Abstract
Cysteine proteases have been shown to be essential virulence factors and drug targets in trypanosomatids and an attractive antidisease vaccine candidate for Trypanosoma congolense. Here, we describe an important amplification of genes encoding cathepsin B-like proteases unique to T. congolense. More than 13 different genes were identified, whereas only one or two highly homologous genes have been identified in other trypanosomatids. These proteases grouped into three evolutionary clusters: TcoCBc1 to TcoCBc5 and TcoCBc6, which possess the classical catalytic triad (Cys, His, and Asn), and TcoCBs7 to TcoCBs13, which contains an unusual catalytic site (Ser, Xaa, and Asn). Expression profiles showed that members of the TcoCBc1 to TcoCBc5 and the TcoCBs7 to TcoCBs13 groups are expressed mainly in bloodstream forms and localize in the lysosomal compartment. The expression of recombinant representatives of each group (TcoCB1, TcoCB6, and TcoCB12) as proenzymes showed that TcoCBc1 and TcoCBc6 are able to autocatalyze their maturation 21 and 31 residues, respectively, upstream of the predicted start of the catalytic domain. Both displayed a carboxydipeptidase function, while only TcoCBc1 behaved as an endopeptidase. TcoCBc1 exhibited biochemical differences regarding inhibitor sensitivity compared to that of other cathepsin B-like proteases. Recombinant pro-TcoCBs12 did not automature in vitro, and the pepsin-matured enzyme was inactive in tests with cathepsin B fluorogenic substrates. In vivo inhibition studies using CA074Me (a cell-permeable cathepsin B-specific inhibitor) demonstrated that TcoCB are involved in lysosomal protein degradation essential for survival in bloodstream form. Furthermore, TcoCBc1 elicited an important immune response in experimentally infected cattle. We propose this family of proteins as a potential therapeutic target and as a plausible antigen for T. congolense diagnosis.
Trypanosoma congolense, the main causative agent of animal trypanosomiasis in Africa, is an extracellular tsetse fly-transmitted protozoan parasite found in the bloodstream of mammalian hosts. Like other trypanosomatids, T. congolense displays a complex life cycle, undergoing important morphological and metabolic changes to adapt to each environment; trypomastigote bloodstream forms proliferate in the blood of the mammalian host, where they express a repertoire of variant surface glycoproteins. Upon uptake during a tsetse blood meal, proliferative trypomastigote procyclic forms differentiate in the insect midgut and then migrate to the salivary glands, where they attach as epimastigote proliferative forms. Eventually, they differentiate into nonproliferative infective metacyclic forms that are transmitted to a new mammalian host during a new blood meal (12, 47, 48). T. congolense affects domestic livestock in sub-Saharan Africa and consequently has a major economic impact. The main pathological features of animal trypanosomiasis are weight loss, anemia, and immunosuppression, but the mechanisms involved are poorly understood.
Attempts to control trypanosomiasis are based mainly on the use of trypanocidal drugs and on vector control. However, because no new drugs have been developed in the last 50 years, drug resistance is increasing. Efforts to develop a vaccine have been hampered by antigenic variation, a mechanism that allows African trypanosomes to escape the host's immune response (23). Alternative or complementary control strategies may be proposed on the basis of the limitation of pathology rather than the prevention of infection. It has been observed that trypanotolerant African taurine cattle, which possess a natural ability to both control trypanosome infection and limit the associated pathology (56), develop a prominent antibody response against a T. congolense cysteine protease (congopain) upon infection (5). The important role played by parasite cysteine proteases in disease processes such as invasion, migration, nutrition, and immune evasion has been extensively documented in recent years (44, 52, 62). Thus, it has been suggested that trypanotolerant cattle control the disease through a more efficient antibody-mediated neutralization of congopain and that immunization against cysteine proteases and other pathogenic factors of the parasite, through the increase of the host's resistance to pathogenic effects of the parasite, are part of control strategies for livestock trypanosomiasis (4). Besides their role in pathogenicity, cysteine proteases are essential to the life cycle of many parasites, since they have functional diversity derived from their unique nucleophilicity, and they are stable in different biological environments. Specific inhibitors currently are being tested as antiparasitic drugs (1, 39, 46, 58), and recombinant proteases have been used as vaccination targets with promising results (20, 38, 42, 60).
Cathepsin L- and cathepsin B-like enzymes, the most extensively studied cysteine proteases, are lysosomal members of the papain superfamily. They are synthesized as inactive precursors that, after the proteolytic removal of the NH2-terminal propeptide, produce a single-chain mature enzyme. The residues involved in the catalytic activity are Cys, His, and Asn, occurring in that order in the sequence. Both types of proteases act as endopeptidases and are involved mainly in the degradation of external (through endocytic or phagocytic processes) or internal proteins (through protein recycling and autophagy) (53).
Cathepsin L-like cysteine proteases have been widely studied in kinetoplastidae, in which they are encoded by multiple genes that usually are organized in tandem arrays in the genome. Trypanosoma cruzi cruzain has been associated with host cell invasion (3, 64), macrophage activation, and immune evasion (29, 66). For Leishmania mexicana, CP2 has been associated with macrophage infection and Th1 suppression (54, 55). For T. congolense, studies have focused on CP1 and CP2 (also known as congopain or C2, for its recombinant catalytic domain), two highly homologous cathepsin L-like proteases with different functional characteristics (17). It was suggested that CP2 plays a role in anemia and immunosuppression during infection (4).
Parasite cathepsin B-like cysteine proteases have been studied less extensively than cathepsin L, as they generally do not represent the major proteolytic activity in parasite extracts. Cathepsin B exhibits endopeptidase but also carboxydipeptidase activity, which involves a unique 20-residue structure named the occluding loop (57), a specific feature of cathepsin B-like proteases. The strategic position of two His residues within this loop (His110 and His111) suggests that they can act as acceptors for the negatively charged carboxylate group of the C terminus of peptides and proteins (35). In kinetoplastidae they are encoded by a unique gene. Trypanosoma brucei TbCatB seems to be essential for the survival of the bloodstream form in vitro (45), and L. mexicana CPC, although not crucial for infectivity, plays a role in the parasite interaction with macrophages in vivo (13).
Here, we describe a novel family of cathepsin B-like cysteine proteases specific to T. congolense. Our findings revealed at least 13 genes coding for cathepsin B-like proteins in T. congolense. Genes are located on different chromosomes and not in a tandem array, as are most of those coding for the cathepsin L-like proteases in other trypanosomatids, and they exhibit an important degree of polymorphism. They all share the typical cathepsin B structures: the occluding loop, a signal peptide, a propeptide, and a catalytic domain representing the mature proteolytically active enzyme. They all lack the C-terminal extension present in cathepsin L-like proteases in trypanosomatids. Several of these genes code for proteins with unusual catalytic sites in which the nucleophilic cysteine is replaced by a serine. We studied representatives of each type of protease biochemically, characterized their expression during the life cycle of the parasite, and analyzed their antigenicity in experimentally infected cattle. These studies might contribute to the identification of new drug targets, the development of antiparasite and antidisease vaccines, and the improvement of diagnostic tools.
MATERIALS AND METHODS
Parasites.
Savannah type T. congolense clones IL-3000 (26) (which induces an acute infection in BALB/c mice) and IL-1180 (28) (which induces a chronic infection) were used. Both clones induce a severe infection in cattle (clone IL-1180 was used previously in experimental bovine infections [5, 7]). T. congolense procyclic forms were grown at 28°C without carbon dioxide and maintained in axenic culture in minimum essential medium Eagle (Sigma) supplemented with 20% (vol/vol) heat-inactivated fetal calf serum (Gibco) and 5 μg/ml hemin (Sigma). Bloodstream forms were obtained from the blood of infected BALB/c mice during the first peak of parasitemia and were purified by centrifugation, followed by chromatography on DEAE-cellulose (Whatman DE-52) (43). Epimastigote forms were obtained by the in vitro differentiation of procyclic forms in cultures by selecting adherent cells in minimum essential medium Eagle supplemented with 8 mM proline (33).
Cloning and site-directed mutagenesis.
Genes were amplified by PCR from genomic DNA preparations of T. congolense IL-1180 using primers designed from consensus sequences selected from the analysis of the 3′ and 5′ untranslated regions (3′UTR and 5′UTR, respectively) of cathepsin B-like genes found in the IL-3000 clone (see Table S1 in the supplemental material). Amplification products were cloned in the TOPO blunt vector (Invitrogen) and sequenced. Proenzyme-coding regions of the TcoCBc1 and TcoCBc6 genes were cloned in the pPICZαA vector (Invitrogen) with six-His N-terminal tags for expression in Pichia pastoris strain X-33. The proenzyme-coding region of the TcoCBs12 gene was cloned in the same vector with and without six-His tags. Mutants were constructed by site-directed mutagenesis (using QuikChange II; Stratagene) to avoid the glycosylation of the mature proteins by replacing the asparagine of the consensus glycosylation sequences with a serine or a glutamine. The mutated sites are indicated in Fig. 2.
FIG. 2.
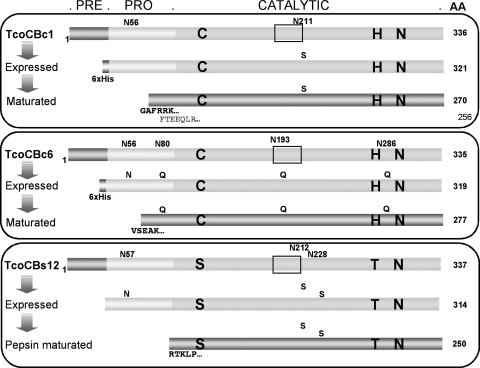
TcoCBc1, TcoCBc6, and TcoCBs12 expression strategies and successfully purified recombinant proteases. The prepeptide (pre), propeptide (pro), and catalytic regions are highlighted. The occluding loop structure identified in cathepsin B-like proteases from other species is framed, and the deduced amino acids of the active site are indicated in large boldface letters. Potential N-glycosylation sites and mutations performed are indicated by small letters at the top of each protein structure. N-terminal sequencing of the matured forms revealed two maturation sites in TcoCBc1, GAFR (major) and FTEE (minor), one for TcoCBc6 (VSEAK), and one for TcoCBs12 (RTKLP).
Expression, purification, and in vitro maturation of TcoCB.
The expression of TcoCBc1 was performed in the EasySelect Pichia expression system (Invitrogen) according to the manufacturer's instructions. Briefly, Pichia pastoris strain X-33 containing the expression plasmid was pregrown overnight at 30°C in YPD (yeast extract, peptone, dextrose) broth until an optical density at 600 nm (OD600) of 6.0 was reached, was diluted in BMMY (buffered minimal medium with methanol and yeast extract), pH 6.0, to an OD of 1.0, and was induced at 30°C with 1% (vol/vol) methanol for 72 to 96 h. The expression of the protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis using a monoclonal anti-poly-His antibody (Sigma). The recombinant proenzyme was purified by immobilized metal ion affinity chromatography (IMAC) using a HisTrap FF column (GE Healthcare) equilibrated in 30 mM Tris-HCl buffer containing 250 mM NaCl and 10 mM imidazole at pH 7.9. The bound proteins were eluted batch-wise with 10 ml of the same buffer containing 500 mM imidazole. The eluted proenzyme was activated by overnight dialysis at 37°C against a 30 mM Na-acetate buffer, pH 4.5, containing 100 mM NaCl and 10 mM 2-mercaptoethanol. The protein then was incubated for an additional 24 h at 37°C to reach maximum maturation. The catalytic domain was purified by anion-exchange chromatography on a HiTrap SP HP column (GE Healthcare) equilibrated in 30 mM Na-acetate buffer, pH 4.5, washed with the same buffer containing 100 mM NaCl, and eluted in 500 mM NaCl.
