a hemocytometer. The cells were concentrated about 10-fold by centrifugation. An aliquot of the concentrated cells was used for serial dilution onto agar-solidified TAP and was kept on low-light shelves to determine the number of viable cells plated. The remaining cells were plated as 500-μl aliquots on medium lacking acetate (either HS agar [10] or TMP agar) to test for photosynthetic competence. The plates were placed under lights (∼40 PPF) on a shelf at room temperature.
DNA isolation, PCR amplification, and sequencing.
Chlamydomonas total genomic DNA was prepared as described previously (11). The rbcL5 (5′-GGCCCTTTCTATGCTCGACTG-3′) and rbcLmid (5′-CCGAATACGTTACCTAC-3′) primers were used in the PCR amplification reactions as the forward and reverse primers, respectively, to amplify a 560-bp segment (including the microsatellite insertion site) from the rbcL genes of the wild-type, OF, IF, and revertant colonies. In some experiments, an ∼180-bp PCR product was generated using a different set of primers, rbcLfor2 (5′-CTACGTAATCAGGTGTGTAG-3′) and rbcLrev (5′-CCGGACAGATTAATTTTAGGA-3′), in order to readily visualize changes in the repeat number (Fig. 2). DNA (1 to 5 ng) was added to individual 100-μl reaction mixtures containing 200 μmol of each dideoxynucleotide, 2.5 units of Taq polymerase (Gibco BRL), 10 pmol of each primer, and 1× PCR buffer (Gibco BRL). Conditions for the 30 cycles of PCR were as follows: denaturation at 95°C for 2 min and 85°C for 5 min, followed by 94°C for 40 s, 30 s at 48°C to anneal the primers with the template, and 30 s at 72°C for DNA extension. A final extension was done for 10 min at 72°C. All of the PCRs were performed with a minicycler model PTC-150-16 (MR Research Inc., Watertown, MA). The PCR-amplified products were separated on 1.5 or 2% agarose gels in Tris-borate-EDTA buffer for the 560-bp and 180-bp products, respectively.
FIG. 2.
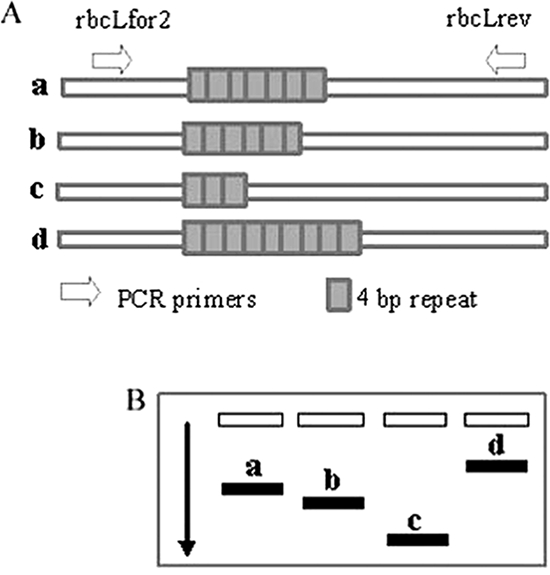
Visualization of DNA changes at the chloroplast microsatellite reporter. (A) Diagram of microsatellite indels with PCR primers. The original out-of-frame construct (OF28) contained seven 4-bp repeats (a). Deletion of one repeat (b) or four repeats (c) or insertion of two repeats (d) made the insertion in frame. (B) Diagram of differential mobility of PCR products.
To check for the presence of the E. coli ΔN recA gene, the recAΔN1 oligonucleotide (7) was used as the forward primer and an oligonucleotide called recA3end (5′-CTGGCATGCTTAAAAATCTTCGTTAGTTTC-3′) was used as the reverse primer to generate a 1-kb PCR product. After the initial denaturation at 95°C for 2 min and 85°C for 5 min, the conditions for the PCR program were repeated for 30 cycles: 94°C for 1 min, 2 min at 36°C to anneal the primers and template, and 2 min at 68°C for DNA extension. The final extension was as described above. The PCR products were run on 1% agarose gels.
PCR products from the rbcL5-rbcLmid amplification were submitted for sequencing by the Michigan State University Genomics Technology and Sequence Facility (http://www.genomics.msu.edu) using the internal primer rbcLfor2. Sequence analyses used Megalign Software from the DNAStar Program (Madison, WI).
Mutation rate.
The microsatellite mutation rate was determined by averaging the frequencies of reversion to photosynthetic competence on parallel plates in two separate experiments and dividing the number of revertants by the number of viable cells calculated to have been spread on each plate.
