Abstract
Aspergillus fumigatus is an important opportunistic fungal pathogen that is responsible for high mortality rates in the immunosuppressed population. CgrA, the A. fumigatus ortholog of a Saccharomyces cerevisiae nucleolar protein involved in ribosome biogenesis, contributes to the virulence of this fungus by supporting rapid growth at 37°C. To determine how CgrA affects ribosome biogenesis in A. fumigatus, polysome profile and ribosomal subunit analyses were performed on both wild-type A. fumigatus and a ΔcgrA mutant. The loss of CgrA was associated with a reduction in the level of 80S monosomes as well as an imbalance in the 60S:40S subunit ratio and the appearance of half-mer ribosomes. The gene expression profile in the ΔcgrA mutant revealed increased abundance of a subset of translational machinery mRNAs relative to the wild type, suggesting a potential compensatory response to CgrA deficiency. Although ΔcgrA conidia germinated normally at 22°C, they swelled excessively when incubated at 37°C and accumulated abnormally high numbers of nuclei. This hypernucleated phenotype could be replicated pharmacologically by germinating wild-type conidia under conditions of reductive stress. These findings indicate that the germination process is particularly vulnerable to global disruption of protein synthesis and suggest that CgrA is involved in both ribosome biogenesis and polarized cell growth in A. fumigatus.
Aspergillus fumigatus is an opportunistic fungal pathogen that has become the predominant mold species responsible for infections in the immunocompromised population (27). Despite the best treatment with recently approved antifungal drugs, invasive aspergillosis continues to have a poor outcome (19, 31, 34, 35, 38, 41), resulting in the highest hospitalization costs among the systemic mycoses (10, 57). The infection is generally acquired through the inhalation of conidia that, in the absence of adequate host defenses, develop into invasive hyphae that cause severe tissue damage (13, 29). Upon entering the lung, the conidia must transition from a metabolically dormant state at ambient temperature to filamentous growth at 37°C. A. fumigatus germinates very rapidly at 37°C (2), and analysis of growth rate variability among clinical isolates has revealed that faster growth correlates with increased virulence in animal models (42). This high rate of growth places considerable demand on the translational machinery, requiring an increase in ribosome production in proportion to the demand for new proteins.
Ribosome biogenesis begins in the nucleolus, a specialized nuclear compartment that is responsible for the transcription of the ribosomal DNA (rDNA) genes, processing of pre-rRNA, and coordinated assembly of pre-rRNAs with ribosomal proteins (17, 18, 53). We previously showed that the ability of A. fumigatus to grow optimally at temperatures above 25°C requires CgrA, the ortholog of a nucleolar protein involved in Saccharomyces cerevisiae ribosome biogenesis (4, 39). CgrA is necessary for wild-type (wt) virulence of A. fumigatus in a mouse model of invasive aspergillosis but is dispensable for virulence in a Drosophila infection model at 25°C. This suggests that CgrA contributes to pathogenesis by providing an adequate pool of ribosomes to sustain rapid growth at mammalian body temperature (4).
Although ribosome biogenesis has been intensively studied in S. cerevisiae, much less is known about the process in filamentous fungi. In this study, polysome profiling and ribosome subunit analyses were performed to determine how CgrA deficiency affects ribosome biogenesis in A. fumigatus. Here we demonstrate that the loss of CgrA creates an imbalance in ribosome subunit stoichiometry and the accumulation of half-mer ribosomes, irrespective of growth temperature. This was associated with an increased abundance of a subset of mRNAs involved in the translational machinery, suggesting that these mRNAs are part of a compensatory response to the ribosome biogenesis defect. Although the ΔcgrA conidia germinated normally when incubated at 22°C, when incubated at 37°C, they showed a delay in polarized growth, excessive isotropic growth, and the accumulation of large numbers of nuclei. Similar observations were made when wt conidia were germinated in the presence of dithiothreitol (DTT), a reducing agent that unfolds proteins and disrupts ribosomes (45). Together, these findings establish a role for CgrA in ribosome biogenesis in A. fumigatus and provide evidence that CgrA contributes to polarized growth during germination at 37°C.
MATERIALS AND METHODS
Strains and culture conditions.
The wt strain used in this study is a clinical isolate designated H237. The ΔcgrA strain was generated by gene disruption following the insertion of a phleomycin resistance cassette (4). This strain was reconstituted to wt (r-wt) by restoring a single copy of the cgrA gene adjacent to the original gene disruption as previously described (4). All conidia were harvested from strains grown on Aspergillus minimal medium (9). For experiments involving extraction of polysomes, conidia were inoculated into liquid cultures of YG (0.5% yeast extract, 2% glucose).
The germination of A. fumigatus conidia involves a series of morphological changes, beginning with a short period of isotropic growth (swelling) that is followed by the establishment of an axis of polarity and the extension of the first germ tube (37). Continued growth of the germling allows the establishment of interconnections between neighboring hyphae, resulting in an asynchronous population of hyphal compartments (54). Thus, all incubation times in this study were adjusted in order to obtain a homogeneous population of young germlings, defined here as conidia with germ tubes approximately 30 μm in length (see Fig. 2A). This allowed a comparison between equivalent growth stages, with the caveat that nuclear abundance differed between the wt and the ΔcgrA mutant. The alternative approach of normalizing to nuclear number was not employed because any comparison between the wt and the slower growing ΔcgrA strain would be confounded by stage-specific effects (7).
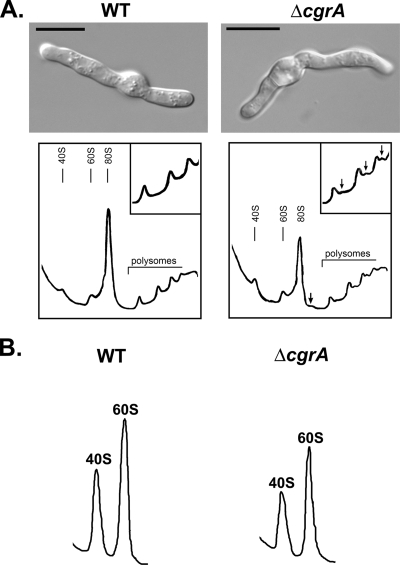
FIG. 2.