The production and initial purification of TcoCBc6 were done as described for TcoCBc1, except that expression was carried out in BMMY buffered at pH 7.3. After the IMAC step, the eluted proenzyme was dialyzed against a 30 mM Na-acetate buffer, pH 4.5, containing 100 mM NaCl and 10 mM 2-mercaptoethanol overnight at 4°C to initiate maturation, further dialyzed against a 20 mM Tris-HCl buffer, pH 8.0, and loaded onto a HiTrap heparin column (GE Healthcare) equilibrated in the same buffer. The bound TcoCBc6 protease was eluted with a linear 0 to 1 M NaCl gradient (2). The active protein concentrations of TcoCBc1 and TcoCBc6 preparations were determined by active-site titration using E-64 (11).
The coding region for the TcoCBs12 proenzyme was cloned into the pPICZαA vector with and without a six-His N-terminal tag to assess its influence on self maturation. The expression and purification of the His-tagged proenzyme was done as described for the TcoCBc1 proenzyme. The expression of the non-His-tagged proenzyme was done as described for TcoCBc1, and purification accomplished with three-phase partitioning by the recovery of the protein fraction that precipitated with 30% (wt/vol of the total volume) ammonium sulfate (69), dialyzed against a 30 mM Na-acetate buffer, pH 4.5, and loaded onto a HiTrap SP column as described for TcoCBc1. After the failure to achieve self maturation in vitro, purified TcoCBs12 was treated with immobilized pepsin (Pierce) as described by the manufacturer to obtain a putative mature protein (61). The catalytic domain was further purified on a HiTrap SP column as described previously.
The maturation site of each protease was determined by N-terminal sequencing (Plateforme D'Analyse et de Microsequençage des Proteines; Institut Pasteur, Paris, France).
Deglycosylation analysis.
N-glycosylation was assessed by SDS-PAGE after the treatment of recombinant proteins or total parasite extracts with endoglycosidase H (New England Biolabs) as described by the manufacturer.
Production of antisera against TcoCBc1 and TcoCBc6 catalytic domains.
Antisera were raised against mature, active nonglycosylated recombinant TcoCBc1 and TcoCBc6 in rabbits and mice. After the collection of preimmune sera, four injections in rabbits and mice with 50 and 10 μg of protein, respectively, were done with complete (primary injection) and incomplete (second, third, and fourth injections) Freund's adjuvant. A rabbit antiserum was raised according to the same protocol against C2, the recombinant catalytic domain of congopain (CP2) produced in Pichia pastoris (A. Boulangé, S. Khamadi, T. Coetzer, and E. Authié, unpublished results).
SDS-PAGE and immunoblotting.
Total protein preparations of 2 × 106 T. congolense procyclic and bloodstream forms were obtained by centrifugation, lysis of living parasites with 2% (wt/vol) SDS, and heating at 100°C in the presence of a protease inhibitor cocktail (Complete Mini, EDTA-free; Roche Diagnostics GmbH). Total protein preparations from IL-1180 epimastigote and metacyclic forms obtained as described previously (30) were kindly provided by Roger Pellé (ILRI, Nairobi, Kenya). Proteins were separated by SDS-PAGE (41), analyzed after being stained with Coomassie blue or transferred onto polyvinyl difluoride (PVDF) membranes (Immobilon-P; Millipore), and processed for Western blotting. Blots were blocked in PBS-T (15 mM phosphate buffer, pH 7.3, containing 150 mM NaCl and 0.05% [vol/vol] Tween 20) supplemented with 5% (wt/vol) nonfat milk for 1 h at room temperature prior to a 1-h incubation with dilutions of the rabbit anti-TcoCBc1 (1:20,000), mouse anti-TcoCBc6 (1:50,000), and rabbit anti-C2 (1:20,000) sera and mouse anti-PFR2 (anti-ParaFlagellar Rod protein 2) monoclonal antibody L8C4 (1:100) (40) or mouse anti-T. brucei tubulin (1:2,000) in PBS-T-milk. After being washed three times in 1 M NaCl, blots were incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG) (both from Sigma) diluted in PBS-T-milk (1:10,000), and antigen-antibody interactions were revealed with the Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Measurement of enzymatic activity and inhibition assays.
TcoCB peptidase activities were determined by the in vitro digestion of native bovine proteins or by a continuous fluorogenic assay using a FluoStar Optima microplate reader (BMG Labtech, Germany). Ten micrograms of purified bovine IgG (Sigma) or bovine serum albumin (BSA) (New England Biolabs) was incubated with 1 μg purified protease in 100 mM Na-acetate buffer, pH 4.5, containing 5 mM dithiothreitol (DTT) for up to 5 h at 37°C. Endopeptidase activities were determined using both benzoyloxycarbonyl-Phe-Arg-7-amido-4-methylcoumarin (Z-Phe-Arg-AMC) and Z-Arg-Arg-AMC substrates (Bachem). Michaelis kinetic parameters were determined (at optimum pHs for each substrate) by using a reaction buffer containing 100 mM Na-acetate, 200 mM NaCl, 5 mM EDTA, 5 mM DTT, pH 4.0, for the Z-Phe-Arg-AMC determinations and a buffer containing 150 mM sodium phosphate, 200 mM NaCl, 5 mM EDTA, 5 mM DTT, pH 6.0, for the Z-Arg-Arg-AMC determinations. The enzymes were preactivated in reaction buffer for 10 min at 37°C, and the reactions were started by adding different concentrations of substrates (0 to 150 mM). The release of AMC was measured at an excitation wavelength of 350 nm and emission wavelength of 460 nm. Initial rates were measured from the linear portion of the progression curves, and the kinetic parameters were determined by nonlinear regression analysis using KaleidaGraph (Synergy Software).
Carboxypeptidase activities were determined using the fluorescence-quenched substrate 2-aminobenzoyl-Phe-Arg-Ala-(2,4-dinitrophenyl)-e-NH2-lysine (Abz-Phe-Arg-Ala-Lys*-OH) (NeoMPS, Strasbourg, France). Michaelis kinetic parameters were determined using a reaction buffer containing 100 mM Na-acetate, 200 mM NaCl, 5 mM EDTA, and 5 mM DTT, pH 4.5. The enzymes were preactivated in reaction buffer for 10 min at 37°C, and the reactions were started by adding different concentrations of substrate (0 to 5 mM). The hydrolysis was monitored by measuring the fluorescence intensity at an excitation wavelength of 320 nm and an emission wavelength of 420 nm. Initial rates were measured from the linear portion of the progression curves, and the kinetic parameters were determined by linear regression analysis using KaleidaGraph.
The peptide bond cleaved by the carboxypeptidase activity of the protease was determined by electrospray ionization-time of flight mass spectrometry analysis (MoBIOS, IECB; Université Bordeaux 1, Talence, France) after the isolation of the products by high-performance liquid chromatography on a C18 column equilibrated with 25% (vol/vol) acetonitrile-0.1% (vol/vol) formic acid and elution using a 25 to 100% linear acetonitrile gradient.
The pH activity profiles of enzymes were determined by measuring the kcat/Km ratio at 0.5 pH intervals over the range of pH 3.5 to 7.5 using the assumption υ = [E][S]kcat/Km at S≪Km, where E is enzyme and S is substrate. The reactions were done in constant ionic-strength acetic acid-morpholinepropanesulfonic acid-Tris buffer (25) containing 200 mM NaCl, 10 mM EDTA, and 5 mM DTT. Data were fitted by nonlinear regression analysis with KaleidaGraph.
To assess the effect of known cysteine protease inhibitors on the activity of TcoCBc1 and C2, 3.5 nM of each protease was incubated at 37°C for 1 h with 350 nM of each inhibitor (all from Calbiochem) in 30 mM acetate buffer, pH 4.5, containing 5 mM DTT and 5 mM EDTA. The residual activity was measured as the initial rate of hydrolysis with 10 μM Z-Phe-Arg-AMC as the substrate and was related to the activity of the noninhibited enzymes.
RNA extraction and RT-PCR analysis.
Total RNA was extracted from procyclic and bloodstream forms of T. congolense IL-3000 using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA from epimastigote and metacyclic forms was kindly provided by Roger Pellé (ILRI, Nairobi, Kenya). The RNA concentration was assessed by spectrophotometry at 260 nm, and the RNA quality was confirmed by running 3 μg of total RNA from each preparation on a 1.2% (wt/vol) agarose gel. Total RNA was treated with RNase-free DNase I (1 U/μg RNA; Promega). The first-strand cDNA was synthesized using an oligo(dT) primer (Promega) and Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's instructions. Briefly, 250 ng of DNase I-treated RNA was preincubated for 5 min at 70°C with the primer and was placed on ice. The reverse transcription-PCR (RT-PCR) mix (1× buffer containing 100 μM deoxynucleoside triphosphates, 200 U Moloney murine leukemia virus reverse transcriptase, and 25 U of the recombinant RNasin RNase inhibitor [Promega]) was added to a final volume of 25 μl, and tubes were incubated at 42°C for 1 h. The cDNA quantity obtained from each parasitic form was normalized using the amplification of the actin gene with specific primers (ACT1 and ACT2) and Paq5000DNA polymerase (Stratagene). Equivalent quantities of cDNA of each form then were amplified according to the same protocol with specific primers for each cathepsin subfamily (see Table S1 in the supplemental material). The forward primers hybridize to the open reading frame (CathBN211S1 for TcoCBc1 to TcoCBc5, CB6ORF513 for TcoCBc6, CBserCom for TcoCBs7 to TcoCBs13, and SerBI4N211QF or SerBI4N227QF for TcoCBs13), and the cathepsins' reverse primers hybridize to the 3′UTR (CathH-TOPO2 for TcoCBc1 to TcoCBc5, CatB3UTR-H for TcoCBc6, CatB3UTR-I for TcoCBs7 to TcoCBs11, CathS-TOPO2 for TcoCBs12, and CatB3UTR-H for TcoCBs13). PCR products were electrophoresed on a 1% (wt/vol) agarose gel; the expected sizes were 471 bp for actin, 731 bp for TcoCBc1 to TcoCBc5, 1,048 bp for TcoCBc6, 589 bp for TcoCBs7 to TcoCBs11, 580 bp for TcoCBs12, and 677, 914, or 961 bp for TcoCBs13 depending on the forward primer used. PCR products were purified and subcloned into the pGEM-T vector (Promega). Positive clones were selected by blue/white screening followed by digestion with NdeI and NcoI (New England Biolabs) to verify the incorporation of an insert of the correct size. Several clones were sequenced with the BigDye Terminator v.1.1 cycle sequencing kit (ABI Prism; Perkin-Elmer Life Sciences) using the universal SP6 or T7 primers, as described by the manufacturer.
Immunoprecipitation of TcoCB from soluble extracts of bloodstream and procyclic forms of T. congolense.
