Abstract
Human astroviruses have a positive-strand RNA genome, which contains three open reading frames (ORF1a, ORF1b, and ORF2). The genomic RNA is translated into two nonstructural polyproteins, nsp1a and nsp1ab, that contain sequences derived from ORF1a and from both ORF1a and ORF1b, respectively. Proteins nsp1a and nsp1ab are thought to be proteolytically processed to yield the viral proteins implicated in the replication of the virus genome; however, the intermediate and final products of this processing have been poorly characterized. To identify the cleavage products of the nonstructural polyproteins of a human astrovirus serotype 8 strain, antisera to selected recombinant proteins were produced and were used to analyze the viral proteins synthesized in astrovirus-infected Caco-2 cells and in cells transfected with recombinant plasmids expressing the ORF1a and ORF1b polyproteins. Pulse-chase experiments identified proteins of approximately 145, 88, 85, and 75 kDa as cleavage intermediates during the polyprotein processing. In addition, these experiments and kinetic analysis of the synthesis of the viral proteins identified polypeptides of 57, 20, and 19 kDa, as well as two products of around 27 kDa, as final cleavage products, with the 57-kDa polypeptide most probably being the virus RNA polymerase and the two ∼27-kDa products being the viral protease. Based on the differential reactivities of the astrovirus proteins with the various antisera used, the individual polypeptides detected were mapped to the virus ORF1a and ORF1b regions.
Human astroviruses (HAstV) have been found to be the second leading cause of acute viral gastroenteritis in young children worldwide, generally responsible for around 4 to 8% of cases (4, 8, 9, 20), although incidences as high as 26% in isolated populations have been reported (15). The HAstV particles are formed by a nonenveloped capsid protein and a polyadenylated positive-strand RNA genome of approximately 7 kb (10, 25). The RNA genome has three open reading frames (ORF1a, ORF1b, and ORF2), each encoding at least one polyprotein.
The virus structural proteins, encoded in ORF2, are synthesized as a polyprotein precursor of approximately 780 amino acid residues (6, 17), which is processed into at least three polypeptides that form the virus capsid. Astroviruses of different serotypes have been reported to contain structural proteins of approximately 34, 29, and 26 kDa (1, 2, 22). A recent characterization of an HAstV serotype 8 (HAstV-8) strain revealed that the capsid initially assembles from a single protein of 70 kDa (VP70), which is later cleaved by trypsin into proteins of approximately 34, 27, and 25 kDa, with the concomitant enhancement of viral infectivity (17). VP70 is synthesized as a precursor of approximately 90 kDa (the primary translation product of ORF2) (17), which is cleaved at its carboxy terminus to yield the mature VP70 protein.
Two nonstructural polyproteins of astrovirus, encoded in the 5′-most ORFs, ORF1a and ORF1b, are believed to be responsible for the replication of the viral RNA (10). Amino acid sequence analysis of the ORF1a-encoded polyprotein predicts four hydrophobic transmembrane regions and viral serine protease and nuclear localization signal motifs, whereas ORF1b includes sequences characteristic of an RNA-dependent RNA polymerase (10, 25). Polyprotein nsp1a, of 103 kDa, includes sequences encoded only in ORF1a, while nsp1ab, of 160 kDa, includes sequences derived from both ORF1a and ORF1b. Protein nsp1ab is produced by a translational frameshift mechanism (12-14, 16), using a signal similar to that described previously for some retroviruses, which is localized close to the ORF1a-ORF1b junction region (10). By analogy with other positive-strand RNA viruses, it is believed that the astrovirus nonstructural polyproteins are cleaved to smaller polypeptides mainly by the viral protease.
The processing of the astrovirus nonstructural polyproteins has not been completely characterized, and some of the reported data are conflicting (5, 7, 11, 24). Willcocks et al. (24) detected products of 75, 34, 20, 6.5, and 5.5 kDa in HAstV-1-infected cells by using antibodies to the carboxy-terminal end of nsp1a (amino acid residues 643 to 940), suggesting that the primary translation product of ORF1a is processed from its amino terminus, and also detected a protein of 59 kDa by using antibodies to nsp1b. Geigenmuller et al. (5) reported proteins of approximately 20 and 27 kDa as final products of nsp1a by transient expression of HAstV-1 cDNA clones and demonstrated that the cleavages at around amino acid residues 410 and 655 of nsp1a, which generate the 27-kDa protein, were dependent on the viral serine protease. Using an in vitro translation assay for HAstV-2 nsp1a and nsp1ab proteins, Gibson et al. (7) could not identify any processed products and suggested that the viral serine protease could require a cellular factor, not present in the reticulocyte lysate, for its activity. In contrast, Kiang and Matsui (11), with a similar approach, identified one viral serine protease-dependent cleavage immediately downstream of the protease motif, which yielded proteins of 64 and 38 kDa, derived from the amino and carboxy termini of nsp1a, respectively. This cleavage site (Gln567/Thr568) identified in nsp1a (11) is different from the two described by Geigenmuller (5) responsible for generating the 27-kDa product.
