Abstract
Norwalk virus (NV), a reference strain of human calicivirus in the Norovirus genus of the family Caliciviridae, contains a positive-strand RNA genome with three open reading frames. ORF1 encodes a 1,789-amino-acid polyprotein that is processed into nonstructural proteins that include an NTPase, VPg, protease, and RNA-dependent RNA polymerase. The N-terminal protein p48 of ORF1 shows no significant sequence similarity to viral or cellular proteins, and its function in the human calicivirus replication cycle is not known. The lack of sequence similarity to any protein in the public databases suggested that p48 may have a unique function in the NV replication cycle or, alternatively, may perform a characterized function in replication by a unique mechanism. In this report, it is shown that p48 displays a vesicular localization pattern in transfected cells when fused to the fluorescent reporter EYFP. A predicted transmembrane domain at the C terminus of p48 was not necessary for the observed localization pattern, but this domain was sufficient to redirect localization of EYFP to a fluorescent pattern consistent with the Golgi apparatus. A yeast two-hybrid screen identified the SNARE regulator vesicle-associated membrane protein-associated protein A (VAP-A) as a binding partner of p48. Biochemical assays confirmed that p48 and VAP-A interact and form a stable complex in mammalian cells. Furthermore, expression of the vesicular stomatitis virus G glcyoprotein on the cell surface was inhibited when cells coexpressed p48, suggesting that p48 disrupts intracellular protein trafficking.
Norwalk virus (NV) is a reference strain of human calicivirus that belongs to the Norovirus genus of the Caliciviridae family. Molecular diagnostic assays show that these viruses are the most important agents of epidemic outbreaks of foodborne viral gastroenteritis worldwide (12, 38). Despite significant advances in the last decade in understanding the epidemiology of human calicivirus infections, much less information is available concerning basic viral replication strategies because of the lack of a cell culture system to propagate infectious virus. The NV genome is a single-stranded positive-sense RNA approximately 7.6 kb in length that encodes three open reading frames (5, 19). ORF1 encodes a 1,789-amino-acid polyprotein that is cleaved by a viral protease into nonstructural proteins necessary for RNA genome replication and generation of progeny virus. The major capsid protein VP1 is encoded in ORF2, and the minor structural protein VP2 is encoded in ORF3.
The nonstructural proteins encoded in the calicivirus ORF1 first were predicted based on sequence similarities with picornavirus nonstructural proteins (25). Amino acid sequence motifs in common with the poliovirus 2C NTPase, 3C protease, and 3D RNA-dependent RNA polymerase (RdRp) were readily identified and provided templates for further characterization of the calicivirus nonstructural proteins. Proteolytic mapping and enzymatic studies of in vitro-translated polyprotein or recombinant protein expression have confirmed the presence of an NTPase (p41), a 3C-like protease (3CLpro), an RdRp, and the location in the polyprotein of the genome-linked protein VPg (3, 9, 22, 28, 34). The proposed six nonstructural proteins encoded in the norovirus ORF1 defined so far proceed N to C terminus, p48-NTPase-p22-VPg-3CLpro-RdRp.
p48 consists of the N-terminal 398 amino acids (numbering from the first in-frame methionine), as defined by in vitro proteolytic processing maps (16, 22). p48 occupies a position in ORF1 that is comparable to the 2A/2B proteins of picornavirus genomes, though there is not sufficient sequence relatedness to suggest that these proteins have similar functions. In contrast to sequence conservation that allowed functional predictions for other NV nonstructural proteins, p48 shows no significant sequence similarity to any protein, viral or cellular, in the public databases that would suggest its function in the replication cycle. Of interest, Hughes and Stanway noted the presence of H-Box/NC sequence motifs in p48 of noroviruses and analogous proteins of human parechoviruses (17). Such motifs have been found in a family of cellular proteins that include H-rev107 (15) and TIG3 (7), both suggested to be involved in control of cell proliferation.
This study was undertaken to characterize the unique NV nonstructural protein p48. The data show that p48 localizes to intracellular vesicles in COS-7 cells when expressed transiently as an EYFP fusion protein. Characterization of a predicted transmembrane (TM) domain at the C terminus of p48 showed that this domain is not required for the localization pattern of p48 but is sufficient to alter localization of a heterologous fluorescent reporter. The vesicle-associated membrane protein (VAMP)-associated protein VAP-A was identified as a p48 binding partner. VAP-A is a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-binding protein shown to play a role in regulated vesicle transport (40). Finally, in cells expressing EYFP-p48, expression of the vesicular stomatitis virus (VSV) G glycoprotein at the cell surface was inhibited, suggesting that p48 disrupts intracellular protein trafficking.
MATERIALS AND METHODS
Yeast two-hybrid screen.
A yeast two-hybrid screen was performed with an oligo-dT-primed MA104 cell cDNA library cloned in the activation domain vector pGADT7 (Clontech). Construction of the library has been described previously (13). p48 was cloned into the pGBKT7 DNA binding domain vector by PCR from a full-length NV cDNA template, pSPNV-F (16), with oligonucleotides 48NEXPF6 (NdeI) (5′-GATCCATATGGCGTCAAAAGACGTC-3′) and 48NEXPR3 (5′-CACGGATCCTCCCTGTAGATGGAAATCTGG-3′), to generate pGB-p48. Restriction enzyme sites are underlined. pGB-p48 was transformed into Saccharomyces cerevisiae AH109 cells by the lithium acetate/PEG method as previously described (11).
pGB-p48 yeast cells (6 × 109) were transformed with 120 μg of the MA104 cDNA library by the lithium acetate-PEG procedure. Transformations were plated on synthetic complete medium lacking leucine, tryptophan, histidine, and adenine (SC -L-W-H-A) and cultured for 2 to 4 days at 30°C. Isolated colonies were streaked onto the same nutrient-deficient medium, and positive interactions were scored by the ability of yeast to grow on SC −L-W-H-A medium and to metabolize the chromogenic substrate 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside. Activation domain plasmids with cDNAs encoding potential interactors were recovered from yeast as described previously (11), and cDNA inserts were sequenced on an ABI 310 genetic analyzer with BigDye Terminator chemistry.
