Abstract
The highly immunogenic mycobacterial proteins ESAT-6, CFP-10, and HspX represent potential target antigens for the development of subunit vaccines and immunodiagnostic tests. Recently, the complete genome sequence revealed the absence of these coding sequences in Mycobacterium ulcerans, the causative agent of the emerging human disease Buruli ulcer. Genome reduction and the acquisition of a cytopathic and immunosuppressive macrolide toxin plasmid are regarded as crucial for the emergence of this pathogen from its environmental progenitor, Mycobacterium marinum. Earlier, we have shown the evolution of M. ulcerans into two distinct lineages. Here, we show that while the genome of M. marinum M contains two copies of the esxB-esxA gene cluster at different loci (designated MURD4 and MURD152), both copies are deleted from the genome of M. ulcerans strains belonging to the classical lineage. Members of the ancestral lineage instead retained some but disrupted most functional MURD4 or MURD152 copies, either by newly identified genomic insertion-deletion events or by conversions of functional genes to pseudogenes via point mutations. Thus, the esxA (ESAT-6), esxB (CFP-10), and hspX genes are located in hot-spot regions for genomic variation where functional disruption seems to be favored by selection pressure. Our detailed genomic analyses have identified a variety of independent genomic changes that have led to the loss of expression of functional ESAT-6, CFP-10, and HspX proteins. Loss of these immunodominant proteins helps the bacteria bypass the host's immunological response and may represent part of an ongoing adaptation of M. ulcerans to survival in host environments that are screened by immunological defense mechanisms.
The emerging pathogen Mycobacterium ulcerans is the causative agent of Buruli ulcer, a mycobacterial disease of skin and soft tissue with the potential to leave sufferers scarred and disabled. While it is endemic in more than 30 countries (26), the major disease burden falls on children living in poor rural communities of West Africa. Buruli ulcer is prevalent in areas neighboring rivers, slow-flowing waters, and swamps, but the exact mode of transmission has remained elusive. This is partly attributable to a clonal population structure and an associated lack of high-resolution genetic fingerprinting methods for microepidemiologic studies.
M. ulcerans seems to have recently evolved via lateral gene transfer and reductive evolution from the fish disease-causing environmental species Mycobacterium marinum (40, 43). Particularly, it has acquired the virulence plasmid, pMUM001, encoding the genes for the synthesis of the macrolide toxin, mycolactone. This toxin has cytopathic and immunomodulatory properties and plays a decisive role in producing an extracellular infection after an initial phase within macrophages (4, 41, 42, 47). In addition, M. ulcerans has undergone extensive gene loss due to DNA deletions, DNA rearrangements, and pseudogene formation, which apparently drives its evolution toward a niche-adapted specialist (27, 34, 39). Previous findings suggest that M. ulcerans lineages from different geographic areas reveal variations in virulence (27, 32; also F. Portaels, unpublished data).
The ESX-1 secretion system is required for the virulence of Mycobacterium tuberculosis and related pathogenic mycobacteria. It comprises the 6-kDa early secretory antigenic target protein (ESAT-6) and the 10-kDa culture filtrate protein (CFP-10), which are among the strongest T-cell response elicitors in tuberculosis patients (7, 8). The genes encoding these proteins are localized on the region of difference 1 (RD1) locus, which is intact in virulent members of the M. tuberculosis complex but absent from the attenuated vaccine strain Mycobacterium bovis BCG (ΔRD1BCG) (21, 29). Similarly, the vole bacillus, Mycobacterium microti, was found to have a natural deletion (ΔRD1microti) overlapping the deletion ΔRD1BCG (6, 18). The so-called extended RD1 encompasses most of the genes that form the ESX-1 secretion apparatus (7, 16, 17) or are crucial for both ESAT-6/CFP-10 secretion and virulence (7, 17, 19, 31). This secretion apparatus enhances virulence in M. tuberculosis and M. marinum infection by secretion of effector proteins into the cytosol of infected macrophages (37), prevention of phagolysosomal maturation (28, 45), and cytolytic activity (24). On the other hand, infected individuals develop strong T-cell responses against these proteins, which seem to be relevant for immune protection (8). The 16-kDa heat shock protein HspX, or α-crystallin-like protein Acr, a dominant protein expressed during static growth in M. tuberculosis, is required for mycobacterial persistence within the macrophage. HspX is yet another potent immune response elicitor and suitable for detecting M. tuberculosis infection (14, 15, 20, 25, 35, 49).