IgG was purified from 2 ml of sera obtained from rabbits immunized with TcoCBc1 or TcoCBc6 using protein G Sepharose fast flow (GE Healthcare) as described by the manufacturer. Purified IgG (5 mg anti-TcoCBc1 and 3 mg anti-TcoCBc6) was coupled to 2 ml CNBr-activated Sepharose fast flow (GE Healthcare) as described by the manufacturer and was used to pull down proteins from soluble extracts of promastigote and bloodstream forms of T. congolense IL-3000. Briefly, 5 × 108 parasites were lysed in 1 ml of 15 mM phosphate buffer, pH 7.4, containing 500 mM NaCl, 0.1% (vol/vol) Triton X-100, and a cocktail of protease inhibitors (Complete mini EDTA-free; Roche Diagnostics GmbH). After centrifugation, the soluble fraction was incubated with 300 μl of resin-coupled IgG for 2 h at room temperature. After two washes, bound proteins were eluted with 1 ml of 0.05% SDS at 100°C, dialyzed against water, lyophilized, separated by SDS-PAGE, and silver stained for mass spectrometry analysis (Pôle Protéomique; Plateforme Génomique Fonctionnelle, Bordeaux, France). The 30-kDa broad band was treated as described elsewhere (59), with a few modifications. After trypsin digestion, peptide mixtures were analyzed by on-line capillary high-performance liquid chromatography coupled to a nanospray LTQ XL IT mass spectrometer. Data were subjected to a BLAST search by SEQUEST through the Bioworks 3.3.1 interface (ThermoFinnigan) against the GeneDB_Tcongolense_Protein_v1.fasta database (from the Wellcome Sanger Centre) and a local CPB_PRT.fasta database containing the IL-1180 cathepsin B sequences. The search parameters were as follows. The mass accuracy of the peptide precursor and peptide fragments was set to 2 and 1 Da, respectively. Only b and y ions were considered for mass calculation. The oxidation of methionines (+16 Da) was considered a differential modification. Two missed trypsin cleavages were allowed. Only peptides with Xcorr values higher than 1.5 (single charge), 2.0 (double charge), and 2.5 (triple charge) were retained. In all cases, the É¢Cn (the delta correlation value) had to be greater than 0.1, and the peptide P value had been less than 1 × 10−3.
Immunofluorescence.
To determine the subcellular localization of TcoCB in T. congolense bloodstream forms, washed trypanosomes were allowed to settle on slides and were fixed for 1 h with methanol at −20°C. After the PBS wash, fixed cells were incubated with dilutions of primary antibodies (mouse anti-TcoCBc1, 1:1,000; and rabbit anti-C2, 1:1,000) in PBS for 1 h, washed again, and incubated for 30 min with secondary antibodies (Alexa Fluor 488 goat anti-mouse IgG and Texas Red-X goat anti-rabbit IgG; both from Molecular Probes). Finally, cells were incubated for 5 min with 1-μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) and mounted in Vectashield (Vector Laboratories).
In vitro antitrypanosomal assays of the cathepsin B-specific cell-permeable inhibitor CA074Me.
T. congolense IL-3000 bloodstream forms were cultured in Iscove's modified Dulbecco's medium, as described previously (9, 16), supplemented with 20% (vol/vol) serum plus (Sigma). Parasites were isolated from the blood of infected mice at the first peak of parasitemia by centrifugation. Cultures (1 ml in 24-well plates) were maintained at 35°C in a humidified atmosphere containing 2% (vol/vol) CO2 (32). CA074Me (Calbiochem) was added at several final concentrations (0 to 250 μM) to 4 × 105 parasites/ml, and cells were counted every 24 h for a 96-h period. Each experiment was repeated three times to determine the minimal CA074Me concentration that affects parasite growth. At that concentration (10 μM), TcoCBc1 protein expression was monitored by immunoblotting as described previously.
To assess the effect of the cell-permeable inhibitor on the degradation of endocytosed proteins, T. congolense bloodstream forms were incubated in PSG buffer (38 mM Na2HPO4, 2 mM NaH2PO4, pH 8.0, 30 mM NaCl, 10 g/liter [wt/vol] glucose) containing 0.5 mg/ml fluorescein isothiocyanate (FITC)-coupled BSA (Sigma) for 3 h at 35°C in a humidified atmosphere containing 2% (vol/vol) CO2 and in the presence or absence of different concentrations of the cathepsin B inhibitor (0.1, 1, and 10 μM). Washed trypanosomes were fixed as previously described and incubated for 1 h with rabbit anti-C2 or rabbit anti-TcoCBc1 (both at a 1:1,000 dilution), followed by incubation with secondary antibody (Texas Red-X goat anti-rabbit IgG) and DAPI in PBS.
Antibody detection by ELISA.
For enzyme-linked immunosorbent assay (ELISA), wells of Maxisorp polystyrene plates (Nunc) were coated overnight at 4°C with 100 μl of a solution containing 2 μg/ml of purified TcoCBc1, deglycosylated pro-TcoCBs12, or C2 in 0.1 M sodium carbonate buffer, pH 9.6. Excess antigen was discarded, and the wells were blocked with a PBS solution containing 0.2% (wt/vol) gelatin at room temperature for 1 h, followed by three washes in PBS-T. Bovine infection sera (100 μl diluted 1:100 in PBS-T) were added to the wells, and the plate was incubated for 2 h at 37°C. After the wells were washed with PBS-T, 100 μl of diluted horseradish peroxidase-conjugated anti-bovine IgG (1:10,000) was added, and the plate was incubated for 10 min at 37°C. The plates were washed in PBS-T, and 100 μl of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS; 0.5 mg/ml; supplemented with 0.03% [wt/wt] hydrogen peroxide) was placed in each well. After 10 min of incubation, the reaction was stopped by adding 100 μl of a 0.1 M hydrofluoric acid solution, pH 3.5, containing 1 mM EDTA. ODs were measured at 405 nm in an ELx800 microplate reader (Bio-Tek). The background value for each well was measured by repeating the experiment in the absence of antigen. Results are expressed as the difference between the OD measured in the presence of antigen and that measured in the absence of antigen.
Nucleotide sequence accession numbers.
Gene sequences reported here were submitted to GenBank and accession numbers EU233643 to EU233655 were assigned to TcoCBc1 to TcoCBs13, respectively.
RESULTS
Identification of TcoCB genes in the T. congolense genome.
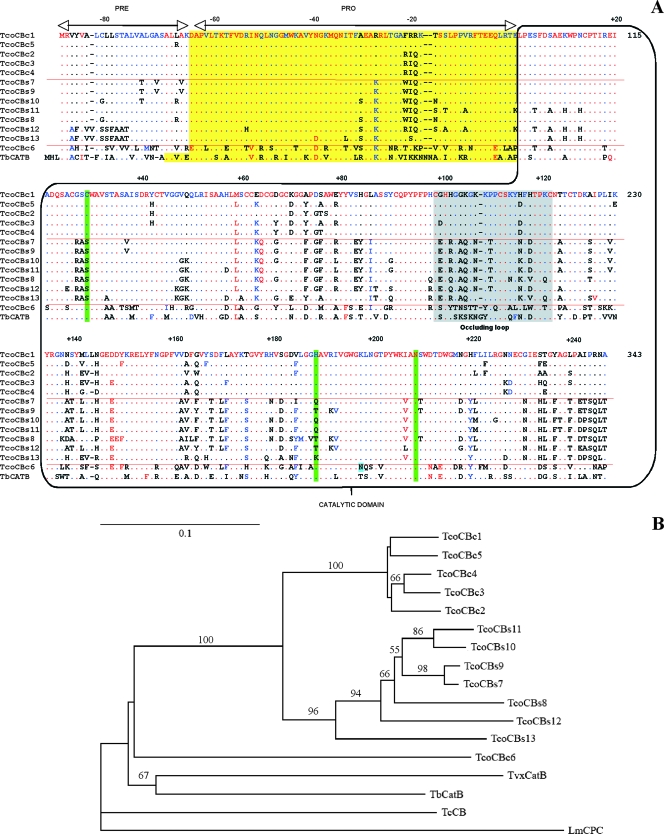
To identify the cathepsin B-like genes from T. congolense, BLAST analyses were performed on the T. congolense IL-3000 genomic database at the Wellcome Sanger Centre (http://www.genedb.org) using, at first, the T. brucei tbcatB gene sequence (accession no. AY508515 [45]). The search identified eight predicted genes with E values between 8.0 e−124 and 1.1 e−73. A second round of BLAST was done using the 1,011-bp open reading frame from the congo886c10.p1k_3 sequence. This identified 14 predicted genes with E values between 2.0 e−220 and 1.0 e−73. Twelve of these genes contained complete coding sequence, encoding proteins of between 335 and 337 amino acids. All identified genes encoded proteins with the expected structure of cathepsin B-like proteins: a prepeptide identified as a signal peptide by SignalP using the hidden Markov model (14), a propeptide, and a catalytic domain. Among these proteases, five contained the conserved catalytic triad of cysteine proteases Cys29, His199, and Asn219 (according to human cathepsin B numbering), whereas seven contained an atypical Ser29, Thr/Lys/Gln199, and Asn219 triad. However, all of these encoded the two conserved motifs that characterize the cathepsin B family of proteases, Gly-Cys-Xaa-Gly-Gly (residues 70 to 74 of human cathepsin B), and the occluding loop motif thought to be responsible for exopeptidase activity (35).
Based on the analysis of the 5′UTR and 3′UTR sequences of the predicted genes, specific primers were designed to amplify the whole family of genes from the T. congolense IL-1180 clone, another savannah type clone used in experimental trypanosomiasis studies of cattle. From the analyzed clones, at least 13 different sequences were identified (GenBank accession numbers EU233643 to EU233655). Six presented the classical catalytic triad, and seven presented the atypical triad described above (Fig. 1A). The open reading frames corresponding to the normal catalytic triad were named TcoCBc1 to TcoCBc6 for T. congolense cathepsin B (with the “c” preceding the number referring to the active-site Cys), and the putative cathepsin B-like proteases with a serine instead of a cysteine in the catalytic triad were named TcoCBs7 to TcoCBs13 (with the “s” preceding the number referring to the active-site Ser). Whereas most of these genes are unique, several closely related sequences encoding isoforms with a high degree of identity (≥99%) were identified for TcoCBc5 (three genes), TcoCBc3 (two genes), and TcoCBs12 (four sequences, of which two are pseudogenes; data not shown). Within the TcoCBc group, the encoded proteins share 92 to 96% identity, with the exception of TcoCBc6, which shares only 58 to 59% identity with TcoCBc1 to TcoCBc5 and 55 to 57% with the TcoCBs group. Within the serine subfamily, proteins share 77 to 98% identity and constitute a more divergent group of proteases. A comparison of the different proteases characterized in clones IL-3000 and IL-1180 revealed a high degree of identity: perfect homologues of TcoCBc1 and TcoCBs8 are found in both clones, protease TcoCBc6 shares 99% with its orthologue in clone IL-3000, TcoCBc1 to TcoCBc5 share more than 90% with their orthologues in clone IL-3000, and the TcoCBs group shares between 77% and 100% identity with its orthologues in clone IL-3000. A phylogenetic analysis illustrated the evolution of the TcoCB family of proteases (Fig. 1B). The TcoCBc1 to TcoCBc5 proteases are very closely related, as are the TcoCBs7 to TcoCBs12 proteases. TcoCBc6 appeared to be the most distantly related protein of the family, almost as divergent as the orthologue proteins from other kinetoplastidae. In order to study this novel family of proteases, we produced, by recombinant technology, one representative of each phylogenetic branch: TcoCBc1, TcoCBc6, and TcoCBs12.