The loss of CgrA impairs ribosome biogenesis. (A) Comparison of the polysome profiles of wt and ΔcgrA germlings. Strains were cultured in YG for 44 to 48 h at 22°C. Extracts were isolated under high-magnesium conditions and fractionated on a 7 to 47% linear sucrose gradient with continuous monitoring at 254 nm. Half-mer ribosomes on the monosome and polysome peaks in the ΔcgrA mutant are indicated by the arrows (insets in the top right corner of each profile show an enlargement of the polysome profiles). A differential interference contrast image showing the stage of growth used for analysis is shown at the top of the figure. Bars, 10 μm. (B) Total subunit profiles from wt and ΔcgrA germlings isolated under low-magnesium conditions. Polysome and subunit profiles were performed at least three times with similar findings.
Polysome profiles and RNA analysis.
For polysome analysis, 1 × 108 conidia from the wt and the ΔcgrA strain were inoculated into 100 ml of YG medium and incubated at 200 rpm. Due to the lower growth rate of the ΔcgrA mutant at higher temperatures, incubation times were adjusted so that each culture was harvested when the conidia had elaborated a germ tube that was approximately 30 μm in length. The wt strain was incubated for 44 h at 22°C and for 11 h at 37°C, whereas the ΔcgrA strain was incubated for 48 h at 22°C and for 24 h at 37°C. For polysome profile analysis, cultures were first treated with cycloheximide (0.1 mg/ml) to halt translation and prevent ribosomes from running off the mRNA. The germlings were then disrupted by crushing in liquid nitrogen and resuspended in a high-Mg2+ lysis buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 30 mM MgCl2, 0.1 mg/ml cycloheximide, and 0.2 mg/ml heparin) to preserve ribosome subunit associations. For ribosome subunit analysis, cycloheximide was omitted to allow polysome runoff. In addition, cultures were treated with 1 mM NaN3 for 15 min before harvesting, and germling extracts were prepared by crushing in liquid nitrogen and resuspending in a low-Mg2+ lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, and 1 mM DTT) to disrupt subunit associations. Each lysis buffer was cleared by centrifugation at 12,000 × g at 4°C for 15 min followed by a second clearing spin for 10 min at the same speed. The supernatant was removed, and the RNA content was quantified at 260 nm. Equal numbers of A260 units were loaded onto a 10-ml linear sucrose gradient (7 to 47%). The composition of the buffer used for the high-Mg2+gradient was 50 mM Tris-acetate, 50 mM NH4Cl, 12 mM MgCl2, 1 mM DTT, and 0.1% diethyl pyrocarbonate. The buffer for low-Mg2+ gradient was prepared with 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 1 mM DTT. The gradients were centrifuged in an Sorvall SW 41Ti rotor at 200,000 × g for 2.5 h at 4°C. Gradient analysis was performed with an ISCO gradient collector with continuous monitoring at 254 nm.
For Northern blot analysis, samples of equal volume were collected from the fractionated ribosome gradient using an ISCO fraction collector system. Total RNA was extracted using TRI reagent LS (Molecular Research Center, Cincinnati, OH), fractionated by formaldehyde gel electrophoresis (9), and visualized by SYBR green staining. The RNA was transferred to BioBond nylon membranes (Sigma) and hybridized to a 32P-labeled DNA probe for A. fumigatus β-actin. The β-actin fragment was PCR amplified from A. fumigatus genomic DNA using the following primers: forward (5′-ATGTCACTGTGCAGATTGTC-3′) and reverse (5′-TTAGAAGCACTTGCGGTGAA-3′).
Transcriptional profiling.
A whole-genome microarray was performed to identify genes that show differential expression in the absence of cgrA during thermal stress, using a strategy that was previously used for the genome reference strain Af293 (40). A total of 5 × 106 conidia from the wt, r-wt, and ΔcgrA strains were cultured at 30°C (minimal thermal stress for the mutant) until they had become germlings (16 h for the wt and r-wt strains and 22 h for the ΔcgrA mutant) in 50 ml of complete minimal medium (Aspergillus minimal medium supplemented with 0.1% yeast extract, 0.2% peptone, and 0.1% tryptone). RNA was extracted from the zero time point at 30°C, which served as the reference sample. The remaining cultures were shifted to 37°C, a temperature at which the ΔcgrA mutant is growth impaired. RNA was extracted after 20, 40, 60, 80, and 100 min of incubation at 37°C by crushing the mycelium in liquid nitrogen and extracting with Trizol reagent (Invitrogen).
The A. fumigatus Af293 DNA amplicon microarray containing 9,516 genes was used in this study (40). The protocols used for microbial RNA aminoallyl labeling (M007) and microbial hybridization of labeled probes (M008) are available at http://pfgrc.tigr.org/protocols.shtml. In order to identify genes exhibiting altered transcription after the temperature shift, the 0-min sample served as the reference upon which up- or down-regulation was measured. All the hybridizations were repeated in dye-swap sets. Hybridized slides were scanned using the Axon GenePix 4000B microarray scanner, and the TIFF images generated were analyzed using TIGR Spotfinder software to obtain relative transcript levels. Hybridization data was normalized using a local regression technique LOWESS (locally weighted scatterplot smoothing) using the software tool MIDAS. The resulting data were averaged from three gene spots on each array and from duplicate flip-dye arrays for each experiment equaling six intensity data points for each gene. Differentially expressed genes at the 95% confidence level were determined using intensity-dependent Z scores (with Z = 1.96) as implemented in MIDAS for all experiments. To identify genes where expression is affected by the cgrA deletion following a shift from 30°C to 37°C, we identified all genes exhibiting differential expression in the wt, ΔcgrA, and r-wt at each time point. Genes that were found to have significantly different expression in each set of experiments were then combined and organized based on similar expression vectors using Euclidean distance and hierarchical clustering with average linkage clustering method with TIGR TM4. All microarray analysis software is available at http://pfgrc.tigr.org/tools.shtml.
Real-time PCR.