Plasmids.
pEYFP-p48 was generated by PCR amplification of sequences encoding p48 from pSPNV-F (16) with sense primer 48NEXPF5 (5′-CCACTCGAGTGATGGCGTCAAAAGACGTC-3′) and antisense primer 48NEXPR3. The PCR fragment was cloned into pEYFP-C1 (Clontech) by standard molecular cloning methods. pEYFP-p48ΔTM encodes amino acids 1 to 360 of the p48 sequence and was constructed with a strategy similar to construction of pEYFP-p48, except that the antisense primer for PCR amplification was 48NEXPR4 (5′-CACGGATCCCGTCCAATCACAGTTTGAAAG-3′).
pEYFP-TM contains the coding sequence of EYFP with amino acids 356 to 398 containing the putative TM domain of p48 fused to the C terminus. pEYFP-TM was constructed with a strategy similar to that for pEYFP-p48, except that the forward primer used in the PCR amplification was 48NTMF2 (5′CCACTCGAGCAAACTGTGATTGGACCGTTC-3′).
pcVAP-A and pGEX-VAP-A were kindly provided by W. S. Trimble (Hospital for Sick Children, Toronto, Ontario, Canada). pcVAP-A encodes VAP-A fused to a c-Myc epitope tag in the plasmid pcDNA3.1(+) (10). pGEX-VAP-A encodes VAP-A fused C-terminally to glutathione S-transferase (GST) to generate a GST-VAP-A fusion protein (10).
pcVSV-G encodes the vesicular stomatitis virus G protein (VSV G) fused to the C-terminal Myc epitope tag in the pcDNA3.1+ vector (Invitrogen). The VSV G gene (Indiana serotype) was amplified by PCR from pTF-G (27) with sense primer VSVG-F (5′-CAGGAATTCACTATGAAGTGCCTTTTG-3′) and antisense primer VSVG-R (5′-CCACTCGAGCTTTCCAAGTCGGTTCAT-3′) and cloned into pcDNA3.1(+) by standard methods.
Transfections and protein analyses.
COS-7 cells were maintained in Dulbecco's modified Eagle's F12 medium supplemented with 5% fetal bovine serum (Atlanta Biologicals). Transfections for biochemical analyses were performed with Lipofectamine (Invitrogen) according to the manufacturer's instructions. Two micrograms of plasmid DNA was transfected into COS-7 cells in six-well plates by mixing the DNA with 10 μl of Lipofectamine reagent in 200 μl of serum-free medium. The transfection mixture was incubated for 45 min at room temperature and then diluted with 800 μl of serum-free medium and added to COS-7 cells. This suspension was replaced after 2 h with 3 ml of medium containing 5% fetal bovine serum. Twenty-four hours posttransfection, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and harvested by scraping into 150 μl of RIPA buffer (150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 10 mM Tris-HCl [pH 7.2]) supplemented with 5 μg of leupeptin/ml and 10 μg of aprotinin/ml (Sigma). Protein expression in transfected cells was analyzed by Western immunoblotting. EYFP fusion proteins were detected with a rabbit polyclonal antibody against green fluorescent protein (GFP) (Abcam) at a dilution of 1:10,000. Myc-tagged VAP-A was detected with an anti-c-Myc monoclonal antibody (Clontech) at a dilution of 1:2,500. Alkaline phosphatase-conjugated secondary antibodies were purchased from Southern Biotechnology Associates.
Transfections in COS-7 cells cultured in four-well microslides for fluorescence microscopy were performed as described above, except that the amounts of transfected DNA and transfection reagent were reduced to 1 μg and 5 μl, respectively, in 100 μl of serum-free medium. Twenty-four hours posttransfection the cells were washed twice with ice-cold PBS and fixed for 20 min with 2% paraformaldehyde. Fixed cells were permeabilized for 5 min with 0.1% Triton X-100 in PBS at room temperature prior to examination with a Nikon Eclipse TE300 microscope.
GST pull-down assays.
GST-VAP-A expression was induced in BL21(DE3) cells cultured to an optical density at 600 nm of 0.6 with 1 mM isopropyl-β-d-thiogalactopyranoside for 2 h at 37°C. Bacteria were collected by centrifugation and suspended in buffer containing 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 0.5% Triton X-100, and 100 μg of lysozyme/ml. A significant proportion of GST-VAP-A was expressed as soluble protein and was purified by methods previously described (6).
p48 mRNA was transcribed from linearized pGB-p48 template with the MEGAscript transcription system (Ambion) following procedures recommended by the manufacturer. In vitro-transcribed p48 mRNA was translated in commercial rabbit reticulocyte lysates under standard reaction conditions recommended by the supplier (Promega) to generate 35S-labeled p48.
GST pull-down assays were performed as previously described with modifications (13). 35S-labeled p48 was mixed with glutathione Sepharose 4B beads bound to GST-VAP-A or GST alone, and the mixtures were rotated end-over-end for 2 h at 4°C. The beads were collected by centrifugation for 5 min at 500 × g at 4°C and then washed five times with buffer containing 40 mM HEPES (pH 7.5), 0.1% Nonidet P-40, 1 mM EDTA, and 100 mM KCl. Bound proteins were eluted by boiling for 5 min in SDS gel-loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol). Proteins were separated by SDS-10% PAGE and visualized by autoradiography.
Coimmunoprecipitations.