To learn about the processing pathway and the final products derived from the nonstructural polyproteins of HAstV-8, virus-infected cells were analyzed with antibodies to different regions of nsp1a and nsp1b. We identified final protein products of 57, 20, and 19 kDa as well as two proteins of around 27 kDa, which were produced from the nsp1a and nsp1ab polyproteins through polypeptide intermediates of 145, 88, 85, and 75 kDa. Our results indicate that the proteolytic processing of the nonstructural polyproteins of astrovirus follows a complex pathway.
MATERIALS AND METHODS
Virus and cells.
Caco-2 cells from the American Type Culture Collection and BHK-21 cells from C. M. Rice (Washington University, St. Louis, Mo.) were used for this work. Cells were cultured in a CO2 atmosphere at 37°C with minimum essential medium (Eagle's salts) (MEM), supplemented with glutamine, and 10% (BHK-21) or 15% (Caco-2) fetal bovine serum (GIBCO/BRL). The HAstV strain Yuc8, isolated from a Mexican child, was adapted to grow in Caco-2 cells (18). Viral stocks from passages 7 to 15 were used in this work, and they were propagated as described previously (17), except that the virus was treated with 200 μg of trypsin/ml for 1 h at 37°C before infection. The recombinant modified vaccinia virus Ankara expressing the T7 RNA polymerase (MVA/T7) (26) was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) and was propagated in BHK-21 cells, basically as described elsewhere (3).
Astrovirus recombinant proteins and antibodies.
Recombinant plasmid vectors expressing selected regions of the astrovirus nonstructural proteins (Table 1) were constructed in pET28 (Novagen) or pGEX4T (Pharmacia) vectors by standard molecular biology techniques. Protein 1a-1 (including amino acid residues 41 to 257 of ORF1a) was expressed in Escherichia coli BL21(DE3) with the vector pET28; proteins 1a-3 (amino acid residues 401 to 638 of ORF1a), 1a-4 (amino acid residues 638 to 857 of ORF1a), 1b-1 (amino acid residues 33 to 171 of ORF1b), and 1b-2 (amino acid residues 201 to 362 of ORF1b) were fused to the carboxy-terminal end of glutathione S-transferase and expressed in E. coli strain JM101. Proteins were purified and used to immunize rabbits as previously described (17). A peptide with the sequence IFLCFMEDSNYVSQIRGLI (named IFLC), corresponding to amino acid residues 328 to 346 of the Yuc8 ORF1a, was synthesized by Research Genetics Co. and coupled to keyhole limpet hemocyanin (KLH; Imject maleimide-activated KLH; Pierce), according to the conditions recommended by the manufacturer. The KLH-conjugated peptide was inoculated into BALB/c mice, according to the same protocol as that used for the recombinant proteins. Sera were collected 2 weeks after the fourth inoculation, after confirmation that antibodies to the astrovirus antigens had been generated.
TABLE 1.
Recombinant Yuc8 proteins and synthetic peptide used to generate sera to astrovirus nonstructural polypeptides
| ORF | Recombinant protein or synthetic peptidea | Amino acid residues includedb |
|---|---|---|
| ORF1a | 1a-1 | 41-257 |
| IFLC | 328-346 | |
| 1a-3 | 401-638 | |
| 1a-4 | 638-857 | |
| ORF1b | 1b-1 | 33-171 |
| 1b-2 | 201-362 |
Recombinant proteins were synthesized in bacteria as fusion polypeptides with either histidine tails or glutathione S-transferase as described in Materials and Methods.
Based on the sequence of the corresponding ORF of Yuc8.
Astrovirus cDNA constructs.
Total RNA from Yuc8-infected cells was extracted with Trizol reagent (GIBCO/BRL) 12 h postinfection (hpi) and used for reverse transcription-PCR to amplify ORF1a and ORF1b. The cDNA fragments obtained were sequenced and compared with the previously reported sequence (18) to confirm their identity. Three nucleotide changes were observed in ORF1a, one conservative and two which resulted in amino acid changes (Leu to His at amino acid positions 3 and 361). Yuc8 sequences derived from ORF1a, ORF1b, and ORF1a-ORF1b were cloned in the pTM1 vector (kindly donated by B. Moss), in which transcription of the astrovirus genes is regulated by the T7 RNA polymerase promoter and their translation is dependent on the internal ribosome entry site element of encephalomyocarditis virus. The pTM1a construct contains the complete ORF1a, pTM1ab contains the complete nonstructural coding region (ORF1a and ORF1b), and pTM1b includes ORF1b starting at amino acid residue 33.