Reconstruction experiment.
Logarithmically growing cultures of two nonphotosynthetic strains, the OF28 reporter construct and the 18-7G rbcL point mutant, were diluted to 1 × 106 cells/ml, as were two photosynthetically competent strains, CC-124 and the IF24 control line. All four strains were taken through a 10-fold dilution series for individual platings on TAP and TMP media. From the dilutions of CC-124 and IF24 that should have had 100, 1,000, and 10,000 cells/ml, 20 μl was added to 2-ml aliquots of the nonphotosynthetic cell lines (1 × 106 cells/ml), and two 500-μl aliquots were plated on TMP medium and placed in the light.
RESULTS
Introduction of replication slippage substrates into the cpDNA of C. reinhardtii.
Wild-type C. reinhardtii cells (CC-125) were transformed biolistically with the plasmids pIF24 and pOF28 (Fig. 1) containing the (AAAG)n repeats in numbers appropriate to give an “in-frame” (24-bp) or “out-of-frame” (28-bp) insertion. The transformants were selected based on their resistance to spectinomycin conferred by the presence of the aadA cassette in the transforming plasmids. Several rounds of subcloning were required to obtain homoplasmic strains, as monitored by PCR amplification. The compositions of the original inserts were verified by sequencing (Fig. 3). In the OF28 line, a premature stop codon in the rbcL gene makes the cells nonphotosynthetic.
FIG. 3.
Sequences and amino acid predictions of the rbcL replication slippage constructs. The wild-type rbcL gene is shown at the top, with boldface characters indicating the 4 bases of the 3′ overhang at the BspMI cut site. The OF28 line has a 28-bp insertion in its rbcL gene, creating a stop codon that renders the cells nonphotosynthetic. The IF24 line has a six-codon insertion relative to the wild type. IF12 and IF36 are two photosynthetically competent lines recovered from the initial OF28 line.
Test for photosynthetic competence in the transformed cells containing the reporter constructs.
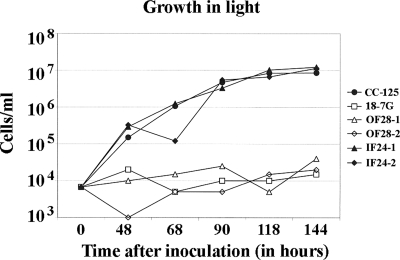
To assess their photosynthetic competence, the in-frame and out-of-frame constructs were inoculated into liquid TAP medium and placed on shakers under low-light and high-light conditions. For comparison, wild-type CC-125 cells and the rbcL mutant 18-7G were grown as controls. As shown in Fig. 4, the wild-type CC-125 and IF24 cells grew well, showing that they were photosynthetically competent. In contrast, the cells of the 18-7G and OF28 strains were light sensitive and did not grow under these conditions. In low light, the cultures all showed similar growth properties (data not shown), growing steadily, although more slowly than the CC-125 and IF24 cultures did in high light.
FIG. 4.
Growth curves. Cultures were inoculated at 104 cells/ml, and cell densities were determined over a 6-day period. The out-of-frame (OF28) and in-frame (IF24) constructs were present in each of two independent transformants. CC-125 is a wild-type control, and 18-7G is an rbcL mutant.
Rates of mutation at the (AAAG)n microsatellites in the cpDNA of the OF28 cells.
Procedures for determining the cell viability and mutation frequency are described in Materials and Methods. The number of viable cells plated was determined from dilution plating of the OF cells on a nonselective medium. In order to monitor the replication slippage events at the (AAAG)n repeats in the rbcL gene, the OF cells were plated on media that selected for restored photosynthetic competence. In 3 to 5 days, the bright-green color of the cells faded and the lawn of cells appeared bleached. One would have thought the cells were dead, but in 4 to 6 weeks, small green colonies appeared. As shown in Table 1, each plate of approximately 50 million viable cells gave rise to hundreds of photosynthetic revertants, with an average frequency of 1.5 × 10−5. These colonies were transferred with sterile toothpicks to fresh HS-plus-spectinomycin plates for maintenance. As described below, all of the colonies arose due to indels in the (AAAG)n microsatellite region.
TABLE 1.
Frequency of mutation of microsatellite reporter
| Expt no. | No. of plates | No. of viable cells/plate | No. of PSa colonies per plate | Mean no. of PS colonies per plate | Median no. of PS colonies per plate | SD for no. of colonies per plate | Mutation frequency (per 107 viable cells) |
|---|---|---|---|---|---|---|---|
| 1 | 15 | 4.6 × 107 | 381-843 | 675 | 749 | 151 | 147 |
| 2 | 11 | 7 × 107 | 958-1,215 | 1,057 | 1,050 | 76 | 151 |
PS, photosynthetic.