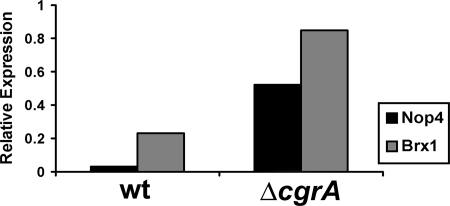
Validation of the microarray data was performed by real-time reverse transcriptase PCR (RT-PCR) on two selected ribosome biogenesis genes, nop4 and brx1 (6, 52). To obtain RNA, the wt and ΔcgrA strains were grown under the same conditions used for microarray analysis. RNA was extracted as described above, from cultures grown at 30°C, which served as the 0-min time point, and at 80 min following the shift to 37°C. Five micrograms of total RNA was converted to cDNA using Superscript II reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed on the cDNA using a SYBR GreenER quantititative PCR supermix universal kit (Invitrogen) using a SmartCycler II instrument (Cepheid, Sunnyvale, CA). The PCR protocol involved 40 cycles, with each cycle consisting of 95°C for 15 s, annealing for 3 s, and extension at 72°C for 30 s. The Ct (cycle threshold) value was determined by the PCR cycle number at which the fluorescent signal crossed the default threshold set at 30 fluorescent units. Ct values were calculated for the two target genes, brx1 and nop4 (6, 52) at the 0- and 80-min time points, as well as for the housekeeping gene fks1 (3). The values for the 80-min time point were then normalized to the values for the 0-min sample and reference gene fks using the method described by Pfaffl (43). The data were presented as a relative expression ratio, using the following formula: ratio = EtargetΔCt (control − sample)/Eref ΔCt (control − sample) where E is the exponential amplification calculated by 10−1/slope and Eref is the E value for the reference sample. The following primer pairs were used for amplification of fks, brx1, and nop4: Fks-forward, 5′-GCCTGGTAGTGAAGCTGAGCGT-3′; Fks-reverse, 5′-CGGTGAATGTAGGCATGTTGTCC-3′; Brx1-forward, 5′-GTTTCACGTCACAAACCTGCA-3′; Brx1-reverse, 5′-CAATCCGCGGTACTCGTTTC-3′; Nop4-forward, 5′-GAGGAGTACCGGAGGTCAAAGG-3′; and Nop4-reverse, 5′-GCGCATGTCGATGTGTGTAGTA-3′.
Confocal microscopy.
To visualize nuclei in live cultures, strains were transfected with a NopA-green fluorescent protein (GFP) expression construct previously shown to localize to nucleoli in A. fumigatus (5). Conidia were inoculated into 5 ml of Aspergillus minimal medium in a 35-mm petri dish containing a 25-mm-diameter coverslip. The cultures were incubated at 22°C or 37°C without shaking until the conidia formed a germ tube that was approximately 30 μm in length. Fluorescent nuclei were observed with a Leica TCS SP2 laser-scanning confocal microscope using a 63× oil objective. For experiments involving nuclear staining with propidium iodide (PI), the germlings were fixed for 3 min at room temperature in a solution containing 3.7% formaldehyde, 0.2% Triton X-100, and 50 mM phosphate buffer, pH 7. The slides were then treated with a 10-mg/ml solution of RNase A for 1 h at 37°C, followed by nuclear staining with PI (12.5 μg/ml). The laser source was an Ar/Kr laser set for GFP detection (excitation, 488 nm; emission, 507 nm) or for PI detection (excitation, 530 nm; emission, 615 nm). Cell walls were stained by incubating in a solution of 0.4 μg/ml calcofluor (fluorescent brightner 28; Sigma) for 5 min at room temperature and visualized by confocal microscopy using a UV laser set for 4′,6′-diamidino-2-phenylindole (DAPI) detection (excitation, 372 nm; emission, 456 nm).
Microarray data accession number.
The full data set for this study has been deposited with ArrayExpress at the EMBL European Bioinformatics Institute (http://www.ebi.ac.uk/) under the accession number E-MEXP-1324.
RESULTS
Polysome profiling in A. fumigatus.
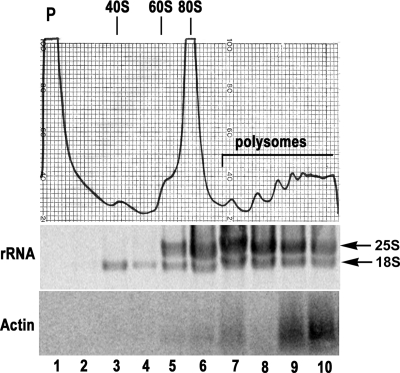
In order to develop techniques for the analysis of ribosome distributions in A. fumigatus, we adapted a protocol from S. cerevisiae (28) and optimized the conditions for analysis of A. fumigatus germlings as described in Materials and Methods. A typical profile showing the normal distribution of ribosome subunits in wt A. fumigatus germlings is shown in Fig. 1 (top panel). The wt profile showed the expected protein peak, followed by peaks representing free 40S subunits, free 60S subunits, the 80S monosomes and polysomes (polyribosomes) corresponding to two or more ribosomes per mRNA. Under these conditions, the 18S rRNA in the 40S subunit was found in fractions 3 and 4, the 60S subunit containing both rRNAs appeared in fraction 5, and the 80S monosome and polysomes were present in fractions 6 to 10 (Fig. 1). The presence of mRNA in the polysome fractions was demonstrated by extracting RNA from each gradient and hybridizing to an actin-specific probe by Northern blot analysis. As expected for actively growing germlings, the majority of the actin mRNA was present in the polysome fractions.
FIG. 1.
Polysome profile analysis in wt A. fumigatus. (Top) Polysome profile of wt A. fumigatus germlings cultured in rich medium at 37°C. The profile shows a protein peak (P), followed by peaks representing free 40S and 60S subunits, the 80S monosome and polysomes representing two or more ribosomes per mRNA. (Middle) SYBR green staining of rRNA in each polysome fraction (10 fractions). (Bottom) Northern blot showing hybridization of RNA in each fraction to an A. fumigatus β-actin probe.
Effects of CgrA disruption on ribosome biogenesis in A. fumigatus.