Pulse-chase experiments.
Caco-2 cells were infected with Yuc8 (multiplicity of infection [MOI] of 5 to 10), previously treated with 200 μg of trypsin/ml (trypsin, 1:250; GIBCO), for 1 h at 37°C to activate the virus infectivity (17). After this treatment, to prevent the cells from detaching from the flasks during the virus adsorption period, the trypsin activity was inhibited by adding 400 μg of soybean trypsin inhibitor (Sigma Co.)/ml, and the virus was added to the cells for 1 h at 37°C. Under these conditions, the viral infectivity was not affected by the presence of the trypsin inhibitor. The cells were washed twice with MEM and kept in MEM at 37°C. Twelve hours postinfection, the cells were pulse-labeled with 50 μCi of [35S]-Express protein labeling mix (NEN Life Science; NEG772)/ml in methionine-free MEM for 1 h at 37°C. After this time, the label was replaced with regular MEM, and the cells were harvested at different times, either in TNS buffer (Tris, 50 mM, pH 7.5; NaCl, 150 mM; sodium dodecyl sulfate [SDS], 0.5%; phenylmethylsulfonyl fluoride, 20 μg/ml; leupeptin, 100 μg/ml) or in TNT buffer (Tris, 50 mM, pH 7.5; NaCl, 150 mM; Triton X-100, 1%; phenylmethylsulfonyl fluoride, 20 μg/ml; leupeptin, 100 μg/ml). These samples were used for protein analysis by immunoprecipitation analysis.
Transient expression from HAstV-8 cDNA clones.
BHK-21 cells grown in 24-well plates were infected with MVA/T7 (MOI of >10). After 1 h at 37°C, the cells were washed, and a mixture of 800 μg of plasmid DNA and 2.5 μl of Lipofectamine 2000 (GIBCO/BRL) in MEM, previously incubated for 30 min at room temperature, was added to each well. This mixture was maintained on the cells for 4 h and then washed twice with MEM, before 50 μCi of [35S]-Express protein labeling mix/ml in methionine-free MEM was added. After 12 h of incubation at 37°C, the cells were harvested in TNS buffer. These samples were used for protein analysis by immunoprecipitation.
Immunoprecipitation.
Twenty microliters of 35S-labeled cell lysates was mixed with 5 μl of a mouse or rabbit antiastrovirus serum in RIPA buffer (Tris, 50 mM, pH 7.5; NaCl, 150 mM; sodium deoxycholate, 1%; Triton X-100, 1%; SDS, 0.1%) and incubated at 4°C. The samples were centrifuged at 13,000 × g, the pellets were discarded, and the supernatants were mixed with 25 μl of 50% protein A-Sepharose (Sigma, catalog no. P3391) in RIPA buffer. After 1 h at room temperature, the resin was washed three times with RIPA buffer and the labeled proteins were analyzed by SDS-polyacrylamide gel electrophoresis.
RESULTS
Few studies have examined the synthesis and processing of the HAstV nonstructural proteins, and many details of these events remain to be elucidated (5, 7, 11, 24). In this study we contribute to the characterization of the processing of the nonstructural polyproteins of astroviruses, using as a model an HAstV-8 strain.
The infection of Caco-2 cells with HAstV at a high MOI does not shut off the cellular protein synthesis, and under these conditions, the capsid precursor polyprotein of 90 kDa (17) is the only viral protein clearly detected in the cell lysate (data not shown and reference 19). Thus, to identify the processing products of the nonstructural polyproteins and to study the pathway of this processing, rabbit antisera to specific regions of the ORF1a- and ORF1b-encoded polyproteins were generated. The proteins were expressed in bacteria as fusion polypeptides with either a histidine tail (His) or glutathione S-transferase (Table 1). To deal with the fact that the region including amino acid residues 250 to 400 of ORF1a is highly hydrophobic, a somewhat hydrophilic synthetic peptide (IFLC) from this region (Table 1) was synthesized and used to generate antibodies in mice.
Antisera to recombinant astrovirus polypeptides recognize specific proteins in Yuc8-infected cells.