Reconstruction experiment.
Because colonies were not seen on the HS medium until many weeks after being plated, we believed that the replication slippage events had not occurred during the growth of the liquid culture or at least had not sorted out prior to the plating. To test this interpretation, a mixing experiment was performed, with small numbers of photosynthetically competent cells (CC-124 and IF24) being added to nonphotosynthetic cells (OF28 and 18-7G) prior to plating them on medium lacking a carbon source. Control platings of the nonphotosynthetic cells alone were observed with a dissecting microscope over several days. Most cells divided into clusters of four before arresting. In 3 to 4 days, the cell lawn had bleached, but after 5 days, small colonies were visible from the wild-type CC-124 cells, and after 8 days, small colonies were visible on the plates with the IF24 cells. Table 2 shows data from the experiment, collected from two plates for each combination.
TABLE 2.
Mixing experimenta
| Expected no. of colonies of photosynthetic strain | Observed no. of green colonies in bleached lawn of cells from strain:
|
|||
|---|---|---|---|---|
| 18-7G
|
OF-28
|
|||
| CC-124 | IF24 | CC-124 | IF24 | |
| 50 | 131 ± 7 | 53 ± 5 | 97 ± 7 | 27 ± 2 |
| 5 | 16 ± 5 | 6 ± 1 | 20 ± 0 | 6 ± 1 |
| 0.5 | 3 ± 1 | 0 | 2 ± 2 | 0 |
Aliquots from a 10-fold dilution series of the photosynthetically competent lines CC-124 (wild type) or IF24 (8-codon insertion in rbcL) were added to nonphotosynthetic cells (1×106 cells/ml) carrying a point mutation in rbcL (18-7G) or an out-of-frame insertion (OF28). The expected colony numbers are based on hemocytometer cell counts. Photosynthetic colonies were scored after 5 to 8 days.
Molecular analysis of slippage events at the (AAAG)n microsatellite site.
Since the OF28 microsatellite reporter construct consisted of a 28-bp insertion containing a stretch of (AAAG)n repeats, a loss of one repeat unit and an insertion of two repeat units were two of the expected slippage events that could restore the reading frame of the disrupted rbcL gene. In addition, a deletion of four repeats (16 bp of the original 28-bp construct) or an insertion of two repeats (creating a 36-bp insert at rbcL) could theoretically also restore photosynthetic competence in the OF28 cells. A subset of 107 “slipped” colonies with restored photosynthetic competence was chosen from three independent experiments to observe the types of slippage that had occurred and also to compare their respective frequencies of occurrence using PCR to visualize the microsatellite. As shown in Fig. 5, PCR amplification of the microsatellite region yielded two types of size variants from the photosynthetically competent colonies: both were smaller than the 169-bp fragment found in the OF28 cells but larger than the amplification product (141 bp) from the wild-type C. reinhardtii cells. Sequencing showed that the two classes of variants had experienced deletions of one or four copies of the 4-bp repeat (Fig. 3); there were 58 of each type of deletion. One variant had a product larger than the PCR fragment from the OF28 cells. In that cell line, the presence of two PCR products (Fig. 6, lane 3) indicated that the cells were heteroplasmic: the upper band shows a larger product than the OF28 PCR product, and the lower band shows a slightly smaller fragment than the 169-bp fragment seen in OF28 cells. Most likely, this condition reflects a recombination event, with the two products representing reciprocal recombinant molecules. In later experiments, a few other colonies were heteroplasmic, but they contained the original-size microsatellite tract, as well as a deleted product (data not shown). One subsequent experiment yielded a 36-bp in-frame variant (Fig. 3, lower line). Like the deletion variants, it was photosynthetically competent but grew more slowly than the wild type.
FIG. 5.