To determine how the loss of CgrA affects the steady-state distribution of ribosomes in A. fumigatus, the polysome profiles of wt and ΔcgrA germlings were compared (Fig. 2A). Monosome levels were reduced in the ΔcgrA mutant by about 40% relative to that in the wt, and half-mer ribosomes were apparent on the ΔcgrA monosome and polysome peaks (Fig. 2A). Comparable defects were previously observed in an S. cerevisiae Δcgr1 mutant, although the reduction in monosomes was somewhat greater in yeast (over 60%) (39). Half-mers, evident as shoulders following the monosome and polysome peaks, represent 43S initiation complexes that are stalled at the AUG and await binding of 60S subunits to form the 80S monosome (30). The presence of half-mers suggest a defect in 60S subunit assembly as has been previously reported in a yeast cgr1 mutant (39). To determine whether the half-mers observed in the ΔcgrA strain were due to an imbalance in subunit stoichiometry, ribosomes were extracted under low-Mg2+ conditions to disrupt subunit association. Total 40S and 60S subunits in the ΔcgrA mutant were approximately 25 to 30% lower than in the wt (Fig. 2B). The ratio of 60S:40S subunits was 1.82 in wt A. fumigatus, similar to wt ratios in S. cerevisiae (11, 12, 39). However, the corresponding ratio in the ΔcgrA strain was 1.66, suggesting imbalanced subunit stoichiometry. The same ribosome defects were evident in the ΔcgrA mutant grown at either 22°C or 37°C, despite the fact that the growth rate of the ΔcgrA mutant was indistinguishable from the growth rate of the wt at 22°C but 65% less than the growth rate of the wt at 37°C (4). This indicates that the loss of CgrA leads to a phenotypic growth defect that is enhanced by elevated temperature, rather than a ribosome biogenesis defect that is temperature dependent.
Transcriptional response to CgrA deficiency.
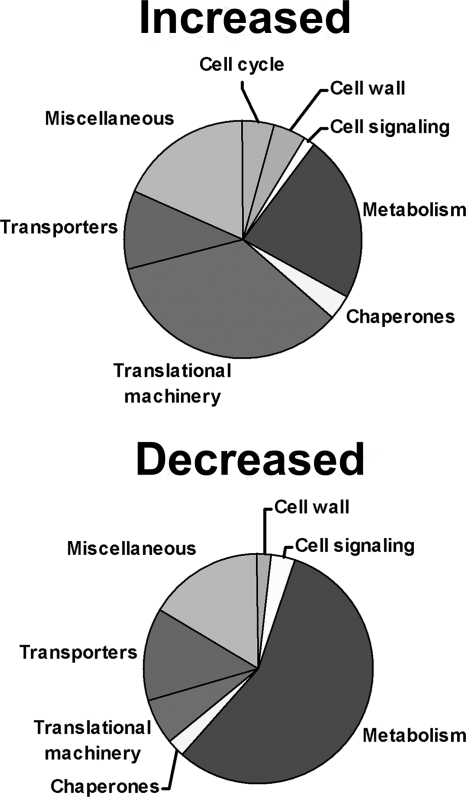
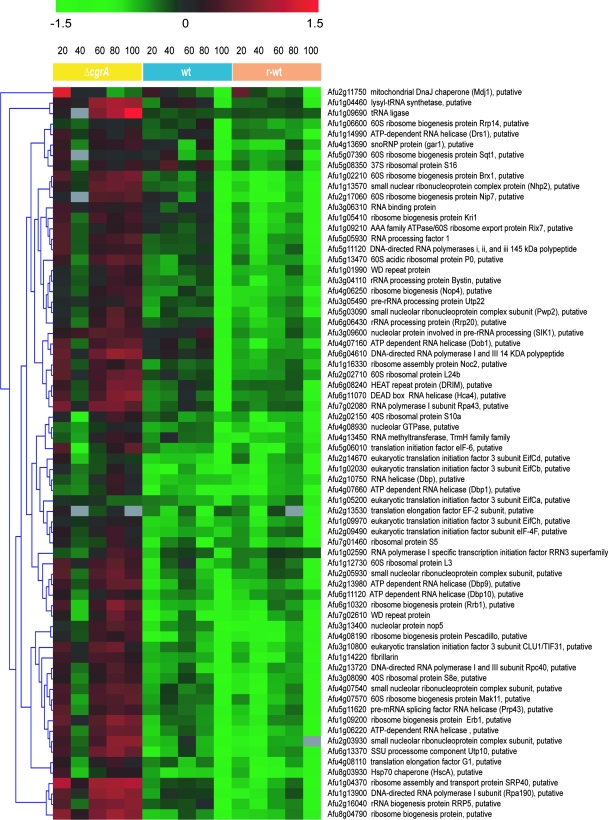
Although the ΔcgrA mutant showed a reduction in 60S:40S subunit stoichiometry, the defect in subunit ratio was less dramatic than what has been previously reported in an S. cerevisiae cgr1 mutant (39). This suggests that A. fumigatus may be able to compensate partially for the absence of CgrA function. To gain insight into potential compensatory pathways, the steady-state expression profile of the ΔcgrA strain was compared to that of the wt following a shift from 30°C to 37°C (4). All mRNAs that showed increased or decreased abundance in the ΔcgrA mutant relative to the level in the wt following the shift are summarized in Fig. 3. The largest category of down-regulated mRNAs in the ΔcgrA mutant was associated with metabolism, a finding consistent with the reduced growth rate of this strain at 37°C. In contrast, the largest number of up-regulated mRNAs in the ΔcgrA mutant belonged in the category of translational machinery, suggesting that these mRNAs constitute an adaptive response to loss of CgrA function. A heat map depicting the translational machinery-associated mRNAs that had increased abundance in the ΔcgrA mutant relative to that in the wt and r-wt strains is shown in Fig. 4. Two of these mRNAs were randomly selected for validation by an independent method. As expected, RT-PCR analysis of the steady-state levels of brx1 and nop4 mRNAs showed increased abundance in the ΔcgrA mutant following a shift from 30°C to 37°C (Fig. 5).
FIG. 3.
Transcriptional response to CgrA deficiency. Summary of functional categories of mRNAs showing increased or decreased abundance in the ΔcgrA mutant relative to the wt, 80 min after a shift from 30°C to 37°C.
FIG. 4.
Expression profile of mRNAs involved in the translational machinery. Clustered display of translational machinery mRNAs that were increased in the ΔcgrA mutant relative to the wt following a shift from 30°C to 37°C. Colors represent the observed expression ratios on a log2 scale at 20, 40, 60, 80, and 100 min following the temperature shift. All measurements are relative to the measurement at time zero. A total of 70 translational machinery genes were clustered based on similar expression patterns using a hierarchical clustering algorithm.