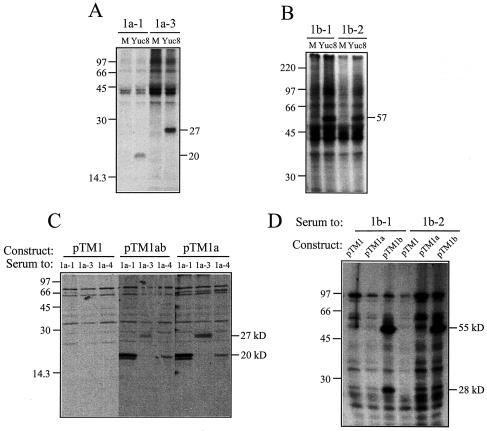
To identify the viral proteins produced during a Yuc8 infection, Caco-2 cells were infected and labeled from 12 hpi until harvesting, 24 h later. The labeled proteins were analyzed by immunoprecipitation with antisera to the Yuc8 recombinant proteins. Products ranging from 19 to 57 kDa were identified in cell lysates from infected but not from noninfected cells (Fig. 1A and B). Antibodies to recombinant proteins 1a-1 and 1a-3 (Table 1 and Fig. 4) were consistently detected in Yuc8-infected cells as broad bands at ∼20 (Fig. 1A) and ∼27 (Fig. 1C) kDa, respectively, which in some gels were each resolved into two polypeptides of similar size (for instance, Fig. 1C, subpanels pTM1a and pTM1ab, and Fig. 2A and C). As expected, based on the predicted size of the putative viral RNA polymerase encoded by ORF1b, antibodies to recombinant proteins 1b-1 and 1b-2 both recognized a protein of approximately 57 kDa in infected cells (Fig. 1B). Antisera to peptide IFLC and to protein 1a-4 did not recognize any viral polypeptides in these assays (data not shown). To confirm these results, clones containing the complete sequences from ORF1a (construct pTM1a) and ORF1a-ORF1b (construct pTM1ab) and a fragment of ORF1b (starting at amino acid residue 33; construct pTM1b) were cloned and transiently expressed in BHK-21 cells by using the MVA/T7-RNA polymerase system (26). In cells transfected with either plasmid pTM1a or plasmid pTM1ab, antibodies to 1a-1 detected two proteins of ∼20 kDa and antibodies to 1a-3 identified a broad band at ∼27 kDa (Fig. 1C). In these experiments, the antiserum to 1a-4 recognized one protein of 20 kDa, which was not observed in lysates from Yuc8-infected cells. This 20-kDa protein would appear to be different from the ∼20-kDa proteins recognized by antibodies to 1a-1, since antisera to 1a-1 and 1a-4 recognize different regions of nsp1a, and the immunoprecipitation experiments were carried out under denaturing conditions. Antibodies to peptide IFLC did not recognize a protein in these experiments (data not shown).
FIG. 1.
Antisera to recombinant nonstructural Yuc8 proteins recognize specific astrovirus products in infected and transfected cells. (A and B) A monolayer of Caco-2 cells infected with Yuc8 was labeled with [35S]-Express labeling protein mix 12 hpi and harvested 24 h later in TNS buffer. (C and D) BHK-21 cells were infected with the recombinant MVA/T7 virus and transfected with the indicated astrovirus constructs, as described in Materials and Methods. Astrovirus proteins were immunoprecipitated with the indicated sera and separated in SDS-15% (A and C) or 11% (B and D) polyacrylamide gels. Mock-infected cells (M) or cells transfected with the vector (pTM1) were included as controls. In panel D, plasmid pTM1a was included as an additional control for the immunoprecipitation with antibodies to nsp1b. The migration of the molecular mass markers (in kilodaltons) (14C-methylated proteins; Amersham Pharmacia; CFA626) and of the viral proteins is indicated.
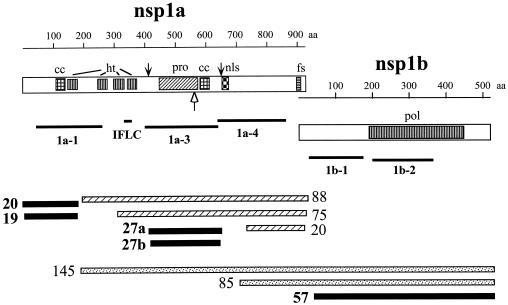
FIG. 4.