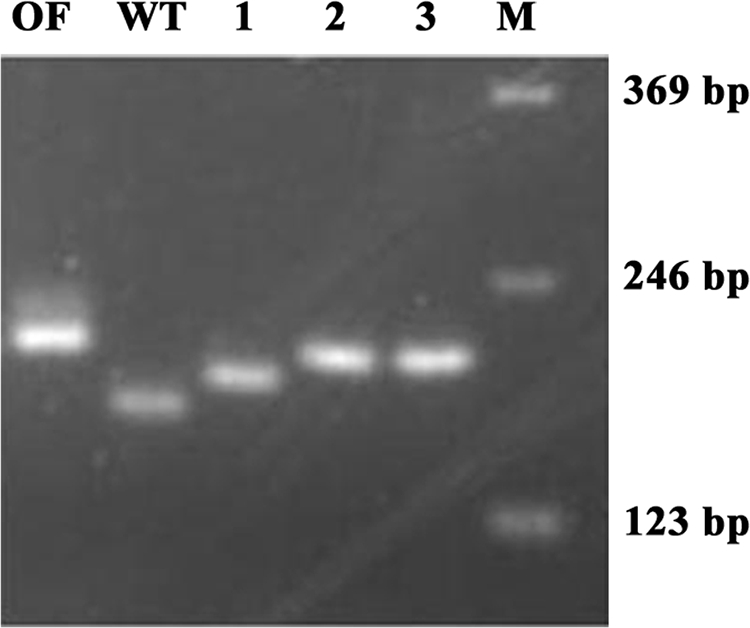
Slippage events at the cpDNA microsatellite reporter. Shown are examples of the PCR-amplified rbcL segment from the original out-of-frame insertion strain (OF28 [OF]), the wild-type C. reinhardtii strain (WT), and several photosynthetically competent colonies derived from the OF28 strain (1 to 3), using primers rbcLfor2 and rbcLrev. Lane M contains a 123-bp DNA ladder (Invitrogen).
FIG. 6.
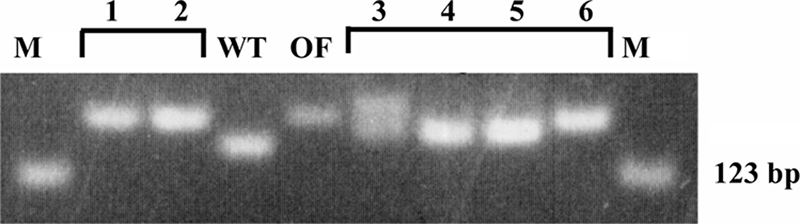
Microsatellite changes in six photosynthetic colonies (1 to 6) isolated from the OF28 cell line. See the legend to Fig. 5 for abbreviations. The PCR products in lane 3 are 4 bp larger and 4 bp smaller than the original OF28 insert.
Rates of change at the microsatellite reporter in recombination-deficient chloroplasts.
In theory, microsatellites containing short direct repeats could undergo intra- or intermolecular recombination, resulting in a change in the copy numbers of the repeats. The recovery of only one colony with a clear reciprocal-recombination event suggested that recombination was not a major contributor to chloroplast microsatellite variability in this study. To test this deduction, we combined the OF28 reporter construct with a ΔN recA construct (7) by classical crossing procedures. The E. coli ΔN recA gene has been shown to inhibit chloroplast recombination when inserted into cpDNA (7). Our first assessment tested the presence and activity of the ΔN recA gene in the original strain, CC-3455, by PCR amplification using appropriate primers to visualize the gene (described in Materials and Methods) and then by confirming the strain's hypersensitivity to 5 mM 5-fluorodeoxyuridine relative to wild-type cells, as described previously (7). This line was crossed to an mt− cell line containing the slippage construct with the linked aadA cassette. The spectinomycin resistance conferred by the aadA gene was used to select biparental progeny from which the desired recombinant could be isolated. Initially, very few biparental progeny were recovered, so the mt+ gametes were exposed to a handheld UV lamp for 2 min prior to mating, and we were able to obtain 5.8% biparental zygotes.
From dissected meiotic progeny, spectinomycin-resistant cells were selected and checked for the presence of both the ΔN recA gene and the slippage construct in the cpDNAs by PCR amplification using the recAΔN1/recA3end and rbcLfor2/rbcLrev primer pairs, respectively. As represented by the meiotic progeny analyzed in Fig. 7B, about half of the biparental meiotic progeny (12 of 27) carried the ΔN recA gene. In contrast, all the progeny that showed resistance to spectinomycin also contained the 28-bp out-of-frame insertion in the closely linked rbcL gene (Fig. 7A). Two meiotic progeny that contained both the slippage construct and the ΔN recA gene were chosen for further experiments. Since the presence of ΔN recA should reduce chloroplast recombination, we tested the cell lines for the frequency of restoration of photosynthetic competence due to insertion/deletion events at the reporter gene. As shown in Table 3, the recovery of photosynthetically competent colonies from the slippage reporter in a recombination-deficient background occurred at a frequency of 5.3 × 10−5, a rate that is approximately three times higher than that of the recombination-proficient lines.