FIG. 5.
Validation of microarray data by real-time PCR. Two mRNAs that showed increased abundance in the ΔcgrA mutant relative to the wt following the shift from 30°C to 37°C by microarray analysis were confirmed by qRT-PCR. Ct values were calculated for each gene 80 min following a shift from 30°C to 37°C. Relative expression values were normalized to the 0-min time point and to the reference gene fks as described in Materials and Methods.
Conidial germination and nuclear duplication.
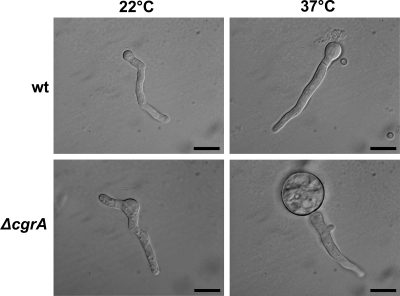
The transcriptional profile of the ΔcgrA strain at 37°C also revealed increases in the levels of some cell cycle-associated mRNAs relative to the levels in the wt (Fig. 3). Since mitosis is normally coordinated with the morphological changes that take place during germination of Aspergillus spp. (23, 37), the germination of wt and ΔcgrA conidia was examined in more detail. Conidia were inoculated into Aspergillus minimal medium and incubated until they had elaborated a germ tube that was approximately 30 μm in length. As expected for wt A. fumigatus, conidia that were germinated at 37°C underwent a period of isotropic growth before establishing a germ tube (Fig. 6, two ungerminated conidia are evident in the top right panel for comparison). In contrast, 90% of the ΔcgrA conidia incubated at 37°C swelled to over three times their normal diameter (Fig. 6), suggesting a defect in the establishment of polarized growth. This increased isotropic growth did not occur in ΔcgrA germlings that were cultured at 22°C (Fig. 6), indicating that the loss of CgrA induces a temperature-dependent polarity defect. However, once polarity was established, the maintenance of polarity in mature hyphae was apparently normal (data not shown). Excessive swelling was never observed in wt conidia at either temperature, although there was a slight increase in size at 37°C relative to 22°C (Fig. 6).
FIG. 6.
Conidial morphogenesis. Conidia from the wt and ΔcgrA strains were inoculated into Aspergillus minimal medium and incubated at 22°C or 37°C until the conidia elaborated a germ tube that was approximately 30 μm in length. (A) Differential interference contrast images of wt and ΔcgrA germlings grown at 22°C for 48 h or at 37°C for 12 h (wt) or 24 h (ΔcgrA). Bars, 10 μm.
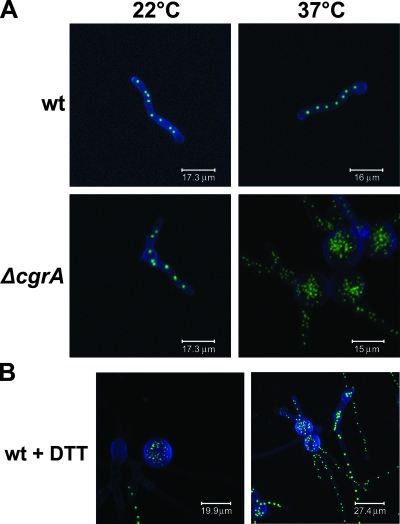
In A. fumigatus, wt conidia undergo the first mitotic division around the time at which polarized growth becomes established, giving rise to a pear-shaped cell with one or two nuclei (37). Nuclei continue to divide as the germ tube extends, and the first septum is usually formed around the fourth mitotic division (37). To determine whether the synchronization between morphogenesis and nuclear division was disrupted in the hyperswollen ΔcgrA germlings, the wt and ΔcgrA strains were transfected with a NopA-GFP expression construct. This GFP fusion encodes a nucleolar protein that fluorescently labels A. fumigatus nuclei (5). Conidia from both strains were germinated until the germ tube was approximately 30 μm in length (Fig. 7A). When cultured at either 22°C or 37°C, wt germlings of this size contained approximately eight nuclei distributed evenly along each hypha (Fig. 7A). At 22°C, the ΔcgrA mutant was indistinguishable from the wt (Fig. 7A). However, at 37°C, the enlarged conidia of the ΔcgrA mutant were filled with multiple nuclei (Fig. 7A). This hypernucleated phenotype was not apparent in mature hyphae (data not shown), suggesting that nuclear accumulation was specific to the early stages of germination. The same results were obtained when wt and ΔcgrA nuclei were stained with PI, indicating that this finding was not an artifact of NopA-GFP expression in the ΔcgrA background (data not shown).
FIG. 7.
Nuclear duplication. (A) Conidia from wt and ΔcgrA strains expressing the nucleolar marker NopA-GFP were inoculated into Aspergillus minimal medium and incubated at 22°C or 37°C until the conidia had elaborated a germ tube that was approximately 30 μm in length. Nuclei were visualized by live-cell imaging with a 63× oil objective on a Leica TCS SP2 laser-scanning confocal microscope set for GFP detection. Sixteen optical sections, ranging in size from 7 to 23 μm in thickness, were obtained through the z plane of each germling and assembled into the single compressed image shown. GFP is shown in green, and the calcofluor-stained cell wall is shown in blue. (B) wt and ΔcgrA conidia expressing NopA-GFP were inoculated into Aspergillus minimal medium containing 1 mM DTT and incubated for 24 h at 37°C.
To determine whether a similar defect in polarized growth could be induced pharmacologically by an agent that unfolds proteins and disrupts ribosomes (45), wt conidia were incubated in the presence of a sublethal concentration of DTT. The DTT delayed the onset of germination and was associated with increased swelling and nuclear accumulation (Fig. 7B). However, as seen in the ΔcgrA mutant, the hypernucleated phenotype was not evident in mature hyphae after they had grown beyond the germling stage (data not shown). This result suggests that the germination process is particularly vulnerable to a global disruption of protein function caused by either the loss of CgrA or DTT stress.