Diagram of the products encoded by ORF1a and ORF1b of HAstV. Long open boxes represent the nsp1a and nsp1b proteins, and the small boxes inside represent the motifs for hydrophobic transmembrane (ht), protease (pro), nuclear localization signal (nls), frameshift (fs), RNA polymerase (pol), and coiled-coil structures (cc) predicted. The diagram is at scale (indicated above the nsp1a and nsp1b proteins), and the relative positions of the astrovirus Yuc8 recombinant proteins 1a-1, 1a-3, 1a-4, 1b-1, and 1b-2 and of the synthetic peptide IFLC used to generate antibodies are shown as black lines. The cleavage sites on nsp1a reported by Geigenmuller et al. (5) and by Kiang and Matsui (11) are indicated by black and open arrows, respectively. The protein products identified in this work (represented by boxes in the lower part of the figure) and their putative localization in the nonstructural polyprotein context are shown. The intermediate protein products are shown as hatched (those derived from nsp1a) or dotted (those derived from nsp1ab), while the identified final products of the processing of Yuc8 nonstructural polyproteins are shown as black boxes. The two proteins of about 27 kDa detected are labeled as 27a and 27b for clarity.
FIG. 2.
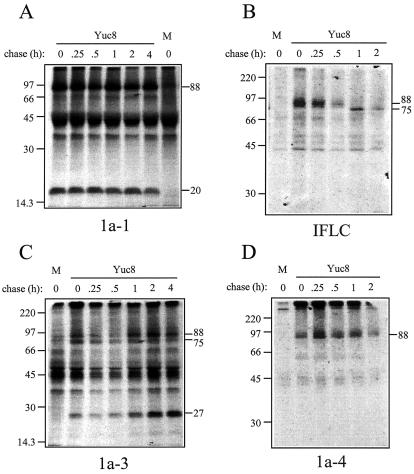
Protein intermediates generated during the processing of nsp1a. A monolayer of Yuc8-infected Caco-2 cells or mock-infected (M) cells was pulse-labeled with [35S]-Express labeling protein mix for 1 h at 12 hpi. The incorporated label was then chased for the indicated times, and the samples were immunoprecipitated with antibodies to 1a-1, IFLC, 1a-3, or 1a-4, as described in Materials and Methods. Proteins were separated in SDS-15% (A and C) or 11% (B and D) polyacrylamide gels. The migration of the molecular mass markers (in kilodaltons) (14C-methylated proteins; Amersham Pharmacia; CFA626) and of the viral proteins is indicated.
When the plasmid construct pTM1b was transiently expressed, antibodies to 1b-1 recognized two proteins, of about 55 and 28 kDa, while the antiserum to 1b-2 detected only the 55-kDa product (Fig. 1D). The smaller size of the product detected in this assay with the antisera to the 1b region than of the product immunoprecipitated from infected cells is probably due to the fact that plasmid pTM1b expresses the viral RNA polymerase starting from amino acid residue 33, while the cleavage site that yields the polymerase from the nsp1ab polyprotein is most probably located upstream of this residue. The 28-kDa protein recognized by antibodies to 1b-1 (Fig. 1D) was not observed in astrovirus-infected Caco-2 cells and may be the result of a spurious cleavage (around amino acid residue 260 of nsp1b) in BHK-21 cells. No protein was detected by the antisera to 1b-1 and 1b-2 when plasmid pTM1ab was transfected, probably because the product of the translational frameshift is expressed at a low level (data not shown). Considering the times at which infected and transfected cells were analyzed, the identified nonstructural astrovirus proteins of 57, 27, 20, and 19 kDa seem to represent the final processing products.
Proteolytic processing of the nsp1a polyprotein.
To determine the protein intermediates generated during the processing of the nonstructural proteins, pulse-chase experiments with Yuc8-infected cells were carried out. The appropriate time for labeling the infected cells was determined based on the detection of the 27-kDa band (recognized by antibodies to 1a-3), since the presence of this product would indicate that the synthesis and processing of the polyproteins had occurred. The 27-kDa band was clearly observed starting at 12 hpi (data not shown).
Based on these results, the cells were infected with Yuc8, radioactively pulsed at 12 hpi for 1 h, and chased for different times. When antibodies to 1a-1 were used to immunoprecipitate the labeled viral proteins, two closely migrating proteins at ∼20 kDa were already detected by the end of the pulse, which remained stable up to 4 h later (Fig. 2A), suggesting that either these products were the result of a cotranslational cleavage or they were generated very rapidly after the synthesis of the full-length polyprotein was completed. These proteins probably correspond to the 20-kDa products identified by the same serum in Fig. 1C. Antibodies to 1a-1 also recognized a protein of ∼88 kDa at different chase times, which probably represents the carboxy-terminal product of nsp1a after the ∼20-kDa polypeptides were released (5). This assumption is supported by the fact that antibodies to 1a-4 recognized a protein of similar size (see below).