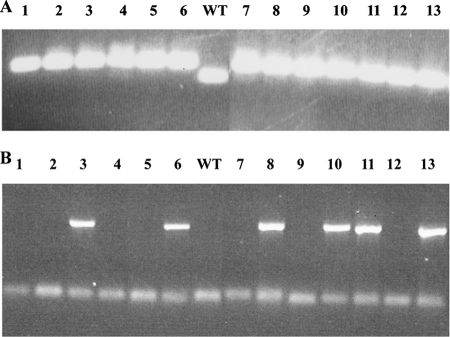
FIG. 7.
PCR analysis to identify meiotic progeny carrying both the cpDNA microsatellite reporter and the E. coli ΔN42RecA gene. (A) PCR screening of spectinomycin-resistant progeny for the microsatellite reporter. PCR products from 13 progeny (1 to13) and the wild type (WT) were amplified with the rbcL5 and rbcLmid primers to allow visualization of the region containing the microsatellite. (B) PCR products indicating the presence of the ΔN42RecA gene. The lanes are labeled as in panel A. The faint lower band is derived from excess oligonucleotides in the reaction mixture.
TABLE 3.
Rates of spontaneous replication slippage events in the cpDNA of C. reinhardtii containing the OF28 construct in a recombination-deficient background
| ΔN recA OF28 strain | Total no. of viable cells plated (106) | No. of cells that showed photosynthetic competence | Rate of mutation to photosynthetic competence per 107 viable cells plated |
|---|---|---|---|
| 1 | 65 | 3,360 | 517 |
| 2 | 104.2 | 5,640 | 541 |
Replication slippage of the cloned microsatellite occurs in E. coli.
To examine the stability of the microsatellite substrate in E. coli, where it has no phenotype, bacteria were grown and plated under nonselective conditions. Single colonies were randomly chosen for small-scale plasmid isolation, and PCR amplifications were performed. Ten out of 22 colonies contained products that were larger than the original pOF28 insert (Fig. 8).
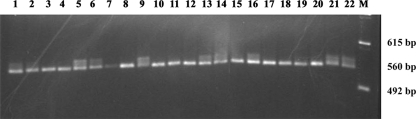
FIG. 8.
PCR products from E. coli subclones carrying the pOF28 plasmid. The subclones were randomly selected; DNA was extracted and amplified with oligonucleotides rbcLfor2 and rbcLrev. Lane M contains the 123-bp DNA ladder.
DISCUSSION
Although the existence of microsatellites and their variability have been well documented in the chloroplast genomes of many plant species, few studies have observed their rates of change and the mechanics of the process in vivo. Phylogenetic comparisons have determined the relative rates of indels at cpDNA microsatellites in an evolutionary context, and results have indicated that the chloroplast DNA has a lower mutation rate for microsatellites than does the nuclear DNA (24, 38). Some evolutionary studies have indicated that indels at cpDNA microsatellites occur at a much higher rate than do base substitutions (8, 24, 29), but other assessments have concluded that indels are less frequent (30, 32). In fact, several traits intrinsic to microsatellites make it difficult for biosystematic comparisons to accurately quantify their variability: deletion or duplication of the same number of repeats could occur independently in two plant lines, causing homoplasy; in addition, multiple sequential changes in the copy number of a repeat within one plant line could be scored as only a single change, thereby distorting their relative frequencies of occurrence. Our objective was to establish a reporter assay that would allow us to determine the rates of cpDNA microsatellite change, ascertain the likelihood of insertion versus deletion, and determine whether replication slippage or recombination was responsible for the variation.
In order to achieve those goals, we used the green alga C. reinhardtii, which frequently serves as a model system for chloroplast studies. Since most cpDNA microsatellites are present in the noncoding regions of the plastid genome, a reporter construct was created to monitor replication slippage events in vivo using an essential gene of the cpDNA, rbcL, in order to provide a phenotypic selection for the slippage event. The rbcL gene product is essential for the fixation of carbon from CO2; an out-of-frame insertion in the gene would disrupt its reading frame, resulting in the death of the algal cells on media without an organic carbon source (acetate). Changes in the number of repeats within the microsatellite would be able to be observed easily in these cells: a deletion of one or four repeat units or an insertion of two repeats would result in photosynthetic competence. Since this assay required that the rbcL gene product be functional with the addition of extra codons, initial experiments were done to test whether an in-frame insertion of additional codons in the rbcL gene of C. reinhardtii would allow the cells to remain photosynthetically competent. As shown in Fig. 4, an in-frame insertion of 24 bp containing AAAG repeats (IF24) in the sixth codon position did not affect the photosynthetic competence or growth properties of the cells. However, an out-of-frame insertion of 28 bp (OF28) caused the cells to be sensitive to high light and to require acetate for growth, traits that are characteristic of rbcL− mutations (33). The OF28 construct thus served as a reporter to monitor microsatellite deletion and duplication events.