FIG. 1.
Amino acid sequence alignment and phylogenetic analysis of the TcoCB family of proteases from T. congolense IL-1180. (A) Alignment of cathepsin B-like enzymes. The boxes denote predicted domains. The predicted catalytic triad is indicated in light green, and the occluding loop structure is in gray. (B) Phylogenetic tree. The phylogeny presented is based on the alignment of the entire amino acid sequence of all the TcoCB family members together with the cathepsin B-like proteases of T. brucei (TbCatB), T. cruzi (TcCB), and L. major (LmCPC) and the predicted cathepsin B-like protease of T. vivax. (TvxCatB). The tree was rooted on the L. major protein. Numbers beside each node indicate bootstrap values as percentages of 100 replicates (determined by the neighbor-joining method). The majority rule bootstrap maximum parsimony tree was in agreement with it but was not as well resolved. The values for branches that are not supported by the >50-bootstrap rule are not indicated.
Production of TcoCBc1, TcoCBc6, and TcoCBs12 recombinant proteases.
Preliminary expression assays of wild-type pro-TcoCB in Pichia pastoris resulted either in the failure of protease production or in the production of highly glycosylated enzymes. Therefore, nonglycosylated mutant forms of the recombinant proteases were engineered by disrupting their potential glycosylation sites, especially those present in the catalytic domains, as these domains also would be used for antiserum production and diagnostic trials. Partially or nonglycosylated mutants were generated by the site-directed mutagenesis of the consensus N-glycosylation sites (Asn-Xaa-Ser/Thr), resulting in the replacement of Asn by Ser or Gln. Figure 2 summarizes the different cloning strategies and the recombinant proteins successfully expressed in Pichia pastoris.
A six-His-tagged version of pro-TcoCBc1 was successfully expressed and purified by IMAC as described in Materials and Methods. An SDS-PAGE analysis of the eluted recombinant protein revealed a prominent band of 39 kDa and a minor protein of 40 kDa (see Fig. S1, lane 1, in the supplemental material). These bands correspond to glycosylated forms of a 35-kDa protein, the expected size of the protease, considering the additional six-His tag, and as deduced from endoglycosidase H treatment (data not shown). The exposure of the proenzyme to an acidic and reducing environment for 48 h generated a major band of 29 kDa (see Fig. S1, lane 2, in the supplemental material). As expected, the deglycosylation of the 29-kDa protein did not reveal additional glycosylation sites (data not shown). The N-terminal sequencing of the processed proteins revealed two sequences: one-third of the analyzed band had FTEEQ at the N terminus, and two-thirds of the band had GAFRR at the N terminus, showing that most of the prodomain is removed 21 amino acids upstream of the predicted N terminus of the catalytic domain (Fig. 2). The catalytic domain was further purified by anion-exchange chromatography. A TcoCBc1_Cys118Ala null mutant also was constructed and tested for maturation; no processing was observed under the same conditions (data not shown), demonstrating that autoprocessing is required for the maturation of the proenzyme.
TcoCBc6 was expressed in Pichia pastoris at pH 7.3. After IMAC purification, the expressed protein migrated as a major band of 40 kDa and as a broad smear above 50 kDa (see Fig. S1, lane 3, in the supplemental material). Again, deglycosylation analysis revealed that these bands correspond to glycosylated forms of a 36-kDa protein. As was the case for pro-TcoCBc1, pro-TcoCBc6 underwent processing at acidic pH under reducing conditions, but it was processed more rapidly than pro-TcoCBc1 (16 h at 4°C, compared to 48 h at 37°C for TcoCBc1). A single nonglycosylated 30-kDa processing product was detected by SDS-PAGE (see Fig. S1, lane 4, in the supplemental material), and N-terminal sequencing resulted in a single sequence (VSEAKR) that corresponds to a catalytic domain beginning at position −31 of the predicted catalytic region (Fig. 2).
Pro-TcoCBs12 was successfully produced in Pichia pastoris with and without an N-terminal six-His tag and was purified as described in Materials and Methods (see Fig. S1, lane 5, in the supplemental material). As expected, a glycosylation site was still present in the prodomain of the protein, as confirmed by endoglycosidase H treatment (data not shown). Attempts to maturate the His-tagged and non-His-tagged proenzymes under the same conditions as those described for TcoCBc1 and TcoCBc6 were unsuccessful. Alternative maturation conditions were tested (e.g., pH, salt concentration, and divalent cations) without success. However, a putative mature protein was obtained by in vitro treatment with pepsin, resulting in a single 29-kDa product as detected by SDS-PAGE (see Fig. S1, lane 6, in the supplemental material). N-terminal sequencing resulted in one sequence (RTKLP) that corresponds almost exactly to the maturation site predicted by sequence homology to those of other known cathepsin B-like proteins (Fig. 1A).
Biochemical characterization of processed recombinant proteases.
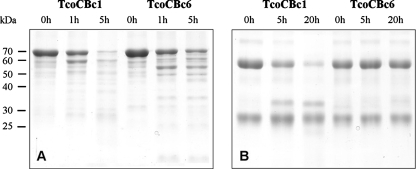
The proteolytic activity of TcoCBc1, TcoCBc6, and pepsin-matured pro-TcoCBs12 (pep-TcoCBs12) was first tested against native bovine proteins in vitro. BSA and IgG were incubated with the mature proteases, and proteolytic products were separated by SDS-PAGE. Only the self-processed proteins (TcoCBc1 and TcoCBc6) exhibited proteolytic activity, as shown in Fig. 3. No activity was observed with the pep-TcoCBs12-purified fraction at acidic, neutral, or basic pH (data not shown). After 5 h of incubation with TcoCBc1, both bovine proteins were partially hydrolyzed. TcoCBc6 was able to hydrolyze BSA but not IgG usually cleaved in the hinge region, suggesting differences in the specificity of each protease. No proteolysis was observed in the absence of recombinant protease (data not shown).
FIG. 3.
SDS-PAGE analysis (with Coomassie blue staining) of in vitro digestions of bovine proteins by recombinant TcoCBc1 and TcoCBc6. Ten micrograms of BSA (A) or bovine IgG (B) was incubated for several hours with 1 μg of recombinant mature proteases in 100 mM Na-acetate buffer, pH 4.5, 5 mM DTT, 5 mM EDTA at 37°C.
Further biochemical analyses were done with two common synthetic cathepsin substrates: Z-Phe-Arg-AMC, a fluorogenic substrate employed for endopeptidase activity tests of both cathepsin L-like and cathepsin B-like enzymes, and Z-Arg-Arg-AMC, a specific substrate of the endopeptidase activity of cathepsin B-like proteases. Proteases from this family are capable of accepting an arginine at position P2 (31), the side chain of which is proposed to form a salt bridge with the side chain of Glu233 (according to TcoCBc1 numbering), a perfectly conserved residue within the T. congolense cathepsin B-like family of proteases.
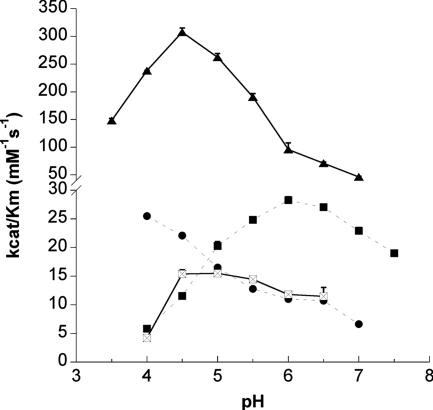
The pH activity profiles of recombinant TcoCBc1 showed maximum activity at pH 4.0 for Z-Phe-Arg-AMC and at pH 6.0 for Z-Arg-Arg-AMC (Fig. 4). Measuring the fluorescence at pH below 4.0 resulted in a partial loss of the substrate, probably due to solubility problems. With these substrates, no endopeptidase activity was detected with recombinant TcoCBc6 or with pep-TcoCBs12, which suggests that these proteases are devoid of endopeptidase activity.
FIG. 4.
pH activity profiles of active recombinant proteases TcoCBc1 and TcoCBc6 with endopeptidase (Z-Phe-Arg-AMC and Z-Arg-Arg-AMC; broken lines) and carboxypeptidase (Abz-Phe-Arg-Ala-Lys*-OH; continuous lines) fluorogenic substrates. Recombinant proteases were assayed in AMT (acetate-MES-Tris) buffers across the indicated pH range in the presence of 5 mM DTT at 37°C after 10 min of preincubation in reaction buffers. •, TcoCBc1 activity with Z-Phe-Arg-AMC; ▪, TcoCBc1 activity with Z-Arg-Arg-AMC; ▴, TcoCBc1 activity with Abz-Phe-Arg-Ala-Lys*-OH; and ⊠, TcoCBc6 activity with Abz-Phe-Arg-Ala-Lys*-OH.
To discriminate the carboxypeptidase activity of the TcoCB family, Abz-Phe-Arg-Ala-Lys*-OH, an internally quenched substrate with a free C-terminal carbonyl group, was used (36). This carboxypeptidase-specific substrate was hydrolyzed by TcoCBc1 and TcoCBc6 at optimum pHs of 4.5 and 5.0, respectively (Fig. 4). Pepsin-treated pro-TcoCBs12 did not hydrolyze the carboxypeptidase substrate in vitro. Mass spectrometry analysis of the hydrolysis products showed that both TcoCBc1 and TcoCBc6 act as carboxydipeptidases, cleaving the Arg-Ala peptidic bond. Table 1 summarizes the kinetic constants for the synthetic substrate hydrolysis by TcoCBc1 and TcoCBc6. These experiments showed that TcoCBc1 functions sixfold more efficiently as a carboxydipeptidase than as an endopeptidase.
TABLE 1.
Michaelis-Menten constants of recombinant TcoCB1c1, TcoCBc6, and pep-TcoCBs12
| Substrate | Michaelis-Menten constant ofa:
|
||||
|---|---|---|---|---|---|
| TcoCBc1
|
TcoCBc6
|
pep-TcoCBs12
|
|||
| kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat/Km (mM−1 s−1) | kcat/Km (mM−1 s−1) | |
| Z-Phe-Arg-AMC | 1.09 | 123.42 | 8.88 | n.h. | n.h. |
| Z-Arg-Arg-AMC | 3.49 | 173.28 | 20.2 | n.h. | n.h. |
| Abz-Phe-Arg-Ala-Lys*-OH | ND | ND | 209.05 | 49.9 | n.h. |
Two endopeptidase substrates, Z-Phe-Arg-AMC and Z-Arg-Arg-AMC, and one carboxypeptidase substrate, Abz-Phe-Arg-Ala-Lys*-OH, were used. Determinations were done in Na-acetate buffer containing 5 mM DTT at the previously determined optimum pH and at 37°C. Values are the means from three replicates; the standard error was always less than 10%. ND, not determined; n.h., not hydrolyzed.
Effect of protease inhibitors on TcoCBc1 activity.