Antibodies to IFLC recognized a protein of ∼88 kDa which was processed to generate a 75-kDa protein, which also seemed to decrease its abundance with time (Fig. 2B). No smaller protein was detected with this serum at later times, probably because it does not recognize the final product(s) of this region.
Antibodies to 1a-3, which should recognize products containing the serine protease motif, detected proteins of approximately 88, 75, and 27 kDa (Fig. 2C); this serum also recognized one protein of 66 kDa at early chase times in an inconsistent manner (data not shown). Two proteins of about 27 kDa, which migrate very close together in Fig. 2C and accumulate at late chase times, seem to represent final products of the polyprotein processing. No precursor-product relationship was clearly observed among the 88-, 75-, and 27-kDa proteins in Fig. 2C; however, these proteins seem to correspond to the products detected by the anti-IFLC and the anti-1a-4 antibodies.
Only one protein of ∼88 kDa was detected by antibodies to 1a-4 in cells infected with Yuc8 (Fig. 2D), which probably corresponded to the ∼88-kDa protein detected by the antisera to 1a-1, 1a-3, and perhaps IFLC. The 20-kDa protein detected by antibodies to 1a-4 in the transient expression experiments (Fig. 1C) was not observed in astrovirus-infected cells.
Proteolytic processing of the nsp1ab polyprotein.
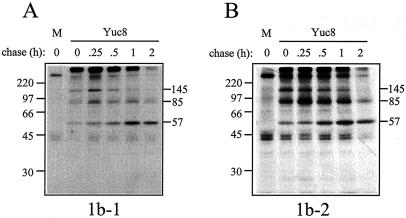
To detect the intermediate proteins that result from the proteolytic cleavage of the nonstructural polyprotein nsp1ab, antisera raised to recombinant proteins 1b-1 and 1b-2 were used. Both sera recognized proteins of about 145, 85, and 57 kDa in astrovirus-infected cells. The first two proteins were more evident at early chase times and were reduced after a chase of 1 or 2 h, while the 57-kDa product accumulated at later times (Fig. 3). This finding suggests that the 145- and 85-kDa proteins represent protein intermediates and that the 57-kDa protein represents the mature viral RNA polymerase.
FIG. 3.
Protein intermediates generated during the processing of nsp1ab. Proteins labeled and chased as described for Fig. 2 were immunoprecipitated with antibodies to either 1b-1 or 1b-2. The proteins were then separated in an SDS-11% polyacrylamide gel. The migration of the molecular mass markers (in kilodaltons) (14C-methylated proteins; Amersham Pharmacia; CFA626) and of the viral proteins is indicated.
DISCUSSION
Although studies of the processing of the nonstructural polyproteins of HAstV have been carried out in several laboratories, this event is still poorly understood (5, 7, 11, 24). The identification of the final and intermediate proteins which result from the proteolytic cleavage of the full-length nonstructural protein precursors has been hampered by the lack of shutoff of the cellular protein synthesis by the virus (19) and because antisera to specific fragments of the nonstructural region have not been available. In this work, the intermediate and final products of processing of the nonstructural polyproteins encoded by ORF1a and ORF1b of Yuc8, an HAstV-8 strain, were analyzed with sera raised to recombinant proteins which spanned most of the nonstructural protein coding region (Fig. 4).
Processing of nsp1a.
The full-length polyproteins nsp1a and nsp1ab, with calculated molecular masses of 103 and 160 kDa, respectively, were not found in cell lysates from Yuc8-infected or plasmid-transfected cells, suggesting that cotranslational processing occurs during their synthesis, as previously shown for HAstV-1 in transient expression experiments (5). Two proteins of around 19 to 20 kDa seem to represent the N-terminal products of the cotranslational processing of polyproteins nsp1a and nsp1b during astrovirus infection. It is unlikely that the cleavages on nsp1a and nsp1ab to generate these amino-terminal products are the result of the action of the viral protease, since they were not observed in a reticulocyte in vitro translation system (7, 11), and the generation of an amino-terminal 20-kDa product has been shown to be independent of the viral serine protease activity by transient expression of nsp1a cDNA clones (5). Of interest, the amino acid sequence GGYA, located at nsp1a amino acid residues 171 to 174, is similar to the junction Erns-E1 site of bovine viral diarrhea virus (GAYA/) cleaved by a cellular protease (21), and cleavage at this nsp1a site would generate a product of approximately 20 kDa (5). The 19- to 20-kDa amino-terminal products of nsp1a may be either the result of a differential cleavage at their carboxy termini or the consequence of the initiation of translation at two different sites, since ORF1a has an additional AUG (inserted into the context ACAATGG), 21 codons downstream of the first AUG (GenBank sequence accession no. AF260508), which is conserved among all other HAstV sequenced so far (GenBank sequence accession numbers Z25771, L13745, and AF141381 for HAstV-1, HAstV-2, and HAstV-3, respectively).