COS-7 cells were cotransfected with pEYFP-p48 or pEYFP-p48ΔTM and with pcVAP-A. Twenty-four hours posttransfection, the cells were washed with PBS and harvested by gentle scraping into 1 ml of buffer containing 50 mM Tris-Cl (pH 8.0), 1 mM EDTA, 120 mM NaCl, and 0.5% Nonidet-P40, supplemented with 5 μg of leupeptin/ml and 10 μg of aprotinin/ml. Cell lysates were precleared with 20 μl of fixed Staphylococcus protein A for 1 h at 4°C. Myc-tagged VAP-A was immunoprecipitated from the precleared cell lysate by incubation for 2 h at 4°C with 5 μg of c-Myc monoclonal antibody. Antigen-antibody complexes then were incubated for 1 h at 4°C with 50 μl of Staphylococcus protein A. The immunoprecipitate was collected by centrifugation and washed four times with NETN buffer (20 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5% Nonidet-P40) containing 900 mM NaCl and then once with NETN buffer supplemented with 100 mM NaCl. Immune complexes were released by boiling for 5 min in 1× SDS gel-loading buffer. Immunoprecipitated proteins were separated by SDS-PAGE and then transferred to nylon membranes and probed with the anti-GFP rabbit polyclonal antibody. The blots were developed with an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G at a dilution of 1:1,000, and proteins were visualized with chromogenic reagents.
Cellular fractionations.
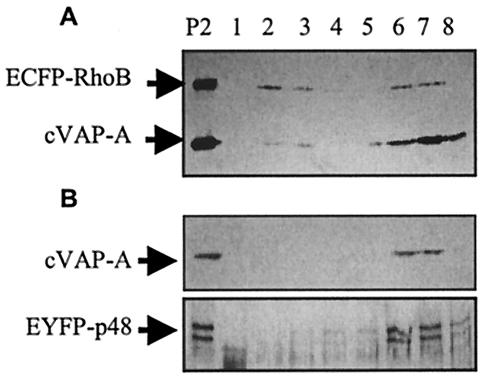
Cellular fractionations were performed by methods described previously (4, 20). COS-7 cells were transfected with plasmids indicated by the experiment and 20 μl of Lipofectamine in 60-mm-diameter cell culture dishes. All fractionation steps were carried out on ice. Twenty-four hours posttransfection, the cells were washed twice with ice-cold PBS and then harvested by gentle scraping into 1 ml of fractionation lysis buffer containing 10 mM HEPES (pH 7.4), 250 mM sucrose, 1 mM EDTA, 5 μg of leupeptin/ml, and 10 μg of aprotinin/ml. The cell suspension was sonicated for three 10-s pulses. Nuclei and unbroken cells were sedimented by centrifugation for 10 min at 800 × g, and the supernatant was ultracentrifuged for 30 min at 46,000 × g (P2) to enrich for large vesicles, including endoplasmic reticulum (ER) and Golgi vesicles, and endosomes. The P2 pellet was suspended in 1 ml of fractionation lysis buffer and was fractionated on 25 to 55% discontinuous sucrose gradients centrifuged for 15 h at 109,000 × g in an SW55 rotor. Eight 640-μl fractions were collected, and proteins in each fraction were separated by SDS-10% PAGE and analyzed by Western immunoblotting. Myc-tagged VAP-A and ECFP-RhoB were detected with the c-Myc monoclonal antibody at a dilution of 1:2,500. EYFP-p48 was detected with a GFP polyclonal antibody.
Membrane protein extractions.
Sodium carbonate extractions were performed as previously described (4). Cells transfected with indicated plasmids were suspended in 0.25 M sucrose-5 mM HEPES (pH 6.8) and disrupted with 30 strokes of a Dounce homogenizer. Intact cells and nuclei were removed by centrifugation for 5 min at 800 × g in a Beckman GH-3.7 rotor. The supernatant was ultracentrifuged for 30 min at 40,000 rpm in a Beckman SW55 rotor. The membrane pellet then was homogenized in 5 ml of 100 mM sodium carbonate (pH 11.3) with 10 strokes of a Dounce homogenizer and then incubated for 30 min on ice. The extracted proteins were separated from membranes by ultracentrifugation as before. The membrane pellet was again suspended in 1 ml of 100 mM sodium carbonate and neutralized to pH 7.0 by addition of 1 N HCl. Extracted proteins were separated by SDS-PAGE and then detected by Western blotting as described above. Treatment of cell lysates with Triton X-100 or Triton X-114 was conducted following the same protocol, except that the membrane pellets were homogenized in 1% of the indicated detergent and the neutralization step was omitted.
Indirect immunofluorescence.
Cells were grown on four-well slides and transfected with the appropriate constructs. Transfections, cell fixation, and permeabilization were performed as described above. Nonspecific protein binding was blocked by a 20-min incubation in PBS containing 0.5% bovine serum albumin, and Myc-tagged VSV G was detected with c-Myc monoclonal antibody followed by tetramethyl rhodamine isocyanate-conjugated goat anti-mouse immunoglobulin G. Images were captured on a Nikon Eclipse TE300 fluorescence microscope.
Glycosidase digestions.
COS-7 cells were cotransfected with pEYFP or pEYFP-p48 and with pcVSV-G. Twenty-four hours posttransfection, cells were labeled with 40 μCi of TRANS 35S-label (ICN) for 2 h and then chased in medium containing 400× unlabeled methionine and cysteine for another 2 h. Myc-tagged VSV G protein was immunoprecipitated from cell lysates with c-Myc monoclonal antibody as described above. Immune complexes were boiled for 10 min in the presence of denaturing buffer containing 0.5% SDS and 1% β-mercaptoethanol and centrifuged briefly, and the clear supernatant was divided in two equal aliquots. One aliquot of each IP reaction was treated with 500 U of either endoglycosidase H or PNGase F (New England Biolabs) for 5 h at 37°C. Proteins were evaluated by SDS-10% PAGE followed by autoradiography.