DISCUSSION
The success of A. fumigatus as an opportunistic fungal pathogen is due in large part to the ability of the fungus to grow rapidly in the host (2). Since infections with A. fumigatus continue to have a poor outcome (31, 44), there is a need for greater understanding of the pathways that sustain the growth of A. fumigatus in the host environment. One of these pathways includes ribosome biogenesis, a complex process that is tightly linked to growth rate (20, 33, 47, 59). We previously showed that the nucleolar protein CgrA is necessary for the growth and virulence of A. fumigatus at 37°C (4), but the contribution of the protein to ribosome biogenesis in this organism was not explored. Here, we reveal how a loss of CgrA function disrupts ribosome biogenesis in A. fumigatus and highlight an unexpected response to such a defect.
The growth rate of wt A. fumigatus is approximately three times greater at 37°C than it is at 22°C (data not shown). In contrast, the ΔcgrA mutant grows normally at 22°C but is unable to increase its growth rate when cultured at 37°C (4). Therefore, we were surprised to find that half-mers were present in the ΔcgrA mutant at 22°C or 37°C. We interpret these data to indicate that the defect in ribosome biogenesis caused by the loss of CgrA is compatible with the limited demands for growth at 22°C but not with the heightened metabolic needs at 37°C. Half-mers, which are not detected under normal conditions, represent 40S preinitiation complexes stalled at the AUG and often arise in response to a defect in ribosome subunit stoichiometry (30). Analysis of total ribosome subunit levels confirmed that the loss of CgrA was associated with a disruption in subunit stoichiometry. This is consistent with the predominant 60S subunit synthesis defect previously reported in an S. cerevisiae cgr1 mutant (39), although the effect was less dramatic in A. fumigatus. The more subtle ribosome defect in A. fumigatus may indicate a level of redundancy in A. fumigatus that is not present in yeast. Alternatively, the presence of half-mers without a large change in stoichiometry could be indicative of a defect in subunit association (16). However, we were unable to identify a stable association of CgrA with fractionated ribosome subunits by Western blot analysis (data not shown), suggesting that if there is a role in subunit association, it is likely to be transient (16, 56).
A whole-genome microarray was used to determine how the loss of CgrA affects the expression profile of the organism under a condition of thermal stress that requires CgrA function to support optimum growth. The data revealed that a subset of translational machinery mRNAs were up-regulated in the ΔcgrA mutant. We hypothesize that these mRNAs are part of an adaptive response to CgrA deficiency, possibly involving the increased nuclear duplication phenotype noted in the ΔcgrA mutant. Interestingly, a number of these mRNAs showed decreased abundance in the wt following the shift to 37°C (Fig. 4), a finding that may reflect the temporary inhibition of the translational machinery previously reported following a heat shock (15, 55). A small number of translational machinery-associated mRNAs showed decreased abundance in the ΔcgrA strain relative to the wt following the temperature shift, including L22, L12, L1, L37, Rps29, S14, and P2 (data not shown). Most of these proteins have functions in ribosome structure or assembly (1, 8, 14, 46, 51). The decreased abundance of these mRNAs in the ΔcgrA strain suggests that their levels are dependent on intact CgrA function, although the mechanism for this is presently unclear.
An unexpected observation from this study was that ΔcgrA conidia swelled excessively and accumulated nuclei when germinated at 37°C, but not when germinated at 22°C. Thus, loss of CgrA function appears to disrupt the normal synchronization between size and nuclear duplication during the germination process, suggesting a defect in polarized growth that is temperature dependent. The multinucleated hyperswollen conidia of the ΔcgrA mutant are reminiscent of the phenotype displayed by polarity mutants of Aspergillus nidulans (22, 24, 26, 32, 48-50). A variety of genes are responsible for these defects in polarized growth, but swoC1 is of particular relevance to this study because it encodes an rRNA pseudouridine synthase (32), suggesting a link between ribosome biogenesis and the establishment of polarity. Further evidence to support this connection has recently been obtained in S. cerevisiae, where a dual function in ribosome biogenesis and polarized growth has been identified for the nucleolar protein Rrp14p (60). Although the mechanism by which ribosome biogenesis can influence polarity is not yet known, it is interesting to note that the rrp14 mutant displays a ribosome defect similar to that of the S. cerevisiae Δcgr1 mutant, including half-mers, decreased 60S subunits, and impaired processing of 27S pre-rRNA to 25S rRNA (39, 60). It is therefore intriguing to hypothesize that certain ribosome defects disproportionately interrupt the translation of mRNAs that encode gene products that influence the establishment of polarized growth. If nuclear duplication continues normally during this period of delayed polarized growth, nuclei would be expected to accumulate in hyperswollen conidia, as was observed in the ΔcgrA mutant. The identities of gene products that could be selectively affected by ribosome disruption remain to be explored.
The presence of multinucleated hyperswollen conidia in the ΔcgrA mutant is also consistent with an alternative model in which nuclear abundance provides a mechanism to regulate ribosome output in proportion to the physiological demand for new proteins. In the vast majority of eukaryotes, the demand for ribosomes is met, in part, by maintaining multiple copies of the rDNA genes (21). However, variability in the number of rDNA genes has been reported within a species, suggesting that rDNA copy number can be modulated depending on the need for protein synthetic capacity. For example, only minor variations in the number of rDNA units are present among different subclones of Candida albicans and S. cerevisiae when grown slowly at 22°C, but increased rDNA content can be selected for by growing these yeasts more rapidly at their optimal temperatures of 37°C and 30°C, respectively (47). In filamentous fungi, the ribosome content of an ungerminated spore is insufficient to support polarized hyphal growth, so ribosome biogenesis is a major synthetic process during the early stages of germination (25, 36, 58). A defect in ribosome biogenesis is therefore likely to impair protein synthesis and disrupt this early stage of growth, particularly at high temperatures when metabolic needs are high. Since nuclei are the source of new ribosomes, the ΔcgrA mutant may accumulate nuclei until there are sufficient ribosomes to meet the demand for germ tube emergence and hyphal growth, a response that would require a parallel increase in size to accommodate the nuclei. Such a response would not be expected to completely rescue the growth defect, however, since each nucleus harbors the same mutation. This is consistent with the fact that the germination of the ΔcgrA mutant is impaired at 37°C (4). Regardless of the exact mechanism involved, the temperature sensitivity of the ΔcgrA phenotype clearly indicates that growth temperature can be an important variable to consider when studying polarity establishment during germination. Since temperature-sensitive alleles are often used for analysis of polarity in filamentous fungi, such experimental approaches should be interpreted with this caveat in mind.