In mycobacterial disease control, highly antigenic proteins serve both as targets for diagnostic tests and as candidate proteins for vaccine development (1, 8, 30). While being present in the sequenced M. marinum strain M (http://www.sanger.ac.uk/cgi-bin/BLAST/submitblast/m_marinum), genes encoding ESAT-6, CFP-10, and HspX are absent from the genome of the sequenced Ghanaian M. ulcerans strain Agy99 (http://genopole.pasteur.fr/Mulc/BuruList.html). However, earlier data showed that some M. ulcerans isolates and other related mycolactone-producing mycobacteria harbor at least segments of these genes (32, 48). Recently, we have identified two distinct genetic lineages of M. ulcerans, with representatives of the ancestral lineage being phylogenetically closer to its progenitor, M. marinum, than members of the M. ulcerans classical lineage (27). Here, we have analyzed a worldwide collection of M. ulcerans strains belonging to these two lineages for the presence of esxA, esxB, and hspX and their surrounding genomic regions.
MATERIALS AND METHODS
Mycobacterial strains and genomic DNA extraction.
M. marinum strain M was used for interspecies comparison. A worldwide strain collection of M. ulcerans had been used earlier for investigation of genomic strain variations (34). Although several attempts to differentiate these strains achieved only low resolution (2, 3, 11, 22, 23, 38, 44), this collection of patient isolates was shown to be divided in two lineages displaying major genomic differences (27). In this study, we used M. ulcerans clinical isolates of both lineages as follows. For the classical lineage, the following strains were used: Ghana Agy99, Ghana ITM (Institute of Tropical Medicine, Antwerp, Belgium) 970321, Ghana ITM 970359, Ghana ITM 970483, Ivory Coast ITM 940662, Ivory Coast ITM 940815, Ivory Coast ITM 940511, Benin ITM 970111, Benin ITM 940886, Benin ITM 940512, Benin ITM 970104, Democratic Republic of Congo (DRC) ITM 5150, DRC ITM 5151, DRC ITM 5155, Togo ITM 970680, Angola ITM 960657, Angola ITM 960658, Papua New Guinea (PNG) ITM 941331, PNG ITM 9537, Malaysia ITM 941328, Australia ITM 941324, Australia ITM 941325, Australia ITM 941327, Australia ITM 9549, Australia ITM 9550, Australia ITM 8849, Australia ITM 940339, Australia ITM 5142, and Australia ITM 5147. For the ancestral lineage, the following strains were used: China ITM 980912, Japan ITM 8756, French Guiana ITM 7922, Surinam ITM 842, and Mexico ITM 5143. The presence of the specific PCR products obtained with primer pairs CH5/CH4 and CH3/CH4 (that exclude each other by design) (see Fig. 1) occurred concomitantly in strains DRC ITM 5151 and PNG ITM 941331. Since also variable-number tandem repeat typing analysis indicated that these strains are impure, we excluded these strains from further analysis.
FIG. 1.
Confirmation of the MURD-specific deletions affecting esxB (CFP-10) and esxA (ESAT-6) in an M. ulcerans worldwide strain collection. (A) Schematic view of an alignment of M. marinum M and M. ulcerans Agy99 genomic sequences displayed by the Artemis comparison tool (9). Regions of conformity are shown in parallel gray plains, an inverted DNA segment is depicted as an inverted surface, and white areas represent unique sequences like MURD152, which is present only in M. marinum M and is deleted from M. ulcerans Agy99. Indicated are the genes esxB and esxA and the PCR primers (CH1 through CH4) used for this experiment. (B) PCR products of 162 bp or 1,712 bp proved the MURD152 deletion of 2.8 kb and the MURD4 deletion of 12 kb, respectively.
Bacterial pellets of about 60 mg (wet weight) were heat inactivated for 1 h at 95°C in 500 μl of extraction buffer (50 mM Tris-HCl, 25 mM EDTA, 5% monosodium glutamate) and sequentially treated with 17 M lysozyme (for 2 h at 37°C) and 0.3 M proteinase K in proteinase K buffer (1 mM Tris-HCl, 5 mM EDTA, 0.05% sodium dodecyl sulfate, pH 7.8; overnight at 45°C). After digestion, the samples were subjected to bead-beater treatment (7 min at 3,000 rpm; Mikro-Dismembrator; B. Braun Biotech International, Melsungen, Germany) with 300 μl of 0.1-mm zirconia beads (BioSpec Products, Bartlesville, OK). DNA was extracted from the supernatants by phenol-chloroform (Fluka, Buchs, Switzerland) extraction and subjected to ethanol precipitation. DNA concentration was measured by determining the optical density at 260 nm (GeneQuant spectrophotometer; Pharmacia Biotech, Cambridge, United Kingdom).
DNA methods.