RESULTS
NV p48 displays a vesicular staining pattern in transfected COS-7 cells.
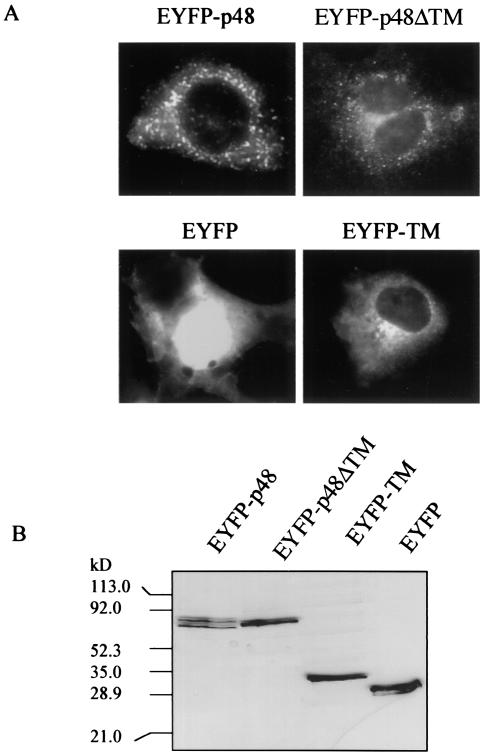
Amino acid sequence analyses of p48 predicted a C-terminal TM domain between amino acids 360 and 379. p48 is the least-conserved protein in ORF1 when human calicivirus strains are compared, yet inspection of multiple sequence alignments of p48 from noroviruses for which sequence is available indicated that this putative TM region was the most conserved (data not shown). Conservation of this region across a protein with low overall sequence similarity suggests that this predicted domain may be functional. We first examined whether the presence of the putative TM domain correlated with a membrane localization pattern in cells expressing p48. COS-7 cells were transfected with the fusion reporter pEYFP-p48. Fluorescent images observed 24 h posttransfection showed a vesicular pattern of expression throughout the cytoplasm and some indication of surface expression, though apparent surface expression was not consistently observed in all cells (Fig. 1A). To determine if the predicted TM domain directed localization of EYFP-p48 to these vesicular structures, we analyzed localization of EYFP-p48ΔTM which does not contain C-terminal amino acids 360 to 398. Expression of EYFP-p48ΔTM in transfected COS-7 cells showed a localization pattern similar to that observed with the full-length protein with respect to a vesicle pattern of fluorescence (Fig. 1A). This observation suggested that this small C-terminal domain was not required for the localization of EYFP-p48 that could be determined at this resolution. However, we extended our analysis of the putative TM domain by fusing amino acids 356 to 398 of p48 to the C terminus of EYFP to determine if this domain alone could confer an expression pattern to EYFP similar to that of EYFP-p48. EYFP alone showed a strong nuclear and diffuse cytoplasmic fluorescence, typical of the expression pattern of GFP and its color-shifted derivatives (Fig. 1A). Fusion of the putative TM domain to the C terminus of EYFP was sufficient to redirect the fluorescent pattern of EYFP to a strong perinuclear distribution consistent with a Golgi staining pattern (Fig. 1A). Taken together, the data suggest that the C-terminal 42 amino acids constituting a putative TM domain of p48 are not necessary for localization of full-length p48 but are sufficient to redirect localization of a heterologous protein in transfected cells.
FIG. 1.
Expression of EYFP-p48 and EYFP-p48ΔTM in COS-7 cells. (A) Cells were transfected with pEYFP-p48, pEYFP-p48ΔTM, or pEYFP-TM and observed 24 h posttransfection. Magnification is ×400. (B) Cell lysates prepared from COS-7 cells transfected with the indicated plasmid were separated by SDS-10% PAGE and subjected to Western immunoblotting with anti-GFP-polyclonal antibody.
Expression of EYFP-p48, EYFP-p48ΔTM, and EYFP-TM in transfected cells was analyzed by Western immunoblotting with an anti-GFP polyclonal antibody. EYFP-p48 was present in two forms with apparent molecular masses of 77 and 72 kDa (Fig. 1B). In contrast, EYFP-p48ΔTM was present in a unique form of 75 kDa. The calculated molecular mass of EYFP-p48 is approximately 73 kDa and that of EYFP-p48ΔTM is approximately 69 kDa. These estimations suggest that the highest-molecular-mass band of the two forms of EYFP-p48 is likely the full-length protein, since several assays for posttranslational modifications including oligosaccharide addition and phosphorylation, were negative (data not shown). The composition of the lower-molecular-mass band detected in cells expressing EYFP-48 currently is not clear, but it may be a cleavage product of full-length protein p48.
p48 binds the SNARE-binding protein VAP-A.
The distinct localization patterns of EYFP-p48 and that of EYFP-TM led us to hypothesize that localization of p48 may be determined through its interaction with a cellular protein. We screened an epithelial cell cDNA library in the yeast two-hybrid interaction trap to identify cellular proteins that interacted with p48. One hundred seventy positive clones were obtained in the two-hybrid screen, and 20 of the cDNAs were sequenced. Sequence analysis revealed five clones with 98% identity across the first 372 nucleotides/126 amino acids (98% nucleotide and 97% amino acid identity) to VAMP-associated protein VAP-A (formerly hVAP-33) (40). VAP-A is the human ortholog of Aplysia californica VAP-33, a protein originally defined through its ability to bind to the vesicle SNAREs (v-SNAREs) VAMP1 and VAMP2 in vitro (33). A. californica VAP-33 is neuron specific and functions in neuronal exocytosis (33). VAP-33 orthologs have since been reported in mouse (32), rat (26), and human (26, 40) and, in contrast to A. californica VAP-33, show more ubiquitous tissue distributions. These proteins play a role in SNARE-mediated vesicle trafficking, though the precise mechanism of how VAP-A regulates vesicle transport is not well understood.