To further investigate the specificity of TcoCBc1, several known inhibitors of cysteine proteases were tested against TcoCBc1 and C2; the latter was used as a T. congolense cathepsin L-like protease control. As shown in Table 2, leupeptin and chymostatin inhibited C2 completely but did not inhibit TcoCBc1, which was completely resistant to both inhibitors. Furthermore, the same result was obtained with a lysinal analogue of leupeptin, acetyl-Leu-Val-lysinal-CHO, which reportedly is a stronger inhibitor of cathepsin B-like cysteine proteases (50). Another potent cathepsin B inhibitor belonging to the N-peptidyl, O-acyl hydroxamate family (Z-Phe-Gly-NHO-Bz-pOMe) was more active against TcoCBc1 than C2, which is in accordance with the reported data (22). As expected, E-64 completely inhibited the activity of both TcoCBc1 and C2. CA074, a specific inhibitor of cathepsin B-like proteases (68), specifically inhibited TcoCBc1 but not C2. As expected, chicken cystatin was a very potent inhibitor of C2 but showed less inhibitory activity against TcoCBc1. This was probably due to the presence of the occluding loop structure (15, 57). No activity was measured in the absence of the reducing agent DTT.
TABLE 2.
Sensitivity of T. congolense recombinant TcoCBc1 and C2 to protease inhibitors in vitroa
| Inhibitor | % Residual activity of:
|
|
|---|---|---|
| TcoCBc1 | C2 | |
| Leupeptin | 100 | 0 |
| Ac-Leu-Val-lysinal-CHO | 85 | 0 |
| Cystatin | 77 | 2 |
| Chymostatin | 100 | 0 |
| E-64 | 0 | 0 |
| CA074 | 0 | 94 |
| Z-Phe-Gly-NHO-Bz-pMe | 13 | 88 |
| Buffer lacking DTT | 0 | 0 |
Protease at a concentration of 3.5 nM was incubated with 350 nM of each inhibitor for 1 h at 37°C in 30 mM Na-acetate buffer, pH 4.5, containing 5 mM DTT and 5 mM EDTA. Assays were performed in the same buffer, and the initial velocities of the hydrolysis of Z-Phe-Arg-AMC were determined and compared to the initial velocity in the absence of inhibitor. The values are the means from three replicates; the standard error was always less than 5%.
TcoCB gene expression analysis.
To assess the expression and localization of the TcoCB proteases in the different life cycle stages of T. congolense, polyclonal antibodies were raised against TcoCBc1 and TcoCBc6 (Fig. 5). Antibodies raised against TcoCBc1 cross-reacted with recombinant TcoCBs12 (Fig. 5A, lanes 5 and 7), TcoCBs9, and TcoCBs13 (data not shown), three closely related proteins from the active-site serine subfamily, which implies that the anti-TcoCBc1 antibodies do not discriminate between TcoCBc1 to TcoCBc5 and TcoCBs7 to TcoCBs13 (due to the close identity of these proteins). The antiserum raised against TcoCBc6 was isoform specific (lanes 5 to 8), as it did not recognize TcoCBc1 and TcoCBs12 (lanes 5 and 7), TcoCBs9, or TcoCBs13 (data not shown). No cross-reactivity was observed between the catalytic domain (C2) of the cathepsin L-like protease CP2 and the TcoCB isoforms using anti-C2 antibodies (17) or the antisera produced against the TcoCB recombinant proteins.
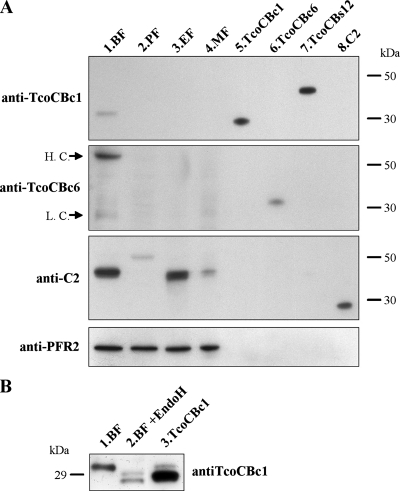
FIG. 5.
Western blot analysis of TcoCB expression in T. congolense. (A) Proteins from 2 × 106 bloodstream, procyclic, epimastigote, and metacyclic forms (lanes 1 to 4, respectively) were separated on SDS-PAGE under reducing conditions together with 10 ng of recombinant proteases TcoCBc1, TcoCBc6, pro-TcoCBs12, and C2 (lanes 5 to 8, respectively), transferred to a PVDF membrane, and probed sequentially with antisera raised against different cysteine proteases. PFR2 was used as a loading control. Arrows indicate mouse IgG heavy chains (H.C.) and light chains (L.C.) recognized by the anti-mouse IgG peroxidase-conjugated secondary antibody. (B) Deglycosylation analysis of T. congolense bloodstream forms. A total of 2 × 106 bloodstream forms were left untreated (lane 1) or were treated (lane 2) with endoglycosidase H, and proteins were separated by SDS-PAGE together with 10 ng of recombinant nonglycosylated TcoCBc1 (lane 3). Proteins were transferred to a PVDF membrane and probed with antiserum raised against TcoCBc1.
Western blot analysis of whole extracts of each of the four life cycle stages of T. congolense (bloodstream, procyclic, epimastigote, and metacyclic forms) using the anti-TcoCBc1 serum (Fig. 5, lanes 1 to 4) showed the recognition of a single 32-kDa band in bloodstream forms only (Fig. 5A, lane 1). Following enzymatic deglycosylation with endoglycosidase H, this protein migrated as two bands of 29 and 30 kDa, which could correspond to several isoforms of the protease and/or products of maturation at alternative sites of the same protein (Fig. 5B). Anti-TcoCBc6 antibodies did not detect this isoform in any of the life cycle stages tested. The two bands revealed in lane 1 corresponded to the heavy and light chains of mouse IgG (as shown by control experiments omitting the primary antibody [data not shown]; the presence of mouse IgG is due to the fact that parasites were isolated from infected mice). These results clearly showed that TcoCBc1 to TcoCBc5 and/or TcoCBs7 to TcoCBs13 are expressed mainly in bloodstream forms of T. congolense, while TcoCBc6 is not expressed at detectable levels in any life cycle stage. CP2 is expressed mainly in bloodstream and epimastigote forms.
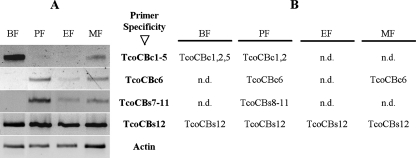
To overcome the limitations resulting from the cross-reactivity of the anti-TcoCBc1 polyclonal antibodies, a genomic approach was adopted by studying the transcription levels of the tcocb genes using a semiquantitative RT-PCR assay (Fig. 6A). The cDNA from each life cycle form was normalized using actin transcripts. The PCR amplification was achieved with specific primers initially tested on genomic DNA to confirm their specificity (see Table S1 in the supplemental material). The sizes of PCR products were checked, and for each sample, the products were cloned and sequenced in order to confirm the specificity of the reaction. The results of the sequencing were always in agreement with the targeted subfamily and allowed the partial identification of the transcript population (Fig. 6B). For the TcoCBc1 to TcoCBc5 subfamily, the amplification was almost exclusively in the bloodstream forms, minor amounts were observed in the metacyclic forms, and hardly any detectable amplification was obtained in the other forms (Fig. 6). In contrast, no amplification was obtained in bloodstream forms for TcoCBc6 or the TcoCBs7 to TcoCBs11 subfamily, but major amplification was seen in the procyclic form and, to a lesser extent, in the epimastigote and metacyclic forms. The TcoCBs12 transcript was the only one for which the signal was constant in all parasitic stages. On the other hand, no amplification could be observed for the TcoCBs13 subfamily. These results indicate that differential expression is at least partly due to transcript abundance regulation, which seems to be in accordance with the protein amounts detected by Western blot analysis. In order to confirm these data, the presence of each subfamily was investigated through immunoprecipitation and mass spectrometry analysis of bloodstream and procyclic forms.
FIG. 6.
RT-PCR analysis of transcripts of the TcoCB gene subgroups expressed during the life cycle of T. congolense. (A) Sets of primers were used to amplify specific TcoCB subfamilies in each of the four stages of the parasite, i.e., the bloodstream (BF), procyclic (PF), epimastigote (EF), and metacyclic (MF) forms. Actin was used as a normalizing control. Specific primers for TcoCBs13 also were used, but no amplification product was obtained (data not shown). (B) Transcript identification. PCR products were cloned and sequenced to identify the transcribed genes. n.d., not done.
Immobilized antibodies directed against TcoCBc1 (which recognize all recombinant TcoCB proteins except TcoCBc6) (Fig. 5) and against TcoCBc6 were used to immunoprecipitate the TcoCB proteases from soluble extracts of T. congolense procyclic and bloodstream forms. Mass spectrometry analysis was used to identify peptides that match both the cysteine and serine subfamilies of TcoCB proteases. Seven peptides matching members of the TcoCBc subfamily (with the exception of TcoCBc6) and eight peptides matching members of the TcoCBs subfamily (with the exception of TcoCBs13) were identified in extracts of bloodstream forms. None of these peptides were identified in T. congolense procyclic-form soluble extracts. The limited number of isoform-specific peptides did not allow the characterization of the expressed isoforms. Nevertheless, out of eight peptides matching the TcoCBs subfamily, four are present in the TcoCBs12 amino acid sequence (see Table S2 in the supplemental material).
Subcellular localization.
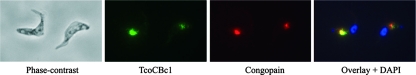
Based on the lysosomal localization of congopain in T. congolense (49), C2 (its catalytic domain) was used as a lysosomal marker in the colocalization analysis. In light of the absence of cross-reactivity of the antisera raised against C2 and TcoCBc1 with denatured proteins (on Western blots) and native proteins (in ELISA [data not shown]), and because of the absence of cathepsin L peptides in pull-down experiments, they were used for colocalization studies. As shown in Fig. 7, both antisera labeled the same compartment in T. congolense bloodstream forms. No labeling was observed in the procyclic forms (data not shown). As the TcoCBc1 antiserum showed cross-reactivity with other isoforms of the TcoCB family (except TcoCBc6), all of the isoforms expressed in bloodstream forms seem to be associated with the same compartment (the lysosome), as is the case for the cathepsin L-like protease congopain.
FIG. 7.
Localization of TcoCB in bloodstream forms of T. congolense by immunofluorescence. Shown are a phase-contrast image of the cells, cells probed with mouse anti-TcoCBc1 antibodies (green), cells probed with rabbit anti-C2 antibody (red), and a merged image showing DAPI staining (blue) and the colocalization of TcoCBc1 and C2 (yellow). Both proteins localize in a defined region between the nucleus and the kinetoplast.
In vitro antitrypanosomal effect of CA074Me.
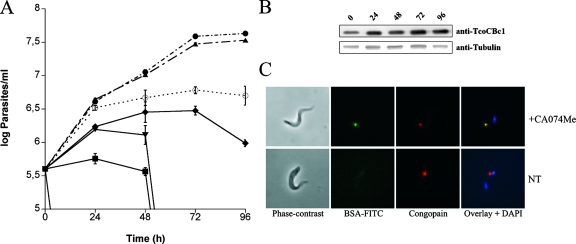
To establish whether cathepsin B-like proteases are essential for trypanosome survival, and in the absence of RNA interference technology in T. congolense bloodstream forms, the effect of a cell-permeable analogue of CA074 was tested on in vitro cultures of T. congolense. As shown in Fig. 8A, concentrations greater than 10 μM significantly affect the growth of parasites in culture compared to that of nontreated cells. Whereas 10 μM CA074Me induced a trypanostatic effect after 24 h, higher concentrations affect early parasite replication and rapidly lead to their death. During the time course of the experiment, a slight time-dependent increase of TcoCB protein expression was observed at an inhibitor concentration of 10 μM, as assessed by Western blot analysis (Fig. 8B); at higher concentrations (25 and 50 μM) an even stronger increase was observed (data not shown). To assess the effect of the inhibitor on protein uptake and degradation, bloodstream forms were incubated for 3 h with FITC-labeled BSA in the presence of 0.1, 1, and 10 μM inhibitor. Cells treated with 1 (data not shown) and 10 μM CA074Me exhibited a marked accumulation of undigested BSA in the lysosomal compartment, as assessed by colocalization with the lysosomal protein CP2 by fluorescence microscopy (Fig. 8C). No accumulation was observed in cells treated with 0.1 μM inhibitor or in the absence of inhibitor. Taken together, these results indicate a role for cathepsin B-like proteases in endocytosed protein degradation and in the survival of T. congolense bloodstream forms in vitro.