Antibodies to 1a-3 recognized two proteins of ∼27 kDa as final products of processing. This finding agrees with a recent report by Geigenmuller et al., who detected two proteins of similar size in HAstV-1-infected cells, using antibodies directed to a protein region equivalent to that recognized by the anti-1a-3 serum used in this work (5). It remains to be determined whether these two proteins are the result of alternative cleavage sites on nsp1a. As mentioned above, proteins of ∼88 and ∼75 kDa were also detected by antibodies to 1a-3; thus, these two proteins seem to represent intermediates of the final products of ∼27 and 20 kDa detected by sera to 1a-3 and 1a-4 proteins, respectively, during the processing of nsp1a. Proteins of >120 and ∼66 kDa were inconsistently detected by antibodies to 1a-3, and they could also represent intermediate cleavage products during the processing of nsp1a and/or nsp1ab (data not shown).
The 20-kDa protein recognized by antibodies to 1a-4 in the transient expression experiments was not observed in astrovirus-infected cells, probably because this region was further processed in the latter case. This idea is supported by the fact that proteins as small as 20, 6.5, and 5.5 kDa have been reported as processing products of the carboxy-terminal region of HAstV-1 nsp1a (24).
Proteins of 75, 34, and 20 kDa (24) and of ∼38 kDa (11) corresponding to the carboxy-terminal end of nsp1a have been reported; however, in this work only the ∼75 (detected by anti-IFLC)- and 20 (detected by anti-1a-4)-kDa proteins were observed. One possible reason for differences in the nonstructural polyprotein processing among astroviruses could be the presence of insertion-deletion regions identified in astroviruses around residue 765 of nsp1a, which have been associated with virus adaptation to cultured cells (23).
Processing of nsp1ab.
Antibodies to 1b-1 and 1b-2 recognized protein products of 145, 85, and 57 kDa in the pulse-chase experiments. The first two proteins most likely represent intermediates of the processing of nsp1ab, while the 57-kDa protein most likely represents the mature viral RNA polymerase. Given the two intermediate protein products of 88 and 85 kDa derived from nsp1a and nsp1ab, respectively, it is conceivable that they might represent the same intermediate product. However, antibodies to IFLC and 1b-2, which recognize these products, would not be able to interact with a single protein of that size, given the relative localization of the region that they recognize in the nonstructural polyproteins. Thus, it is more likely that these proteins represent in fact two intermediates of the nonstructural polyprotein processing, as indicated in Fig. 4.
Conclusions.
Based on the protein products observed in this and other works, some general conclusions about the proteolytic processing of HAstV nonstructural polyproteins can be made. (i) Processing at the amino terminus of nsp1a and nsp1ab is likely to occur cotranslationally, and it probably occurs at residue Ala174. (ii) Although functional in an in vitro translation assay, the cleavage site dependent on the viral protease at Gln567/Thr568 of nsp1a described by Kiang and Matsui for HAstV-1 (11) does not seem to be used during processing of nsp1a in Yuc8-infected cells, since it would prevent the generation of the ∼27-kDa polypeptides, which contain the protease motif; of interest, a 27-kDa protein has also been detected in HAstV-1-infected cells (5). (iii) The cleavage site to yield the viral RNA polymerase of 57 kDa seems to occur upstream of amino acid residue 33 of nsp1b. (iv) The end cleavage proteolytic products of the nonstructural polyproteins are the 57-kDa polypeptide, which probably represents the RNA polymerase; two proteins of ∼27 kDa that might have the viral serine protease activity; and the 19- to 20-kDa proteins at the amino terminus of nsp1a, of unknown function.
The putative localization of the nonstructural intermediate and final proteins detected in this work relative to the ORF1a and ORF1b regions was assigned based on their reactivities with the antisera raised to the specific regions of the polyproteins (Fig. 4). Further studies are required to determine the precise astrovirus nonstructural polyprotein cleavage sites, to define the role of the viral and possibly cellular proteases in these cleavages, and to determine the function of the virus nonstructural proteins that result from this complex proteolytic cleavage pathway of large polyprotein precursors.