PCR was performed using FirePol 10× BD buffer and 0.5 μl of FirePol Taq polymerase (Solis BioDyne, Tartu, Estonia), 2.5 ng of genomic DNA or the equivalent volume of nuclease-free water as a negative control, a 0.6 μM concentration of each forward and reverse primer, 1.7 mM MgCl2, and a 0.3 mM concentration of each deoxynucleoside triphosphate in a total volume of 30 μl. PCRs were run in a GeneAmp PCR system 9700 PCR machine. The thermal profile for PCR amplification of M. ulcerans genomic DNA included an initial denaturation step of 95 to 98°C for 5 min, followed by 32 cycles of 95°C for 20 s, annealing at 58 to 65°C for 20 s, and elongation at 72°C for 30 s up to 4 min. The PCRs were finalized by an extension step at 72°C for 10 min. For experiments with more than 30 samples, Hot Star Taq (Qiagen AG, Hombrechtikon, Switzerland) was used according to the manufacturer's protocol. In order to retrieve PCR products that were subsequently subjected to sequencing, iProof high-fidelity DNA polymerase (Bio-Rad Laboratories, Hercules, CA) was used. PCR products were analyzed on 1 to 2% agarose gels by gel electrophoresis using ethidium bromide staining and the AlphaImager illuminator and AlphaImager software (Alpha Innotech, San Leandro, CA). Primers as summarized in Table 1 were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). PCR fragments produced for analysis of unknown genomic sequences were purified using a NucleoSpin purification kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) and subjected to direct sequencing or cloned using a TOPO TA cloning kit (Invitrogen Corp., Carlsbad, CA), transformed into JM109 (Sigma Aldrich, Buchs, Switzerland) bacterial cells, and sequenced after DNA preparation (Miniprep Kit; Sigma Aldrich, Buchs, Switzerland). Variable-number tandem repeat typing analysis undertaken for confirmation of strain identities was performed according to the method of Stragier et al. (44). Sequencing was performed using a BigDye kit and an ABI Prism 310 genetic sequence analyzer (Perkin-Elmer, Waltham, MA). All gene sequences were reproduced and subjected to alignment and comparison with an ABI Prism Autoassembler, version 1.4.0 (Perkin-Elmer, Waltham, MA).
TABLE 1.
Primers used in this study and description of their respective PCR products
| RD | Locus | Description of PCR product | Product size (bp) | Primer 1
|
Primer 2
|
||
|---|---|---|---|---|---|---|---|
| Name | Sequence (5′-3′) | Name | Sequence (5′-3′) | ||||
| 13/14 | MURD4/MURD152 | Presence of esxB-esxA cluster in MURD4 and/or MURD152 | 610 | CH1 | TGAAGACCGATGCCGCTAC | CH2 | AACATCCCCGTGACGTTG |
| 13 | MURD152 | MURD152 deletion as in Agy99 | 162 | CH3 | CGTTGGGGTGAATTTCTTTG | CH4 | AGTCTGACGGCGACTCATCT |
| 13 | MURD152 | Presence of esxB-esxA cluster in MURD152 | 968 | CH5 | TTGGCGAGGAAAGAAAGAGA | CH4 | AGTCTGACGGCGACTCATCT |
| 14 | MURD4 | Presence of esxB-esxA cluster in MURD4 | 810 | CH6 | GACCCAAAGAGATAGAGAGTCCA | CH7 | TCATCGGTGTCGGTGTAGTG |
| 14 | MURD4 | MURD4 deletion as in Agy99 | 1,712 | CH8 | GACCCAGACGATGTGAATTG | CH9 | GGAGCATGTTCACGATGTTG |
| 13 | MURD152 | Deletion ΔRD13A | 2,354 | CH18 | CAGTTATCGTGCGGGAATTT | CH19 | ATCGGGAGAAAGACCGAAGT |
| 13 | MURD152 | Deletion ΔRD13B | 1,650 | CH10 | CTGGCGGAAACAACAACC | CH11 | TCCTGGTCAAGTTGGAGACC |
| 13 | MURD152 | MURD152 deletion as in Agy99 | 3,198 | CH10 | CTGGCGGAAACAACAACC | CH12 | GCCGCTAACTTGAAGAATCG |
| 13 | MURD152 | MURD152 deletion as in Agy99 | 1,662 | CH13 | TTCTCGCTCAATCTCCCCTA | CH11 | TCCTGGTCAAGTTGGAGACC |
| 15 | MURD92 | Presence of hspX in MURD92 | 791 | CH14 | GGCGCTTAAACCGGTCGTTG | CH15 | CGCCAAACCCAGGACAATCA |
| 15 | MURD92 | MURD92 deletion as in Agy99 | 469 | CH16 | AGCTGGCTAGCGTCGTACC | CH17 | CCCAAAGCTCGTAGATCAGC |
Data analyses and bioinformatics.