FIG. 8.
Effect of the cathepsin B cell-permeable inhibitor CA074Me on T. congolense bloodstream forms. (A) Parasites were cultured in the presence of several concentrations of CA074Me, and cells were counted every 24 h (•, 0 μM; ▴, 1 μM; ○, 10 μM; ♦, 25 μM; ▾, 100 μM; ▪, 250 μM). Each value represents the mean from three experiments. Standard errors are indicated. (B) TcoCB protein expression was monitored during treatment with 10 μM inhibitor by Western blotting. Tubulin was used as a loading control. (C) Immunofluorescence analysis of parasites incubated for 3 h with FITC-labeled BSA in PSG buffer in the presence (+CA074Me) or absence (NT) of 10 μM inhibitor. Note the accumulation of undigested BSA (green) in the lysosomal compartment of inhibitor-treated parasites as assessed by colocalization with the lysosomal marker congopain (red).
Antigenicity of TcoCB in infected cattle.
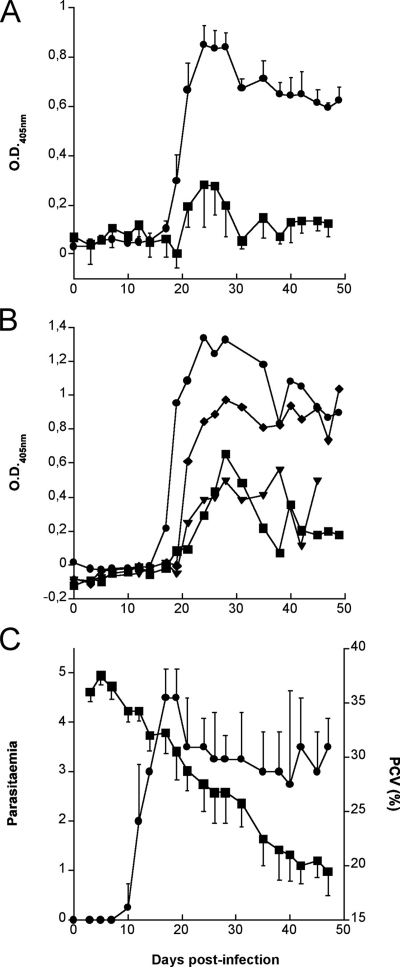
To determine if antibodies to TcoCB isoforms are elicited in T. congolense-infected cattle, the time course of humoral responses to TcoCBc1, deglycosylated pro-TcoCBs12, and C2 was studied in four trypanosusceptible Boran cattle (Bos indicus) following primary infection with T. congolense IL-1180. Sera were collected twice weekly and tested by ELISA for the presence of antibodies to TcoCB. In all four cattle, antibodies against TcoCBc1 consistently appeared 2 weeks after infection, reached a peak between 3 and 4 weeks, and thereafter declined but remained at a significant level until treatment (Fig. 9A). A more variable response was observed with TcoCBs12, as two individuals developed a prominent response similar to that against TcoCBc1, whereas the other two gave less prominent responses (Fig. 9B). In addition, there were important fluctuations in antibody levels during the course of infection. As expected from previous results (5), only low levels of antibody to the catalytic domain of congopain were detected in those cattle. Parasitemia and packed red cell volume, as a measure of anemia, were monitored as described earlier (5) (Fig. 9C).
FIG. 9.
Kinetics of antibody responses to TcoCBc1, deglycosylated pro-TcoCBs12, and C2 in trypanosusceptible cattle infected with T. congolense. Boran cattle (four animals) were infected at day 0 with T. congolense IL-1180. (C) The cattle were monitored for parasitemia (•) and packed red-cell volume (PCV) (▪) before treatment. Collected sera were tested by ELISA for reactivity to TcoCBc1 (•) and C2 (▪) (A) as well as pro-TcoCBs12 (B). For panels A and C, each point represents the mean result from four cattle; standard errors are indicated. The data shown in panel B are from individual animals.
DISCUSSION
Proteolytic enzymes play key roles in the development of parasitic diseases. They are essential for parasite survival and likely contribute to pathogenic processes. Previous work has established that cathepsin L-like cysteine proteases, namely congopain and its isoforms, are the main proteolytic activities in T. congolense bloodstream forms. These enzymes are antigenic in infected cattle, and there is an apparent correlation between the level of specific IgG responses and the degree of natural resistance to disease, termed trypanotolerance (5, 6, 49). It remains difficult to establish a direct relationship between these proteolytic enzymes and pathogenicity, although congopain was detected in an active form in plasma from infected animals, encouraging further research in the field. Little information is available concerning other cysteine proteases in T. congolense.
In a search for new cysteine proteases in T. congolense, we identified a family of genes composed of at least 13 genes coding for cathepsin B-like proteases, which confirms that the cysteine protease repertoire is more complex than initially thought. The number of genes identified may represent only part of the total repertoire, as the sequencing of clone IL-3000 is not yet completed. This genomic amplification is unique to T. congolense among the pathologically important kinetoplastidae (there is only one gene in T. brucei, two highly homologous genes in T. cruzi and Trypanosoma vivax, and one gene in L. major). Two subfamilies were identified in T. congolense IL-1180, six genes encoding cysteine proteases with the classical catalytic triad of clan CA (Cys, His, and Asn) and seven genes encoding proteins in which the catalytic cysteine is replaced by serine (Fig. 1A). The phylogenetic analysis of this family of proteases (Fig. 1B) shows that each subgroup seems to have evolved independently from a common cathepsin B-like ancestor. Three individual groups emerged: the TcoCBc1 to TcoCBc5 subfamily, the TcoCBs7 to TcoCBs13 subfamily, and TcoCBc6, which represents an independently evolved protein within the family. The unique cathepsin B-like gene of T. brucei seems to be essential for the survival of the bloodstream form in vitro; therefore, an amplification of the cathepsin B-like protease repertoire in T. congolense probably is the result of a functional specialization.
In order to determine the expression profile of the TcoCB protease family during the life cycle of T. congolense, and as this parasite exhibits complex posttranscriptional regulation mechanisms (18), several approaches were used: transcript analysis by RT-PCR, protein expression by immunoblotting, and mass spectrometry after immunoprecipitation. Immunoblot profiling showed that members of the TcoCB family are specifically expressed in bloodstream forms of T. congolense and that no TcoCBc6 is present at detectable levels in any stage of the life cycle of the parasite (neither at the protein nor at the transcript level). The absence of the specificity of the antiserum raised against TcoCBc1 did not allow differentiation between the expression of TcoCBc1 to TcoCBc5 and TcoCBs7 to TcoCBs13. However, since the only transcript from the active-site serine subfamily found at detectable amounts in bloodstream forms was TcoCBs12, and as the immunoprecipitation analysis allowed the identification of four specific peptides of the active-site serine subfamily in whole-protein extracts (see Table S2 in the supplemental material), there is sufficient evidence to suggest that at least one member of this subfamily (TcoCBs12) is expressed in bloodstream forms of T. congolense. However, three other peptides matching members of this subfamily that do not match TcoCBs12 also were identified, suggesting that other genes not identified in this study are expressed. One or more proteins from the TcoCBc1 to TcoCBc5 group also are expressed in bloodstream forms, but the specific identity of the expressed proteases cannot be confirmed, as mass spectrometry analysis mainly identified peptides common to all five members of the subfamily (see Table S2 in the supplemental material).
Representatives of each phylogenetically distinct group were recombinantly expressed: TcoCBc1 from the TcoCBc1 to TcoCBc5 branch, TcoCBs12 from the TcoCBs7 to TcoCBs13 branch, and TcoCBc6, the most divergent protein of the family. Like other cathepsin B-like cysteine proteases, TcoCBc1 and TcoCBc6 were produced as precursor proteins that were able to undergo autoproteolytic cleavage to generate active proteases. Proteins were always self processed upstream of the predicted site (Fig. 2), as assessed by N-terminal sequencing. Pro-TcoCBs12 was not able to undergo self maturation in the conditions tested. Therefore, trans-maturation using other proteases was tested. As reported for other cathepsin B-like proteases, pepsin treatment allowed the cleavage of the propeptide within the predicted maturation region (position −3 from the predicted site).
The proteolytic activity of mature proteases was tested on native proteins, and kinetic parameters were determined using known fluorogenic substrates. Proteolytic profiles of bovine proteins digested by TcoCBc1 and TcoCBc6 show that these two enzymes have different substrate specificities. This was confirmed by in vitro characterization using synthetic substrates. Pepsin-treated pro-TcoCBs12 showed no activity against native proteins or fluorogenic substrates. The biochemical characterization of the two active representatives of the family (TcoCBc1 and TcoCBc6) showed that both optimum pH and sequence specificity are different for each protein, suggesting that they do not represent a redundant system of protein degradation and that they could play a role outside of lysosomes, as they are still active at neutral pH, as reported previously for other cathepsins (8, 21, 24, 37). Michaelis-Menten constants for both TcoCBc1 and TcoCBc6 were determined using endopeptidase and carboxypeptidase fluorogenic substrates (Table 1). TcoCBc1 displayed poor activity against both endopeptidase substrates compared to that of other cathepsin B-like proteases (kcat/Km values of 8.88 and 20.2 mM−1 s−1 for Z-Phe-Arg-AMC and Z-Arg-Arg-AMC, respectively, compared to 1,400 and 900 mM−1 s−1 of human cathepsin B for Z-Phe-Arg-AMC and Z-Arg-Arg-AMC, respectively [10]) and TcoCBc6 showed no activity against the endopeptidase substrates tested. Both TcoCBc1 and TcoCBc6 exhibit carboxypeptidase activity toward the fluorogenic substrate employed, and kcat/Km values were, as for the endopeptidase substrates, lower than expected compared to those of other cathepsin B-like enzymes (kcat/Km values of 210 and 50 mM−1 s−1 for TcoCBc1 and TcoCBc6, respectively, compared to 1,944 mM−1 s−1 for human cathepsin B [36]). As demonstrated for most of the cathepsin B-like enzymes studied in the past, TcoCBc1 and TcoCBc6 behave mostly as carboxydipeptidases, and endopeptidase activity would only be an evolutionary remnant of an ancestral papain-like cysteine protease (35).