Acknowledgments
We thank Elizabeth Mata and Graciela Cabeza for assistance with animal care. We also acknowledge enriching discussions with U. Geigenmuller.
This work was partially supported by grants MENSE31739 and G37621-N from the National Council for Science and Technology-Mexico, grants IN200999 and IN227602 from DGAPA-UNAM, and grants 55003662 and 55000613 from the Howard Hughes Medical Institute.
REFERENCES
- 1.Bass, D. M., and S. Qiu. 2000. Proteolytic processing of the astrovirus capsid. J. Virol. 74:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belliot, G., H. Laveran, and S. Monroe. 1997. Capsid protein composition of reference strains and wild isolates of human astroviruses. Virus Res. 49:49-57. [DOI] [PubMed] [Google Scholar]
- 3.Carrol, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell lines. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 4.Dennehy, P. H., S. M. Nelson, S. Spangenberger, J. S. Noel, S. S. Monroe, and R. I. Glass. 2001. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 184:10-15. [DOI] [PubMed] [Google Scholar]
- 5.Geigenmuller, U., T. Chew, N. Ginzton, and S. M. Matsui. 2002. Processing of nonstructural protein 1a of human astrovirus. J. Virol. 76:2003-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geigenmuller, U., N. H. Ginzton, and S. M. Matsui. 2002. Studies on intracellular processing of the capsid protein of human astrovirus serotype 1 in infected cells. J. Gen. Virol. 83:1691-1695. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, C. A., J. Chen, S. A. Monroe, and M. R. Denison. 1998. Expression and processing of nonstructural proteins of the human astroviruses. Adv. Exp. Med. Biol. 440:387-391. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero, M., J. Noel, D. Mitchell, J. Calva, A. Morrow, J. Martinez, G. Rosales, F. Velazquez, S. Monroe, R. Glass, L. Pickering, and G. Ruiz-Palacios. 1998. A prospective study of astrovirus diarrhea of infancy in Mexico City. Pediatr. Infect. Dis. J. 17:723-727. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann, J., D. Taylor, P. Echeverria, and N. Blacklow. 1991. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 324:1757-1760. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, B., S. Monroe, E. Koonin, S. Stine, and R. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiang, D., and S. M. Matsui. 2002. Proteolytic processing of a human astrovirus nonstructural protein. J. Gen. Virol. 83:25-34. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, T., and S. Matsui. 1995. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch. Virol. 140:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis, T., and S. Matsui. 1996. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J. Virol. 70:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis, T., and S. Matsui. 1997. Studies of the astrovirus signal that induces (−1) ribosomal frameshifting. Adv. Exp. Med. Biol. 412:323-330. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado, Y., M. Cantwell, M. Old, D. Hill, M. L. Sanchez, L. Logan, F. Millan-Velasco, J. L. Valdespino, J. Sepulveda, and S. Matsui. 1998. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J. Infect. Dis. 178:334-339. [DOI] [PubMed] [Google Scholar]
- 16.Marczinke, B., A. Bloys, T. Brown, M. Willcocks, M. Carter, and I. Brierley. 1994. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 68:5588-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méndez, E., M. T. Fernández, S. López, M. Méndez-Toss, and C. F. Arias. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 76:7996-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendez-Toss, M., P. Romero-Guido, M. E. Munguia, E. Mendez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 19.Monroe, S., S. Stine, L. Gorelkin, J. Herrmann, N. Blacklow, and R. Glass. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J. Virol. 65:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Baez, N., R. O'Brien, S. Q. Qiu, and D. M. Bass. 2002. Astrovirus, adenovirus, and rotavirus in hospitalized children: prevalence and association with gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 35:64-68. [DOI] [PubMed] [Google Scholar]
- 21.Rumenapf, T., G. Unger, J. H. Staruss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67:3288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Fauquier, A., A. Carrascosa, J. Carrascosa, A. Otero, R. Glass, J. Lopez, C. S. Martin, and J. Melero. 1994. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201:312-320. [DOI] [PubMed] [Google Scholar]
- 23.Willcocks, M. M., N. Ashton, J. B. Kurtz, W. D. Cubitt, and M. J. Carter. 1994. Cell culture adaptation of astrovirus involves a deletion. J. Virol. 68:6057-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willcocks, M. M., A. S. Boxall, and M. J. Carter. 1999. Processing and intracellular location of human astrovirus non-structural proteins. J. Gen. Virol. 80:2607-2611. [DOI] [PubMed] [Google Scholar]
- 25.Willcocks, M. M., T. D. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 75:1785-1788. [DOI] [PubMed] [Google Scholar]
- 26.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]