The question remains as to why thermal stress induces ΔcgrA germlings to increase in size and accumulate nuclei but does not affect nuclear number or size in mature vegetative hyphae. A possible explanation for this difference is that the mechanism that coordinates nuclear duplication with size and septation is fundamentally different between these two stages of growth. For example, a germling that has not yet formed a septum, defined as “predivisional” (23), undergoes about four nuclear duplications before it lays down the first septum (37). This requires an uncoupling of cell division (septation) from mitosis in order to allow the uninucleated conidium to develop into a multinucleated hypha (23). In contrast, “postdivisional” hyphae are composed of multinucleated cellular compartments that are delimited by septa. These compartments exhibit autonomous nuclear duplication cycles that are tightly coordinated with size and septation (23). Thus, it is conceivable that the intracellular environment of a predivisional germling allows for nuclear accumulation, but the more rigid coupling between cell division and mitosis in postdivisional hyphae may be more restrictive. Interestingly, we found that the effects of CgrA deficiency on conidial swelling and nuclear duplication could be replicated by germinating wt conidia in the presence of DTT, a reducing agent that disrupts many cell functions, including the translational machinery (45). As with the ΔcgrA mutant, continued growth allowed the nuclei to distribute normally in mature hyphal compartments, suggesting that it is the germination process that is particularly vulnerable to a widespread loss of protein function caused by either CgrA deficiency or DTT.
Taken together, the data outlined in this study establish a role for CgrA in ribosome biogenesis in A. fumigatus and provide new evidence that this is linked to the establishment of polarized growth during germination at 37°C. It will be of interest in future studies to determine the mechanism by which CgrA affects polarity and to elucidate the mechanism by which temperature influences this function.
Acknowledgments
This work was supported by NIH grant R01AI48746 to D.S.A. and an American Heart Association Predoctoral Fellowship to R.B.
We thank J. Woolford, Carnegie Mellon University, for providing technical advice.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Antunez de Mayolo, P., and J. L. Woolford, Jr. 2003. Interactions of yeast ribosomal protein rpS14 with RNA. J. Mol. Biol. 333697-709. [DOI] [PubMed] [Google Scholar]
- 2.Araujo, R., and A. G. Rodrigues. 2004. Variability of germinative potential among pathogenic species of Aspergillus. J. Clin. Microbiol. 424335-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauvais, A., J. M. Bruneau, P. C. Mol, M. J. Buitrago, R. Legrand, and J. P. Latge. 2001. Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 1832273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhabhra, R., M. D. Miley, E. Mylonakis, D. Boettner, J. Fortwendel, J. C. Panepinto, M. Postow, J. C. Rhodes, and D. S. Askew. 2004. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect. Immun. 724731-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhabhra, R., W. Zhao, J. C. Rhodes, and D. S. Askew. 2006. Nucleolar localization of Aspergillus fumigatus CgrA is temperature-dependent. Fungal Genet. Biol. 431-7. [DOI] [PubMed] [Google Scholar]
- 6.Bogengruber, E., P. Briza, E. Doppler, H. Wimmer, L. Koller, F. Fasiolo, B. Senger, J. H. Hegemann, and M. Breitenbach. 2003. Functional analysis in yeast of the Brix protein superfamily involved in the biogenesis of ribosomes. FEMS Yeast Res. 335-43. [DOI] [PubMed] [Google Scholar]
- 7.Breakspear, A., and M. Momany. 2007. The first fifty microarray studies in filamentous fungi. Microbiology 1537-15. [DOI] [PubMed] [Google Scholar]
- 8.Briones, E., C. Briones, M. Remacha, and J. P. Ballesta. 1998. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 27331956-31961. [DOI] [PubMed] [Google Scholar]
- 9.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 11351-56. [DOI] [PubMed] [Google Scholar]
- 10.Dasbach, E. J., G. M. Davies, and S. M. Teutsch. 2000. Burden of aspergillosis-related hospitalizations in the United States. Clin. Infect. Dis. 311524-1528. [DOI] [PubMed] [Google Scholar]
- 11.Daugeron, M. C., and P. Linder. 1998. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 171128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26781-805. [DOI] [PubMed] [Google Scholar]
- 14.Deshmukh, M., J. Stark, L. C. Yeh, J. C. Lee, and J. L. Woolford, Jr. 1995. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 27030148-30156. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisinger, D. P., F. A. Dick, and B. L. Trumpower. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 175136-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14313-318. [DOI] [PubMed] [Google Scholar]
- 18.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 31317-42. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda, T., M. Boeckh, R. A. Carter, B. M. Sandmaier, M. B. Maris, D. G. Maloney, P. J. Martin, R. F. Storb, and K. A. Marr. 2003. Risks and outcomes of invasive fungal infections in recipients of allogeneic hematopoietic stem cell transplants after nonmyeloablative conditioning. Blood 102827-833. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-y-Merchand, J. A., M. J. Garcia, S. Gonzalez-Rico, M. J. Colston, and R. A. Cox. 1997. Strategies used by pathogenic and nonpathogenic mycobacteria to synthesize rRNA. J. Bacteriol. 1796949-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadjiolov, A. A. 1985. The nucleolus and ribosome biogenesis, vol. 12. Springer-Verlag Wien, New York, NY.