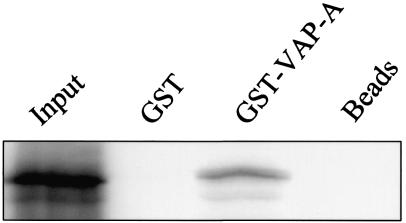
A GST pull-down assay was performed to assess whether the interaction between p48 and VAP-A detected by the two-hybrid assay also occurred in a complementary in vitro assay. VAP-A was expressed as a GST fusion protein in bacteria and tested for its ability to bind p48 that was synthesized in rabbit reticulocyte lysates. p48 bound to GST-VAP-A but not to GST or to beads alone (Fig. 2). These data suggested that the p48-VAP-A interaction was not unique to the yeast two-hybrid assay and led us to further investigate the interaction in mammalian cells.
FIG. 2.
In vitro-translated p48 binds GST-VAP-A in a pull-down assay. 35S-p48 was incubated with glutathione-Sepharose 4B beads only, bead-immobilized GST, or GST-VAP-A. The bound radiolabeled protein was eluted and analyzed by SDS-10% PAGE and autoradiography. The amount of 35S-p48 in the Input lane represents the total amount of protein added to each pull-down reaction.
p48 and VAP-A form a stable complex in transfected COS-7 cells.
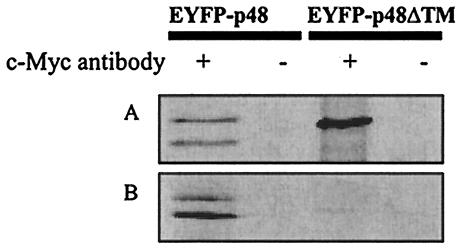
To determine if p48 and VAP-A interacted in mammalian cells, COS-7 cells were cotransfected with plasmids encoding EYFP-p48 and Myc-tagged VAP-A. Immunoprecipitation of transfected cell lysates with an anti-c-Myc monoclonal antibody, followed by Western immunoblotting with an anti-GFP antibody, showed that both forms of EYFP-p48 coimmunoprecipitated with the Myc-tagged VAP-A (Fig. 3A). Similarly, EYFP-p48ΔTM also coimmunoprecipitated with Myc-tagged VAP-A, suggesting that the interacting domain on p48 was upstream of amino acid 360. These data are consistent with the hypothesis that a cellular protein directs localization of p48. Neither EYFP-p48, EYFP-p48ΔTM, nor EYFP was detected when these plasmids were transfected into cells in the absence of Myc-tagged VAP-A (data not shown).
FIG. 3.
EYFP-p48 and EYFP-p48ΔTM coimmunoprecipitate with Myc-tagged VAP-A. (A) EYFP-p48 or EYFP-p48ΔTM and Myc-tagged VAP-A were coexpressed in COS-7 cells. Twenty-four hours posttransfection, the cell lysates were subjected to immunoprecipitation in the presence or the absence of anti-c-Myc monoclonal antibody. The immunoprecipitates were separated by SDS-PAGE and then subjected to immunoblotting with rabbit anti-GFP antibody. (B) pEYFP-p48 or pEYFP-p48ΔTM and pcVAP-A were transfected in separate cultures of COS-7 cells. The cell lysates were mixed and then subjected to immunoprecipitation as described above.
The coimmunoprecipitation data suggested that p48 interacted with VAP-A in cotransfected cells. To determine whether the interaction required coexpression or if the observed interaction between EYFP-p48 (or EYFP-p48ΔTM) and Myc-tagged VAP-A occurred subsequent to cell lysis, COS-7 cells were transfected either with pEYFP-p48 or with pcVAP-A, and then the lysates were mixed prior to immunoprecipitation with the anti-c-Myc monoclonal antibody. Under these conditions, both forms of EYFP-p48 coimmunoprecipitated with Myc-tagged VAP-A (Fig. 3B). In contrast, scarce but detectable amounts of EYFP-p48ΔTM were coimmunoprecipitated with Myc-tagged VAP-A. These data suggest that the TM domain of p48 does play some role in the interaction with VAP-A, but this role extends beyond a simple protein-protein interaction that could occur in mixed cell lysates.
EYFP-p48 behaves as an integral membrane protein.
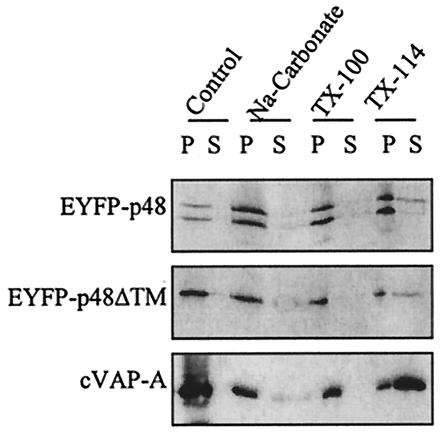
Type II integral membrane proteins are anchored in lipid bilayers through TM domains located at the C terminus, and the majority of the protein is cytoplasmic (39). VAP-A and SNAREs are type II or tail-anchored membrane proteins (20). Given the prediction of a C-terminal TM domain in p48 and its interaction with VAP-A, we examined whether p48 also was a tail-anchored integral membrane protein. Separation of cellular fractions showed both EYFP-p48 and EYFP-p48ΔTM in membrane fractions (Fig. 4). Treatment of cell lysates with sodium carbonate (pH 11.3), which releases peripheral, but not integral, membrane proteins, did not significantly release EYFP-p48 or EYFP-p48ΔTM from membrane pellets, nor did treatment with Triton X-100 (Fig. 4). Similar results were obtained for VAP-A. The only treatment that more conclusively released EYFP-p48, EYFP-p48ΔTM, and VAP-A was Triton X-114. This extraction pattern is consistent with that reported previously for VAP-A (20). These data suggested that p48 is an integral membrane protein. However, the behavior of EYFP-p48ΔTM in these experiments complicates a straightforward interpretation of the data. EYFP-p48ΔTM has a deletion of the putative C-terminal TM domain, and no other potential TM domains were predicted across the length of the protein. Both EYFP-p48 and EYFP-p48ΔTM behaved as integral membrane proteins in extraction experiments, evidenced by susceptibility to membrane release only by Triton X-114. One possibility as suggested before is that p48 and VAP-A form a stable membrane complex that is independent of the predicted TM domain in p48, and thus, the behavior of both EYFP-p48 and EYFP-p48ΔTM in extraction experiments is dictated by the integral membrane properties of VAP-A. This possibility is supported by the coimmunoprecipitation and fluorescence microscopy data, which show that the TM domain is not necessary for interaction with VAP-A when these two proteins are coexpressed.