In experiments analogous to the Luria-Delbruck fluctuation tests (19), cells from large logarithmically growing cultures were subdivided among many selective plates. As shown in Table 1, an average of 150 revertants arose for every 10 million viable cells plated (a frequency of 1.5 × 10−5). A mixing experiment (Table 2) showed that if photosynthetic cells were present in the large culture, they would have produced green colonies within a week, whereas in our tracking of the microsatellite reporter, 4 weeks elapsed before the first green, photosynthetically competent colonies were observed. Because cell division stops soon after the cells are plated on the selective medium, the cpDNA microsatellite variability must occur within the nondividing cells. A change in the microsatellite that reestablishes the rbcL reading frame should be dominant; hence, the reversion would not have to reach fixation to produce a photosynthetically competent cell. However, if the microsatellite variation occurs through replication slippage, cpDNA replication must occur in spite of an arrest of cell division. This idea is not as unorthodox at it might at first seem, since the replication of Chlamydomonas cpDNA is not synchronized with its nuclear replication, but rather, occurs steadily during the growth of liquid cultures (reviewed in reference 27). Furthermore, evidence exists for turnover of the Chlamydomonas cpDNA (29).
Precautions were taken to exclude any preexisting event that might have been present in the starter cells of the assay: these included plating of the early preculture cells on media without acetate to test for the presence of early revertants and checking the sizes of the microsatellite insert with PCR amplification at the beginning of the culture. If anything, our calculation of the frequency is an underestimate, since only a third of the possible deletion or insertion events (those that reestablished the reading frame) would be detected by our assay. Multiplying our quantifications by 3 produces an estimated indel mutation rate for the 4-bp microsatellites of 4.5 × 10−5. This value is equivalent to estimates from evolutionary comparisons (24) for rates of cpDNA replication slippage of mononucleotide repeats 10 or more bases long in intergenic spacers. However, in our experimental system, mononucleotide repeats (a stretch of 13 As) varied at such a high frequency that as soon as we recovered a biolistically transformed cell line through the linked selectable marker, some cells in the colony had become photosynthetically competent due to indels within the microsatellite. Hence, we were unable to quantify the rate of mononucleotide repeat variation, but it must be several orders of magnitude higher than for the 4-bp microsatellite.
In our PCR screens of a subset of the photosynthetic revertants of the AAAG microsatellite, only one colony appeared to contain reciprocal-recombination products (Fig. 5). Hence, replication slippage seemed to be responsible for the production of the microsatellite indels within the chloroplast, as is true for other biological systems. However, to more thoroughly assess the contribution of recombination to microsatellite variability, the microsatellite reporter was combined with a chloroplast-located dominant-negative allele of the E. coli recA gene. To our surprise, the frequency of variation resulting in photosynthetically competent colonies in the recombination-deficient strain was about three times higher than in the recombination-proficient lines (Table 3). Since slippage that restores the rbcL reading frame should produce a dominant allele, we believe that this result indicates that the majority of these mutations are eliminated through recombination (gene conversion). Reducing recombination among the highly polyploid cpDNAs would enhance the survival and recovery of the revertant alleles.
With one exception, the slipped colonies that we recovered experienced deletion of one or four repeat units; the two deletion types occurred with equal frequencies. In one later experiment, we recovered a two-repeat-unit duplication that restored photosynthetic competence (IF36). Although photosynthetically competent, IF36 grows somewhat more slowly than the wild type, and hence, there may be some bias against the recovery of longer forms. Nonetheless, the fact that we were able to recognize and recover a duplication variant leads us to conclude that replication slippage in the chloroplast DNA of Chlamydomonas has a bias toward deletions rather than insertions. This is in marked contrast to what occurs to this same substrate when it is maintained in a plasmid in E. coli. As shown in Fig. 8, expansions rather than deletions typified the changes in the bacteria.
The observation that deletions are the prevalent form of replication slippage in the Chlamydomonas chloroplast suggests that the template strand of the cpDNA is more prone to slippage than is the daughter strand. A deletion bias during replication slippage has been reported for other organisms, including cpDNA of a number of plant species, such as petunia and alfalfa (1), mitochondrial microsatellites of yeast (31), and nuclear genomes of animals, such as snails (39). In the Chlamydomonas chloroplast genome, “short dispersed repeats” have been reported to have proliferated to a great abundance (20), and therefore, we were surprised to find a strong deletion bias. However, the short dispersed repeats described earlier (20) are significantly longer (30 to 36 bp) than those we analyzed and are rarely tandem direct repeats, so it is unlikely that they are substrates for replication slippage.