- 22.Harispe, L., C. Portela, C. Scazzocchio, M. A. Peñalva, and L. Gorfinkiel. 2008. Ras GTPase-activating protein regulation of actin cytoskeleton and hyphal polarity in Aspergillus nidulans. Eukaryot. Cell 7141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, S. D. 1997. The duplication cycle in Aspergillus nidulans. Fungal Genet. Biol. 221-12. [DOI] [PubMed] [Google Scholar]
- 24.Harris, S. D., A. F. Hofmann, H. W. Tedford, and M. P. Lee. 1999. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 1511015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horikoshi, K., and Y. Ikeda. 1968. Studies on the conidia of Aspergillus oryzae. VII. Development of protein synthesizing activity during germination. Biochim. Biophys. Acta 166505-511. [PubMed] [Google Scholar]
- 26.Kaminskyj, S. G., and J. E. Hamer. 1998. hyp loci control cell pattern formation in the vegetative mycelium of Aspergillus nidulans. Genetics 148669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffman, C. A. 2006. Fungal infections. Proc. Am. Thorac. Soc. 335-40. [DOI] [PubMed] [Google Scholar]
- 28.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 177283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, J. H., T. V. Pestova, B. S. Shin, C. Cao, S. K. Choi, and T. E. Dever. 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. USA 9916689-16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32358-366. [DOI] [PubMed] [Google Scholar]
- 32.Lin, X., and M. Momany. 2003. The Aspergillus nidulans swoC1 mutant shows defects in growth and development. Genetics 165543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maleszka, R., and G. D. Clark-Walker. 1990. Magnification of the rDNA cluster in Kluyveromyces lactis. Mol. Gen. Genet. 223342-344. [DOI] [PubMed] [Google Scholar]
- 34.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34909-917. [DOI] [PubMed] [Google Scholar]
- 35.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33641-647. [DOI] [PubMed] [Google Scholar]
- 36.Mirkes, P. E. 1974. Polysomes, ribonucleic acid, and protein synthesis during germination of Neurospora crassa conidia. J. Bacteriol. 117196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Momany, M., and I. Taylor. 2000. Landmarks in the early duplication cycles of Aspergillus fumigatus and Aspergillus nidulans: polarity, germ tube emergence and septation. Microbiology 1463279-3284. [DOI] [PubMed] [Google Scholar]
- 38.Morgan, J., K. A. Wannemuehler, K. A. Marr, S. Hadley, D. P. Kontoyiannis, T. J. Walsh, S. K. Fridkin, P. G. Pappas, and D. W. Warnock. 2005. Incidence of invasive aspergillosis following hematopoietic stem cell and solid organ transplantation: interim results of a prospective multicenter surveillance program. Med. Mycol. 43(Suppl. 1)S49-S58. [DOI] [PubMed] [Google Scholar]
- 39.Moy, T. I., D. Boettner, J. C. Rhodes, P. A. Silver, and D. S. Askew. 2002. Identification of a role for Saccharomyces cerevisiae Cgr1p in pre-rRNA processing and 60S ribosome subunit synthesis. Microbiology 1481081-1090. [DOI] [PubMed] [Google Scholar]
- 40.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 4381151-1156. [DOI] [PubMed] [Google Scholar]
- 41.Pagano, L., M. Caira, A. Candoni, M. Offidani, L. Fianchi, B. Martino, D. Pastore, M. Picardi, A. Bonini, A. Chierichini, R. Fanci, C. Caramatti, R. Invernizzi, D. Mattei, M. E. Mitra, L. Melillo, F. Aversa, M. T. Van Lint, P. Falcucci, C. G. Valentini, C. Girmenia, and A. Nosari. 2006. The epidemiology of fungal infections in patients with hematologic malignancies: the SEIFEM-2004 study. Haematologica 911068-1075. [PubMed] [Google Scholar]
- 42.Paisley, D., G. D. Robson, and D. W. Denning. 2005. Correlation between in vitro growth rate and in vivo virulence in Aspergillus fumigatus. Med. Mycol. 43397-401. [DOI] [PubMed] [Google Scholar]
- 43.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaller, M. A., P. G. Pappas, and J. R. Wingard. 2006. Invasive fungal pathogens: current epidemiological trends. Clin. Infect. Dis. 43S3-S14. [Google Scholar]
- 45.Rand, J. D., and C. M. Grant. 2006. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell 17387-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remacha, M., A. Jimenez-Diaz, C. Santos, E. Briones, R. Zambrano, M. A. Rodriguez Gabriel, E. Guarinos, and J. P. Ballesta. 1995. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 73959-968. [DOI] [PubMed] [Google Scholar]
- 47.Rustchenko, E. P., T. M. Curran, and F. Sherman. 1993. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J. Bacteriol. 1757189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw, B. D., C. Momany, and M. Momany. 2002. Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot. Cell 1241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw, B. D., and M. Momany. 2002. Aspergillus nidulans polarity mutant swoA is complemented by protein O-mannosyltransferase pmtA. Fungal Genet. Biol. 37263-270. [DOI] [PubMed] [Google Scholar]
- 50.Shaw, B. D., and S. Upadhyay. 2005. Aspergillus nidulans swoK encodes an RNA binding protein that is important for cell polarity. Fungal Genet. Biol. 42862-872. [DOI] [PubMed] [Google Scholar]
- 51.Shu-Nu, C., C. H. Lin, and A. Lin. 2000. An acidic amino acid cluster regulates the nucleolar localization and ribosome assembly of human ribosomal protein L22. FEBS Lett. 48422-28. [DOI] [PubMed] [Google Scholar]
- 52.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 133127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33261-311. [DOI] [PubMed] [Google Scholar]
- 54.Vinck, A., M. Terlou, W. R. Pestman, E. P. Martens, A. F. Ram, C. A. van den Hondel, and H. A. Wosten. 2005. Hyphal differentiation in the exploring mycelium of Aspergillus niger. Mol. Microbiol. 58693-699. [DOI] [PubMed] [Google Scholar]
- 55.Warner, J. R., and C. Gorenstein. 1977. The synthesis of eucaryotic ribosomal proteins in vitro. Cell 11201-212. [DOI] [PubMed] [Google Scholar]
- 56.West, M., J. B. Hedges, A. Chen, and A. W. Johnson. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 253802-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, L. S., C. M. Reyes, M. Stolpman, J. Speckman, K. Allen, and J. Beney. 2002. The direct cost and incidence of systemic fungal infections. Value Health 526-34. [DOI] [PubMed] [Google Scholar]
- 58.Winther, M. D., and L. Stevens. 1981. RNA synthesis during the germination of conidia of Aspergillus nidulans. Microbios 30153-162. [PubMed] [Google Scholar]
- 59.Woolford, J. L., Jr., and J. R. Warner. 1991. The ribosome and its synthesis, p. 587-626. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Yamada, H., C. Horigome, T. Okada, C. Shirai, and K. Mizuta. 2007. Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity. RNA 131977-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]