FIG. 4.
p48 behaves as an integral membrane protein in COS-7 cells. Lysates of COS-7 cells transfected with EYFP-p48, EYFP-p48ΔTM, or VAP-A were treated with sodium carbonate, Triton X-100, or Triton X-114, as described in Materials and Methods. Proteins in the Control lane received no treatment. Pellets (P) and supernatants (S) from each sample were separated by SDS-PAGE, followed by Western blotting with the appropriate antibody.
EYFP-p48 and VAP-A cofractionate with large Golgi vesicles and endosomes in transfected COS-7 cells.
Replication of viruses with positive-strand RNA genomes occurs anchored to cellular membrane structures. Poliovirus and feline calicivirus infections both cause reorganization of cellular membranes and accumulation of intracellular vesicles (2, 23, 35, 36), presumably to provide structures competent to promote productive replication complex assembly. VAP-A localizes to the ER-Golgi intermediate compartment in COS-7 cells (41), and we have shown an interaction between VAP-A and p48. We analyzed the distribution of VAP-A and EYFP-p48 by subcellular fractionation to determine the nature of the endomembrane structures suggested by fluorescence microscopy. As a cellular marker, we cotransfected cells with the fluorescent reporter pECFP-Endo (hereafter referred to as pECFP-RhoB). This plasmid contains the coding sequence of the RhoB GTPase fused to ECFP. It originally was reported that RhoB localized to late endosomes (1), but subsequent studies showed that RhoB localized to the plasma membrane, Golgi, and peri-Golgi vesicles (24). Figure 5 shows the distribution of EYFP-p48, VAP-A, and ECFP-RhoB in fractionated sucrose gradients. All three of these proteins were found in fractions containing endosomes and ER and Golgi vesicles, suggesting that the fluorescent pattern of EYFP-p48 accurately reflects an association with vesicles.
FIG. 5.
p48 and VAP-A cofractionate with ECFP-RhoB. COS-7 cells transfected with pcVAP-A and pEYFP-p48 (A) or pcVAP-A and pECFP-RhoB (B) were subjected to subcellular fractionation on sucrose gradient as described in Materials and Methods. Sucrose gradient fractions were analyzed by Western immunoblotting with c-Myc monoclonal antibody for cVAP-A and ECFP-RhoB and GFP polyclonal antibody for EYFP-p48. Sedimentation is from left to right (lanes 1 to 8). The lane labeled P is the membrane pellet not subjected to fraction procedures.
Trafficking of VSV G protein to the cell surface is interrupted by p48 expression.
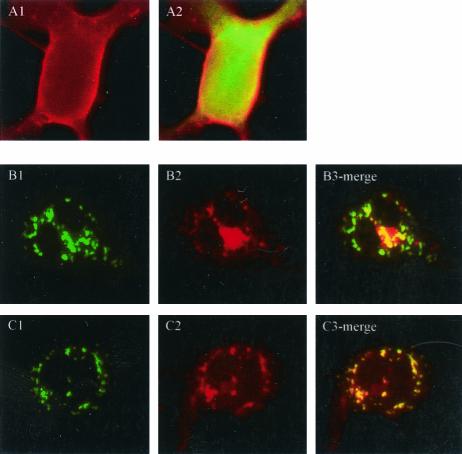
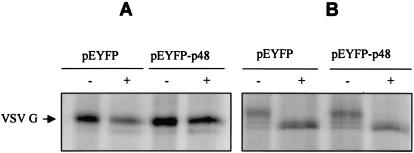
The precise mechanistic role of VAP-A in regulation of membrane trafficking through its interactions with SNAREs is not clear. Several studies have suggested a role for VAP-A in regulation of vesicle transport (10), neurotransmitter release (33), and interaction with microtubule network and tight junctions (20, 32). The interaction between p48 and VAP-A led us to analyze localization of the G protein of VSV in the presence and in the absence of p48. VSV G is a glycoprotein used extensively to study secretory protein trafficking (21). VSV G successfully reaches the outer membranes of transfected cells in the absence of EYFP-p48 (Fig. 6A1 and A2). In contrast, in the presence of EYFP-p48, VSV G protein shows a disrupted localization pattern that in some areas overlaps that of EYFP-p48 (Fig. 6B). Similar results were obtained when VSV G was expressed in the presence of EYFP-p48ΔTM except that, interestingly, colocalization was more extensive than that observed with EYFP-p48 and was nearly complete (Fig. 6C). Coexpression of VSV G and EYFP had no effect on VSV G trafficking (Fig. 6A2). These data suggested that p48 disrupts intracellular protein trafficking that normally would result in cell surface expression of VSV G. To determine at what step trafficking of VSV G to the cell surface was blocked, cells were cotransfected with VSV G in the presence and in the absence of EYFP-p48. 35S-labeled VSV G was immunoprecipitated and subjected to digestion with endoglycosidase H (Fig. 7A) or with PNGase F (Fig. 7B). VSV G expressed in the presence of EYFP-p48 was endoglycosidase H resistant but sensitive to digestion with PNGaseF. These data show that VSV G reaches the Golgi, and therefore, the block to cell surface expression must occur at a post-Golgi step in the protein trafficking pathway.