We have previously shown that base substitution at a particular target site in the cpDNA occurs at a rate of 0.15 × 10−9 to 11 × 10−9 cells in C. reinhardtii (11). This means that cpDNA replication slippage at a 4-base repeat occurs 1,000 to 100,000 times more frequently than base substitution. Although our results have the advantage of providing base substitution and replication slippage rates in the same organism, the values may not be directly comparable, since the base substitution rates were observed for the 16S rRNA gene within the cpDNA inverted repeat, while the slippage events occurred in a reporter construct in the single-copy region. Others have found that copy correction in the inverted repeats reduces the mutation rate by half (reviewed in reference 21).
In bacteria, the mismatch repair system defined by the MutHLS repair system is known to be the key surveillance system for correcting mismatches generated by replication slippage events (4). During replication through microsatellites, strand slippage results in the formation of a looped structure with unpaired bases on either the template or the daughter strand. Such a looped structure is recognized by the MutS protein of the mismatch repair pathway and targeted for excision (18). It has been hypothesized that in prokaryotes, a looped structure containing one to three unpaired bases in the loop is more readily recognized and repaired than is a repeat of four or more nucleotides (18, 22). Our chloroplast data paint a different picture; variations due to slippage of the AAAG repeats were definitely recovered less frequently than changes in a mononucleotide repeat, which were so frequent that we could not quantify them. The observation that the same 4-bp-repeat microsatellite in E. coli experiences daughter strand slippage at an extremely high rate suggests that endosymbiosis may have benefited the chloroplast by providing its genetic system with repair enzymes that minimize replication slippage of the daughter strand.
Acknowledgments
We thank Nick Gillham and John Boynton for hosting research leave for B.B.S. and Heriberto Cerutti for strains and suggestions.
This work was supported by grant MCB 9982600 from the National Science Foundation to B.B.S. and was carried out in compliance with current laws governing genetic experimentation in the United States.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Aldrich, J., B. W. Cherney, E. Merlin, and L. Christopherson. 1988. The role of insertions/deletions in the evolution of the intergenic region between psbA and trnH in the chloroplast genome. Curr. Genet. 14137-146. [DOI] [PubMed] [Google Scholar]
- 2.Alou, A. H., A. Azaiez, M. Jean, and F. J. Belzile. 2004. Involvement of the Arabidopsis thaliana AtPMS1 gene in somatic repeat instability. Plant Mol. Biol. 56339-349. [DOI] [PubMed] [Google Scholar]
- 3.Azaiez, A., E. F. Bouchard, M. Jean, and F. J. Belzile. 2006. Length, orientation, and plant host influence the mutation frequency in microsatellites. Genome 491366-1373. [DOI] [PubMed] [Google Scholar]
- 4.Bichara, M., J. Wagner, and I. B. Lambert. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598144-163. [DOI] [PubMed] [Google Scholar]
- 5.Boynton, J. E., and N. W. Gillham. 1993. Chloroplast transformation in Chlamydomonas. Methods Enzymol. 217510-536. [DOI] [PubMed] [Google Scholar]
- 6.Broun, P., and S. D. Tanksley. 1996. Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol. Gen. Genet. 25039-49. [DOI] [PubMed] [Google Scholar]
- 7.Cerutti, H., A. M. Johnson, J. E. Boynton, and N. W. Gillham. 1995. Inhibition of chloroplast DNA recombination and repair by dominant negative mutants of Escherichia coli RecA. Mol. Cell. Biol. 153003-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clegg, M. T., B. S. Gaut, G. H. Learn, and B. R. Morton. 1994. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 916795-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dron, M., M. Rahire, and J.-D. Rochaix. 1982. Sequence of the chloroplast DNA region of Chlamydomonas reinhardii containing the gene of the large subunit of ribulose bisphospohate carboxylase and parts of its flanking genes. J. Mol. Biol. 162775-793. [DOI] [PubMed] [Google Scholar]
- 10.Gorman, D. A., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 541665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GuhaMajumdar, M., and B. B. Sears. 2005. Chloroplast base substitutions: an experimental assessment. Mol. Genet. Genomics 273177-183. [DOI] [PubMed] [Google Scholar]
- 12.Gur-Arie, R., C. J. Cohen, Y. Eitan, L. Shelef, E. M. Hallerman, and Y. Kashi. 2000. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 1062-71. [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, E. H. 1989. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 14.Ishii, T., and S. R. McCouch. 2000. Microsatellites and microsynteny in the chloroplast genomes of Oryza and eight other Gramineae species. Theor. Appl. Genet. 1001257-1266. [Google Scholar]