Commonly used inhibitors, such as E-64, a specific cysteine protease inhibitor, CA074, a cathepsin B-specific inhibitor, and Z-Phe-Gly-NHO-Bz-pOMe (known to be a strong inhibitor of cathepsins), inhibit recombinant TcoCBc1 in vitro as expected for cathepsin B-like proteases. On the other hand, chymostatin, leupeptin, and its lysinal analogue, Ac-Leu-Val-Lysinal-CHO, show no or a poor inhibitory effect on the recombinant protease. These inhibitors, although not specific for cysteine proteases, strongly inhibit cathepsin B-like enzymes (22, 50, 58, 68). These results indicate that the TcoCBc1 specificity differs from that of other known cathepsin B-like proteases. In this context, even if little information about the real kinetic behavior of the proteases in their natural environment can be obtained using nonnatural synthetic substrates, results from kinetic and inhibition studies demonstrate that TcoCBc1 is an atypical cathepsin B-like protease in terms of substrate specificity and inhibitor sensitivity.
The serine subfamily represents the first example of cathepsin B-like enzymes with a serine replacing the catalytic cysteine. Intriguingly, this substitution is perfectly conserved in all seven genes of the TcoCBs subfamily, as are the cathepsin B-like signatures, i.e., the occluding loop motif and the conserved Gly-Cys-Xaa-Gly-Gly motif. Furthermore, pull-down experiments showed that at least one of these atypical cathepsin B-like proteins is expressed in bloodstream forms of T. congolense. All of this evidence strongly suggests that these proteins play a role in the life cycle of this extracellular parasite. Plasmodium serine type serine repeat antigens (SERA) represent another unique and unusual example of a papain-like protease in which the canonical catalytic cysteine is replaced by a serine (34). At least one member of this protease family (SERA5) retains catalytic function and plays an important role in the blood stage function of the parasite (51). In T. congolense, TcoCB are expressed as maturated enzymes as shown by Western blot analysis. Whether TcoCB are self matured, like TcoCBc1 and TcoCBc6, or whether they are matured by the action of other parasitic proteases (a cathepsin B from Schistosoma mansoni is trans activated by an endogenous asparaginyl endopeptidase [63], and cathepsin X, which displays strict carboxypeptidase activity, needs trans maturation to become active [65]) or by the action of host proteases remains unclear. The pepsin-matured TcoCBs12 protein was not active in any of the tested conditions, suggesting that the enzyme has narrow specificity that cannot be detected with the substrates tested or that the recombinant protease is misfolded, which also would avoid self maturation. Finally, another alternative is that this atypical family of proteins plays another role in T. congolense, such as diverting the host immune system during infection (like members of the complex sialidase family in T. cruzi) (19, 27, 67). However, the striking conservation within the pocket suggests that the protein preserves ligand-binding properties and that its biological role implies a peptidic partner.
To assess the function of TcoCB, and in the absence of RNA interference technology in T. congolense bloodstream forms, the effect of a cathepsin B-specific inhibitor was tested on the viability of bloodstream forms. As the cathepsin B inhibitor CA074 showed a marked effect on recombinant TcoCBc1 activity in vitro, its cell-permeable analogue, CA074Me, was used in in vitro cultures. Results showed a deleterious effect of the inhibitor at concentrations greater than 10 μM after 48 h of treatment. After only 3 h of treatment with 1 or 10 μM inhibitor, parasites exhibited a marked impairment in external protein degradation following endocytosis. This was revealed by the accumulation of undigested BSA in the lysosomal compartment, showing that these proteins are involved in lysosomal protein degradation, as was shown for T. brucei by RNA interference studies (45).
We demonstrated that TcoCBc1, despite the relative conservation of cathepsin B between the parasite and its hosts, is a potent antigen that elicits prominent antibody responses in T. congolense-infected cattle. Four individuals of the trypanosusceptible Boran breed consistently developed high levels of specific IgG during infection. This is in contrast to congopain, which appears to elicit prominent IgG responses only in trypanotolerant cattle (5). Immune responses to TcoCBs12 were more heterogeneous, with only two out of the four individuals developing high levels of specific IgG. Furthermore, antibody levels fluctuated during the course of infection, a phenomenon that occurs frequently in trypanosomiasis and may result from the coexistence in plasma of antigens, corresponding antibodies, and immune complexes.
Preliminary results indicate that antibodies against TcoCBc1 and TcoCBs12 are specific to T. congolense. Indeed, sera from cattle infected with other trypanosomatids, such as T. brucei or T. vivax, or with other common African pathogens, such as Babesia, Anaplasma, or Theileria, did not react with TcoCB (data not shown). This suggests that antigens from the TcoCB repertoire are suitable for the development of species-specific diagnostic tests.
Finally, we have provided evidence for a diverse repertoire of cathepsin B-like enzymes implicated in lysosomal protein degradation in T. congolense, some members of which have very special characteristics compared to those of mammalian cathepsin B. Not only is there an entire group that presents a unique serine-containing catalytic triad but also the substrate specificity and susceptibility to inhibitors of those that possess the normal cysteine-containing triad seem to be unique among cathepsin B proteins. Their unique specificity, together with the evidence that they are essential for the survival of T. congolense bloodstream forms in vitro, suggests that this family is a very interesting therapeutic target.
Supplementary Material
Acknowledgments
This work was supported by the Commission of the European Communities' Sixth Framework Programme, priority INCO-DEV, project Trypadvac2, contract PL003716, the Centre National de la Recherche Scientifique, and the Université Victor Segalen Bordeaux 2.
We are grateful to F. Bringaud for help in phylogenetic analysis, to F. Vellieux, D. Robinson, and N. Boucher for their critical reading of the manuscript, to G. Lalmanach, I. Lascu, and M. Castroviejo for useful discussions, to S. Claverol for liquid chromatography-mass spectrometry/mass spectrometry analyses, and to R. Pellé for providing us with T. congolense material.
Footnotes
Published ahead of print on 15 February 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abdulla, M. H., K. C. Lim, M. Sajid, J. H. McKerrow, and C. R. Caffrey. 2007. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 4e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, P. C., I. L. Nantes, J. R. Chagas, C. C. Rizzi, A. Faljoni-Alario, E. Carmona, L. Juliano, H. B. Nader, and I. L. Tersariol. 2001. Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. J. Biol. Chem. 276944-951. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, I. M., J. Scharfstein, and A. P. Lima. 2004. A new cruzipain-mediated pathway of human cell invasion by Trypanosoma cruzi requires trypomastigote membranes. Infect. Immun. 725892-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Authié, E., A. Boulange, D. Muteti, G. Lalmanach, F. Gauthier, and A. J. Musoke. 2001. Immunisation of cattle with cysteine proteinases of Trypanosoma congolense: targetting the disease rather than the parasite. Int. J. Parasitol. 311429-1433. [DOI] [PubMed] [Google Scholar]
- 5.Authié, E., G. Duvallet, C. Robertson, and D. J. Williams. 1993. Antibody responses to a 33 kDa cysteine protease of Trypanosoma congolense: relationship to ‘trypanotolerance’ in cattle. Parasite Immunol. 15465-474. [DOI] [PubMed] [Google Scholar]
- 6.Authié, E., D. K. Muteti, Z. R. Mbawa, J. D. Lonsdale-Eccles, P. Webster, and C. W. Wells. 1992. Identification of a 33-kilodalton immunodominant antigen of Trypanosoma congolense as a cysteine protease. Mol. Biochem. Parasitol. 56103-116. [DOI] [PubMed] [Google Scholar]
- 7.Authié, E., D. K. Muteti, and D. J. Williams. 1993. Antibody responses to invariant antigens of Trypanosoma congolense in cattle of differing susceptibility to trypanosomiasis. Parasite Immunol. 15101-111. [DOI] [PubMed] [Google Scholar]
- 8.Authier, F., J. S. Mort, A. W. Bell, B. I. Posner, and J. J. Bergeron. 1995. Proteolysis of glucagon within hepatic endosomes by membrane-associated cathepsins B and D. J. Biol. Chem. 27015798-15807. [DOI] [PubMed] [Google Scholar]
- 9.Baltz, T., D. Baltz, C. Giroud, and J. Crockett. 1985. Cultivation in a semi-defined medium of animal infective forms of Trypanosoma brucei, T. equiperdum, T. evansi, T. rhodesiense and T. gambiense. EMBO J. 41273-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baricos, W. H., Y. Zhou, R. W. Mason, and A. J. Barrett. 1988. Human kidney cathepsins B and L. Characterization and potential role in degradation of glomerular basement membrane. Biochem. J. 252301-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett, A. J., and H. Kirschke. 1981. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80535-561. [DOI] [PubMed] [Google Scholar]
- 12.Barrett, M. P., R. J. Burchmore, A. Stich, J. O. Lazzari, A. C. Frasch, J. J. Cazzulo, and S. Krishna. 2003. The trypanosomiases. Lancet 3621469-1480. [DOI] [PubMed] [Google Scholar]
- 13.Bart, G., M. J. Frame, R. Carter, G. H. Coombs, and J. C. Mottram. 1997. Cathepsin B-like cysteine proteinase-deficient mutants of Leishmania mexicana. Mol. Biochem. Parasitol. 8853-61. [DOI] [PubMed] [Google Scholar]
- 14.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 15.Björk, I., E. Pol, E. Raub-Segall, M. Abrahamson, A. D. Rowan, and J. S. Mort. 1994. Differential changes in the association and dissociation rate constants for binding of cystatins to target proteinases occurring on N-terminal truncation of the inhibitors indicate that the interaction mechanism varies with different enzymes. Biochem. J. 299219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boucher, N., D. Dacheux, C. Giroud, and T. Baltz. 2007. An essential cell cycle-regulated nucleolar protein relocates to the mitotic spindle where it is involved in mitotic progression in Trypanosoma brucei. J. Biol. Chem. 28213780-13790. [DOI] [PubMed] [Google Scholar]
- 17.Boulangé, A., C. Serveau, M. Brillard, C. Minet, F. Gauthier, A. Diallo, G. Lalmanach, and E. Authie. 2001. Functional expression of the catalytic domains of two cysteine proteinases from Trypanosoma congolense. Int. J. Parasitol. 311435-1440. [DOI] [PubMed] [Google Scholar]
- 18.Clayton, C., and M. Shapira. 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 15693-101. [DOI] [PubMed] [Google Scholar]
- 19.Cremona, M. L., O. Campetella, D. O. Sanchez, and A. C. Frasch. 1999. Enzymically inactive members of the trans-sialidase family from Trypanosoma cruzi display beta-galactose binding activity. Glycobiology 9581-587. [DOI] [PubMed] [Google Scholar]
- 20.Dalton, J. P., P. J. Brindley, D. P. Knox, C. P. Brady, P. J. Hotez, S. Donnelly, S. M. O'Neill, G. Mulcahy, and A. Loukas. 2003. Helminth vaccines: from mining genomic information for vaccine targets to systems used for protein expression. Int. J. Parasitol. 33621-640. [DOI] [PubMed] [Google Scholar]
- 21.Dehrmann, F. M., T. H. Coetzer, R. N. Pike, and C. Dennison. 1995. Mature cathepsin L is substantially active in the ionic milieu of the extracellular medium. Arch. Biochem. Biophys. 32493-98. [DOI] [PubMed] [Google Scholar]
- 22.Demuth, H. U., A. Schierhorn, P. Bryan, R. Hofke, H. Kirschke, and D. Bromme. 1996. N-peptidyl, O-acyl hydroxamates: comparison of the selective inhibition of serine and cysteine proteinases. Biochim. Biophys. Acta 1295179-186. [DOI] [PubMed] [Google Scholar]
- 23.Donelson, J. E., K. L. Hill, and N. M. El-Sayed. 1998. Multiple mechanisms of immune evasion by African trypanosomes. Mol. Biochem. Parasitol. 9151-66. [DOI] [PubMed] [Google Scholar]
- 24.Dunn, A. D., H. E. Crutchfield, and J. T. Dunn. 1991. Thyroglobulin processing by thyroidal proteases. Major sites of cleavage by cathepsins B, D, and L. J. Biol. Chem. 26620198-20204. [PubMed] [Google Scholar]
- 25.Ellis, K. J., and J. F. Morrison. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87405-426. [DOI] [PubMed] [Google Scholar]
- 26.Fish, W. R., C. W. Muriuki, A. M. Muthiani, D. J. Grab, and J. D. Lonsdale-Eccles. 1989. Disulfide bond involvement in the maintenance of the cryptic nature of the cross-reacting determinant of metacyclic forms of Trypanosoma congolense. Biochemistry 285415-5421. [DOI] [PubMed] [Google Scholar]
- 27.Frasch, A. C. 2000. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol. Today 16282-286. [DOI] [PubMed] [Google Scholar]
- 28.Geigy, R., and M. Kauffmann. 1973. Sleeping sickness survey in the Serengeti area (Tanzania) 1971. I. Examination of large mammals for trypanosomes. Acta Trop. 3012-23. [PubMed] [Google Scholar]
- 29.Giordanengo, L., N. Guinazu, C. Stempin, R. Fretes, F. Cerban, and S. Gea. 2002. Cruzipain, a major Trypanosoma cruzi antigen, conditions the host immune response in favor of parasite. Eur. J. Immunol. 321003-1011. [DOI] [PubMed] [Google Scholar]
- 30.Gray, M. A., H. Hirumi, and P. R. Gardiner. 1987. The salivarian trypanosomes: insect forms, p. 118-152. In A. E. R. Taylor and J. R. Baker (ed.), In vitro methods for parasite cultivation. Academic Press, London, United Kingdom.