FIG. 6.
Expression of EYFP-p48 prevents cell surface expression of VSV G protein. COS-7 cells were transfected with pcVSV-G and pEYFP (A), pcVSV-G and pEYFP-48 (B), or pcVSV-G and pEYFP-p48ΔTM (C). VSV G was detected 24 h posttransfection by indirect immunofluorescence with an anti-c-Myc monoclonal antibody, followed by tetramethyl rhodamine isocyanate-conjugated secondary antibody. Magnification is ×400.
FIG. 7.
VSV G protein acquires endoglycosidase H resistance in the presence of EYFP-p48. COS-7 cells were cotransfected with pEYFP or pEYFP-p48 and pcVSV-G. The cells were labeled for 2 h with TRANS 35S-label followed by a 2-h chase period. VSV G protein was immunoprecipitated from cell lysates with c-Myc monoclonal antibody and treated with either endoglycosidase H (A) or PNGase F (B).
DISCUSSION
We have shown that the nonstructural protein p48 of NV, when expressed in cells as a fusion protein, targets to intracellular vesicles, forms a complex with the SNARE binding protein VAP-A, and disrupts trafficking of the VSV G protein to the cell surface. The C-terminal 42 amino acids of p48 that include a predicted TM domain between amino acids 360 and 379, though not necessary for vesicle localization or for VAP-A interaction, are sufficient to redirect localization of the heterologous fluorescent reporter EYFP to structures consistent with the Golgi apparatus. The inability to examine the role of p48 in an NV-infected cell precludes a direct assessment of the function of this protein in the NV replication cycle. However, some predictions can be made based on the data presented here, interpreted in the context of what is known for other RNA viruses.
p48 forms a complex with the SNARE binding protein VAP-A. SNAREs function in intracellular protein trafficking by mediating fusion events between vesicles and target membranes (18). Though several functions of VAP-A have been suggested, its precise role in vesicle trafficking with regard to its interactions with SNARE molecules is not well understood. VAP-A has been localized to the ER-Golgi intermediate compartment in nonpolarized cells, and VAP-A binds promiscuously to the VAMPs and to SNARE proteins involved in ER-to-Golgi trafficking, including Rbet1 and Rsec22 (41). Based on the number of interactions between VAP-A and SNARE proteins, several possible roles for VAP-A in vesicle fusion were suggested (41). VAP-A could mediate fusion between vesicles and target membranes. Alternatively, VAP-A could function in a regulatory capacity in vesicle docking and fusion, as opposed to a mechanical role in the fusion event itself. In support of the latter idea, overexpression of VAP-A prevented export of the insulin-dependent glucose transporter 4 to the cell surface in L6 myoblasts, and cooverexpression of VAMP-2 relieved the inhibition, suggesting that levels of VAP-A available for SNARE protein interactions are critical for vesicle trafficking (10). We have shown that in COS-7 cells expressing p48, VSV G transport to the cell surface is inhibited. As a potential mechanism analogous to that suggested for regulated glucose transporter 4 trafficking, p48 could titrate VAP-A into complexes that are incapable of regulating or mediating vesicle fusion, resulting in disruption of the cellular trafficking pathway. Alternatively, if VAP-A does play an active role in SNARE-mediated membrane fusion, p48 may interfere with this process by competing with cellular partners of VAP-A for binding. Subcellular fractionation assays showed EYFP-p48 cofractionated with ECFP-RhoB, which is known to be associated with the Golgi and peri-Golgi vesicles (24). The modification of VSV G to an endoglycosidase H-resistant form when expressed in the presence of EYFP-p48 suggests that trafficking of VSV G is disrupted at a post-Golgi step. These data are consistent with the cosedimentation of EYFP-p48 with ECFP-RhoB in subcellular fractionation experiments. Ongoing studies will determine if the interaction between p48 and VAP-A is directly responsible for the disruption in protein trafficking observed here.
Poliovirus infection inhibits the cell secretory pathway, and nonstructural proteins 2B and 3A are sufficient to mediate this effect (8). Though the mechanism of how protein trafficking in cells expressing EYFP-p48 is inhibited is not yet known, it is tempting to suggest a common mechanism shared between poliovirus and the human caliciviruses. Interference with SNARE-mediated vesicle docking and fusion, or with the functions of cellular proteins that regulate bidirectional vesicle trafficking, may illustrate a common pathway even if the precise mechanism by which this interference is manifested is different. Both 2B and 3A of poliovirus inhibit secretory vesicle trafficking, yet they do so by different mechanisms (8). The suggestion was made that one mechanism may involve interference with, or interaction with, a cellular protein involved in membrane fusion, a prediction that is at least partially supported by the data shown here for p48 interactions with VAP-A.