- 15.Kindle, K. L., K. L. Richards, and D. B. Stern. 1991. Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 881721-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4203-221. [DOI] [PubMed] [Google Scholar]
- 17.Li, Y.-C., A. B. Korol, T. Fahima, A. Beiles, and E. Nevo. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol.r Ecol. 112453-2465. [DOI] [PubMed] [Google Scholar]
- 18.Lovett, S. T. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 521243-1253. [DOI] [PubMed] [Google Scholar]
- 19.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maul, J. E., J. W. Lilly, L. Cui. C. W. dePamphilis, W. Miller, E. H. Harris, and D. B. Stern. 2002. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 142659-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer, J. D. 1987. Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. Am. Nat. 130s6-s29. [Google Scholar]
- 22.Parker, B. O., and M. G. Marinus. 1992. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl. Acad. Sci. USA 891730-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provan, J., G. Corbett, J. W. McNicol, and W. Powell. 1997. Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome 40104-110. [DOI] [PubMed] [Google Scholar]
- 24.Provan, J., N. Soranzo, N. J. Wilson, D. B. Goldstein, and W. Powell. 1999. A low mutation rate for chloroplast microsatellites. Genetics 153943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provan, J., W. Powell, and P. M. Hollingsworth. 2001. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol. 16142-147. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 27.Sears, B. B. 1998. Replication, recombination, and repair in the chloroplast genetic system of Chlamydomonas, p. 115-138. In J. D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria of Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Sears, B. B., J. E. Boynton, and N. W. Gillham. 1980. The effect of gametogenesis regimes on the chloroplast genetic system of Chlamydomonas reinhardtii. Genetics 9695-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears, B. B., and K. P. VanWinkle-Swift. 1994. The salvage-turnover-repair (STOR) model for uniparental inheritance in Chlamydomonas: DNA as a source of sustenance. J. Hered. 85366-376. [DOI] [PubMed] [Google Scholar]
- 30.Shaw, J., E. Lickey, J. Beck, S. Farmer, W. Liu, J. Miller, K. Siripun, C. Winder, E. Schilling, and R. Small. 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92142-166. [DOI] [PubMed] [Google Scholar]
- 31.Sia, E. A., C. A. Butler, M. Dominska, P. Greenwell, T. D. Fox, and T. D. Petes. 2000. Analysis of microsatellite mutations in the mitochondrial DNA of Sacharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small, R., J. Ryburn, R. Cronn, T. Seelanan, and J. Wendel. 1998. The tortoise and the hare: choosing between noncoding plastome and nuclear ADH sequences for phylogeny. Reconstruction in a recently diverged plant group. Am. J. Bot. 851301-1315. [PubMed] [Google Scholar]
- 33.Spreitzer, R. J., and L. Mets. 1981. Photosynthesis-deficient mutants of Chlamydomonas reinhardii with associated light-sensitive phenotypes. Plant Physiol. 67565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spreitzer, R. J., M. Goldschmidt-Clermont, M. Rahire, and J. D. Rochaix. 1985. Nonsense mutations in the Chlamydomonas chloroplast gene that codes for the large subunit of ribulosebiphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA 825460-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toth, G., Z. Gaspari, and J. Jurka. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Udupa, S. M., and M. Baum. 2001. High mutation rate and mutational bias at (TAA)n microsatellite loci in chickpea (Cicer arietinum L.) Mol. Genet. Genomics 2651097-1103. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez, J. F., T. Perez, J. Albornoz, and A. Dominguez. 2000. Estimation of microsatellite mutation rates in Drosophila melanogaster. Genet. Res. 76323-326. [DOI] [PubMed] [Google Scholar]
- 38.Viguera, E., D. Canceill, and S. D. Ehrlich. 2001. Replication slippage involves DNA polymerase pausing and disassociation. EMBO J. 202587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weetman, D., L. Hauser, and G. R. Carvalho. 2002. Reconstruction of microsatellite mutation history reveals a strong and consistent deletion bias in invasive clonal snails, Potamopyrgus antipodarum. Genetics 162813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Y., J. E. Strassman, and D. C. Queller. 2000. Insertions, substitutions, and the origin of microsatellites. Genet. Res. 76227-236. [DOI] [PubMed] [Google Scholar]