- 31.Hasnain, S., T. Hirama, C. P. Huber, P. Mason, and J. S. Mort. 1993. Characterization of cathepsin B specificity by site-directed mutagenesis. Importance of Glu245 in the S2-P2 specificity for arginine and its role in transition state stabilization. J. Biol. Chem. 268235-240. [PubMed] [Google Scholar]
- 32.Hesse, F., P. M. Selzer, K. Muhlstadt, and M. Duszenko. 1995. A novel cultivation technique for long-term maintenance of bloodstream form trypanosomes in vitro. Mol. Biochem. Parasitol. 70157-166. [DOI] [PubMed] [Google Scholar]
- 33.Hirumi, H., and K. Hirumi. 1991. In vitro cultivation of Trypanosoma congolense bloodstream forms in the absence of feeder cell layers. Parasitology 102225-236. [DOI] [PubMed] [Google Scholar]
- 34.Hodder, A. N., D. R. Drew, V. C. Epa, M. Delorenzi, R. Bourgon, S. K. Miller, R. L. Moritz, D. F. Frecklington, R. J. Simpson, T. P. Speed, R. N. Pike, and B. S. Crabb. 2003. Enzymic, phylogenetic, and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5. J. Biol. Chem. 27848169-48177. [DOI] [PubMed] [Google Scholar]
- 35.Illy, C., O. Quraishi, J. Wang, E. Purisima, T. Vernet, and J. S. Mort. 1997. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 2721197-1202. [DOI] [PubMed] [Google Scholar]
- 36.Judice, W. A., L. Puzer, S. S. Cotrin, A. K. Carmona, G. H. Coombs, L. Juliano, and M. A. Juliano. 2004. Carboxydipeptidase activities of recombinant cysteine peptidases. Cruzain of Trypanosoma cruzi and CPB of Leishmania mexicana. Eur. J. Biochem. 2711046-1053. [DOI] [PubMed] [Google Scholar]
- 37.Jutras, I., and T. L. Reudelhuber. 1999. Prorenin processing by cathepsin B in vitro and in transfected cells. FEBS Lett. 44348-52. [DOI] [PubMed] [Google Scholar]
- 38.Kesik, M., L. Jedlina-Panasiuk, M. Kozak-Cieszczyk, A. Plucienniczak, and H. Wedrychowicz. 2007. Enteral vaccination of rats against Fasciola hepatica using recombinant cysteine proteinase (cathepsin L1). Vaccine 253619-3628. [DOI] [PubMed] [Google Scholar]
- 39.Knox, D. P. 2007. Proteinase inhibitors and helminth parasite infection. Parasite Immunol. 2957-71. [DOI] [PubMed] [Google Scholar]
- 40.Kohl, L., T. Sherwin, and K. Gull. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46105-109. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 42.Lalmanach, G., A. Boulange, C. Serveau, F. Lecaille, J. Scharfstein, F. Gauthier, and E. Authie. 2002. Congopain from Trypanosoma congolense: drug target and vaccine candidate. Biol. Chem. 383739-749. [DOI] [PubMed] [Google Scholar]
- 43.Lanham, S. M., and D. G. Godfrey. 1970. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 28521-534. [DOI] [PubMed] [Google Scholar]
- 44.Lecaille, F., J. Kaleta, and D. Bromme. 2002. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chem. Rev. 1024459-4488. [DOI] [PubMed] [Google Scholar]
- 45.Mackey, Z. B., T. C. O'Brien, D. C. Greenbaum, R. B. Blank, and J. H. McKerrow. 2004. A cathepsin B-like protease is required for host protein degradation in Trypanosoma brucei. J. Biol. Chem. 27948426-48433. [DOI] [PubMed] [Google Scholar]
- 46.Mahmoudzadeh-Niknam, H., and J. H. McKerrow. 2004. Leishmania tropica: cysteine proteases are essential for growth and pathogenicity. Exp. Parasitol. 106158-163. [DOI] [PubMed] [Google Scholar]
- 47.Matthews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews, K. R., J. R. Ellis, and A. Paterou. 2004. Molecular regulation of the life cycle of African trypanosomes. Trends Parasitol. 2040-47. [DOI] [PubMed] [Google Scholar]
- 49.Mbawa, Z. R., P. Webster, and J. D. Lonsdale-Eccles. 1991. Immunolocalization of a cysteine protease within the lysosomal system of Trypanosoma congolense. Eur. J. Cell Biol. 56243-250. [PubMed] [Google Scholar]
- 50.McConnell, R. M., J. L. York, D. Frizzell, and C. Ezell. 1993. Inhibition studies of some serine and thiol proteinases by new leupeptin analogues. J. Med. Chem. 361084-1089. [DOI] [PubMed] [Google Scholar]
- 51.McCoubrie, J. E., S. K. Miller, T. Sargeant, R. T. Good, A. N. Hodder, T. P. Speed, T. F. de Koning-Ward, and B. S. Crabb. 2007. Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect. Immun. 755565-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKerrow, J. H., C. R. Caffrey, B. Kelly, P. Loke, and M. Sajid. 2006. Proteases in parasitic diseases. Annu. Rev. Pathol. Mech. Dis. 1497-536. [DOI] [PubMed] [Google Scholar]
- 53.Mort, J. S., and D. J. Buttle. 1997. Cathepsin B. Int. J. Biochem. Cell Biol. 29715-720. [DOI] [PubMed] [Google Scholar]
- 54.Mottram, J. C., G. H. Coombs, and J. Alexander. 2004. Cysteine peptidases as virulence factors of Leishmania. Curr. Opin. Microbiol. 7375-381. [DOI] [PubMed] [Google Scholar]
- 55.Mottram, J. C., A. E. Souza, J. E. Hutchison, R. Carter, M. J. Frame, and G. H. Coombs. 1996. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc. Natl. Acad. Sci. USA 936008-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray, M., W. I. Morrison, and D. D. Whitelaw. 1982. Host susceptibility to African trypanosomiasis: trypanotolerance. Adv. Parasitol. 211-68. [DOI] [PubMed] [Google Scholar]
- 57.Musil, D., D. Zucic, D. Turk, R. A. Engh, I. Mayr, R. Huber, T. Popovic, V. Turk, T. Towatari, N. Katunuma, et al. 1991. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 102321-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nkemgu, N. J., R. Grande, E. Hansell, J. H. McKerrow, C. R. Caffrey, and D. Steverding. 2003. Improved trypanocidal activities of cathepsin L inhibitors. Int. J. Antimicrob. Agents 22155-159. [DOI] [PubMed] [Google Scholar]
- 59.Pyndiah, S., J. P. Lasserre, A. Menard, S. Claverol, V. Prouzet-Mauleon, F. Megraud, F. Zerbib, and M. Bonneu. 2007. Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from Helicobacter pylori. Mol. Cell. Proteomics 6193-206. [DOI] [PubMed] [Google Scholar]
- 60.Redmond, D. L., and D. P. Knox. 2006. Further protection studies using recombinant forms of Haemonchus contortus cysteine proteinases. Parasite Immunol. 28213-219. [DOI] [PubMed] [Google Scholar]
- 61.Ren, W. P., R. Fridman, J. R. Zabrecky, L. D. Morris, N. A. Day, and B. F. Sloane. 1996. Expression of functional recombinant human procathepsin B in mammalian cells. Biochem. J. 319793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenthal, P. J. 2004. Cysteine proteases of malaria parasites. Int. J. Parasitol. 341489-1499. [DOI] [PubMed] [Google Scholar]
- 63.Sajid, M., J. H. McKerrow, E. Hansell, M. A. Mathieu, K. D. Lucas, I. Hsieh, D. Greenbaum, M. Bogyo, J. P. Salter, K. C. Lim, C. Franklin, J. H. Kim, and C. R. Caffrey. 2003. Functional expression and characterization of Schistosoma mansoni cathepsin B and its trans-activation by an endogenous asparaginyl endopeptidase. Mol. Biochem. Parasitol. 13165-75. [DOI] [PubMed] [Google Scholar]
- 64.Scharfstein, J., V. Schmitz, V. Morandi, M. M. Capella, A. P. Lima, A. Morrot, L. Juliano, and W. Muller-Esterl. 2000. Host cell invasion by Trypanosoma cruzi is potentiated by activation of bradykinin B(2) receptors. J. Exp. Med. 1921289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivaraman, J., D. K. Nagler, R. Zhang, R. Menard, and M. Cygler. 2000. Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine. J. Mol. Biol. 295939-951. [DOI] [PubMed] [Google Scholar]
- 66.Stempin, C., L. Giordanengo, S. Gea, and F. Cerban. 2002. Alternative activation and increase of Trypanosoma cruzi survival in murine macrophages stimulated by cruzipain, a parasite antigen. J. Leukoc. Biol. 72727-734. [PubMed] [Google Scholar]
- 67.Tarleton, R. L. 2007. Immune system recognition of Trypanosoma cruzi. Curr. Opin. Immunol. 19430-434. [DOI] [PubMed] [Google Scholar]
- 68.Towatari, T., T. Nikawa, M. Murata, C. Yokoo, M. Tamai, K. Hanada, and N. Katunuma. 1991. Novel epoxysuccinyl peptides. A selective inhibitor of cathepsin B, in vivo. FEBS Lett. 280311-315. [DOI] [PubMed] [Google Scholar]
- 69.Troeberg, L., R. N. Pike, R. E. Morty, R. K. Berry, T. H. Coetzer, and J. D. Lonsdale-Eccles. 1996. Proteases from Trypanosoma brucei brucei. Purification, characterisation and interactions with host regulatory molecules. Eur. J. Biochem. 238728-736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.