Replication of positive-strand RNA viruses occurs anchored to the cytoplasmic face of cellular membranes (2, 29, 31). How these complexes are assembled on specific membranes into functional replication machinery is not completely understood, but it may involve selective targeting by viral proteins. Poliovirus 2BC was consistently present in ER-derived vesicles containing Sec13 and Sec31 components of COPII coats, and it therefore was suggested that 2BC may function as an anchoring or targeting protein for poliovirus replication complexes to these specific vesicle structures (30). Likewise, Shaad and colleagues (31) have determined that the 6-kDa protein of tobacco etch virus is sufficient to target a heterologous reporter molecule to vesicle structures in transfected plant cells. Based on these examples, one function of p48 may be to target calicivirus replication complexes to specific membranes. In support of this prediction, Green and colleagues showed the presence of p32, the feline calicivirus p48 analogue, in active membrane-associated replication complexes isolated from feline calicivirus-infected cells (14). The ability of the C-terminal 42 amino acids of p48, which includes the putative TM domain, to localize a heterologous protein to structures consistent with the Golgi apparatus suggests a mechanism by which calicivirus replication complexes may be anchored to cellular membranes. A C-terminal TM domain characteristic of type II integral membrane proteins (39) suggests that p48 is inserted into membranes posttranslationally, with the majority of the protein oriented on the cytoplasmic face. Such an orientation could serve as scaffolding for assembly of functional replication complexes through interactions with other NV nonstructural proteins. Of interest, the hepatitis C virus nonstructural proteins NS5A and NS5B also interact with VAP-A, and it was suggested that VAP-A may serve as a membrane receptor for hepatitis C virus replication complexes (37). This also may be true for NV p48. Ongoing studies are examining the organization and topography of assembly or association of the NV nonstructural proteins with intracellular membranes.
Acknowledgments
This work was supported by Public Health Service grant AI-43450 to M.E.H and the Montana Agricultural Experiment Station (MAES 2003-13).
We extend appreciation to W. S. Trimble for sharing the VAP-A plasmids.
REFERENCES
- 1.Adamson, P., H. F. Paterson, and A. Hall. 1992. Intracellular localization of the p21rho proteins. J. Cell Biol. 119:617-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 3.Boniotti, B., C. Wirblich, M. Sibilia, G. Meyers, H. J. Thiel, and C. Rossi. 1994. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J. Virol. 68:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrere-Kremer, S., C. Montpellier-Pala, L. Cocquerel, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 76:3720-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, I. N., and P. R. Lambden. 2000. Organization and expression of calicivirus genes. J. Infect. Dis. 181(Suppl. 2):S309-S316. [DOI] [PubMed] [Google Scholar]
- 6.Daughenbaugh, K., C. S. H. J. W. Fraser, and M. E. Hardy. 2003. The genome linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 22:2852-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiSepio, D., C. Ghosn, R. L. Eckert, A. Deucher, N. Robinson, M. Duvic, R. A. Chandraratna, and S. Nagpal. 1998. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc. Natl. Acad. Sci. USA 95:14811-14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunham, D. M., X. Jiang, T. Berke, A. W. Smith, and D. O. Matson. 1998. Genomic mapping of a calicivirus VPg. Arch. Virol. 143:2421-2430. [DOI] [PubMed] [Google Scholar]
- 10.Foster, L. J., M. L. Weir, D. Y. Lim, Z. Liu, W. S. Trimble, and A. Klip. 2000. A functional role for VAP-33 in insulin-stimulated GLUT4 traffic. Traffic 1:512-521. [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and R. A. Woods. 1998. Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method, p. 53-66. In A. J. P. Brown and M. F. Tuite (ed.), Methods in microbiology. Academic Press, San Diego, Calif.
- 12.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 13.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. F. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, K. Y., A. Mory, M. H. Fogg, A. Weisberg, G. Belliot, M. Wagner, T. Mitra, E. Ehrenfeld, C. E. Cameron, and S. V. Sosnovtsev. 2002. Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J. Virol. 76:8582-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajnal, A., R. Klemenz, and R. Schafer. 1994. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene 9:479-490. [PubMed] [Google Scholar]
- 16.Hardy, M. E., T. J. Crone, J. E. Brower, and K. Ettayebi. 2002. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 89:29-39. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, P. J., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 81:201-207. [DOI] [PubMed] [Google Scholar]
- 18.Jahn, R., and T. C. Sudhof. 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68:863-911. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 20.Lapierre, L. A., P. L. Tuma, J. Navarre, J. R. Goldenring, and J. M. Anderson. 1999. VAP-33 localizes to both an intracellular vesicle population and with occludin at the tight junction. J. Cell Sci. 112:3723-3732. [DOI] [PubMed] [Google Scholar]
- 21.Lippincott-Schwartz, J., T. H. Roberts, and K. Hirschberg. 2000. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 16:557-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love, D. N., and M. Sabine. 1975. Electron microscopic observation of feline kidney cells infected with a feline calicivirus. Arch. Virol. 48:213-228. [DOI] [PubMed] [Google Scholar]
- 24.Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush, and M. R. Philips. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17:145-160. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura, Y., M. Hayashi, H. Inada, and T. Tanaka. 1999. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 254:21-26. [DOI] [PubMed] [Google Scholar]
- 27.Pattnaik, A. K., and G. W. Wertz. 1991. Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc. Natl. Acad. Sci. USA 88:1379-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfister, T., and E. Wimmer. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 75:1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaad, M. C., A. D. Lellis, and J. C. Carrington. 1997. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71:8624-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skehel, P. A., R. Fabian-Fine, and E. R. Kandel. 2000. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl. Acad. Sci. USA 97:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skehel, P. A., K. C. Martin, E. R. Kandel, and D. Bartsch. 1995. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science 269:1580-1583. [DOI] [PubMed] [Google Scholar]
- 34.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 35.Studdert, M. J., and J. D. O'Shea. 1975. Ultrastructural studies of the development of feline calicivirus in a feline embryo cell line. Arch. Virol. 48:317-325. [DOI] [PubMed] [Google Scholar]
- 36.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 38.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 39.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 40.Weir, M. L., A. Klip, and W. S. Trimble. 1998. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem. J. 333:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir, M. L., H. Xie, A. Klip, and W. S. Trimble. 2001. VAP-A binds promiscuously to both v- and tSNAREs. Biochem. Biophys. Res. Commun. 286:616-621. [DOI] [PubMed] [Google Scholar]