Retrieved sequences were compared to the BuruList (http://genopole.pasteur.fr/Mulc/BuruList.html) and the M. marinum (http://www.sanger.ac.uk/cgi-bin/BLAST/submitblast/m_marinum) BLAST servers and analyzed using the Sequence Manipulation Suite (http://bioinformatics.org/sms/index.html), the sequence alignment tool BLAST 2 sequences (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi), the multiple sequence alignment website Multalin (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html), and the Artemis software program, release 9 (The Wellcome Trust Sanger Institute, Hinxton, United Kingdom) (36). The sequences for M. tuberculosis were retrieved from the following Web page: http://www.sanger.ac.uk/Projects/M_tuberculosis. Linear genomic comparison was performed using the Artemis comparison tool software, release 6 (9).
Nucleotide sequence accession numbers.
The sequences of the indicated genes from M. ulcerans strains have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under the following accession numbers (the associated protein is shown in parentheses): for hspX (HspX), accession numbers EU257156, EU257157, EU257158, EU257159, and EU257160; for esxA (ESAT-6), accession numbers EU257151, EU257152, EU257153, EU257154, and EU257155; and for esxB (CFP-10), accession numbers EU257146, EU257147, EU257148, EU257149, and EU257150. Accession numbers correspond to genes from the Japan 8756, China 980912, Surinam 842, French Guiana 9722, and Mexico 5143 strains, in respective order. Note that the annotated hspX gene in M. ulcerans Agy99 is an orthologue of M. tuberculosis htpX and that the M. tuberculosis hspX orthologue is not present in strain Agy99.
RESULTS
Presence of esxB-esxA in M. ulcerans strains of the ancestral lineage.
Blast searches of the partially annotated genome of M. marinum M (http://www.sanger.ac.uk/cgi-bin/BLAST/submitblast/m_marinum) showed that this strain contains two copies of the esxB-esxA (CFP-10-ESAT-6) gene cluster. Both copies are deleted from the genome of the African M. ulcerans isolate Agy99 (43). The corresponding two RDs between the genome sequences of the two mycobacterial species have been designated MURD152 (M. marinum genome position 6489253 to 6592034) and MURD4 (M. marinum genome position 218302 to 230285) (43).
Compared to M. marinum M, the M. ulcerans Agy99 genome has a 2.8-kb deletion in MURD152, which is associated with a large inversion at the 5′ end of the deletion (Fig. 1A). To test whether all M. ulcerans lineages share this genome constellation in MURD152, we screened a comprehensive M. ulcerans strain collection of worldwide origin by PCR analysis using a primer pair (CH3 and CH4) that yields a PCR product of 162 bp only when MURD152 is deleted and flanked by the inverted sequence (Fig. 1A and B). Whereas members of the ancestral lineage (strains from Asia, South America, and Mexico) were negative, members of the classical lineage (strains from Africa, Papua New Guinea, Malaysia, and Australia) were positive, except for strain Australia 9549, which has a larger deletion in this region (see below). Likewise, a PCR using a primer pair (CH8 and CH9) specific for the sequence constellation of strain Agy99 in MURD4 revealed a PCR product of 1,712 bp only for representatives of the classical and not for members of the ancestral lineage (Fig. 1B), demonstrating genomic diversity between the two M. ulcerans lineages in this locus.
A PCR with primers (CH1 and CH2) corresponding to the 5′ end of the esxB coding sequence and the 3′ end of the esxA coding sequence (Fig. 1A) yielded a PCR product of the expected size of 610 bp with genomic DNA from the M. marinum control and in all M. ulcerans strains belonging to the ancestral lineage (Fig. 2). Primers corresponding to the flanking regions of either the MURD4- or the MURD152-associated esxB-esxA gene cassette were used to determine the localization of this cluster in the genomes of these M. ulcerans strains (Fig. 2). Results indicated that esxB-esxA of the Asian and South American strains is located in MURD152, whereas in the Mexican strain the gene cluster is located in MURD4 (Fig. 2). These localizations were verified by PCR analyses extending several kilobases further into the flanking regions. While in the Asian and South American haplotypes the respective M. marinum MURD152 genome constellations were found, the cluster was flanked in the case of the Mexican haplotype by the MURD4-associated sequences of M. marinum.
FIG. 2.
Localization of the two esxB-esxA clusters in the genomes of strains of the M. ulcerans ancestral lineage. Positions of the corresponding primers are indicated for the PCR product of the esxB-esxA cluster, where CH1 and CH2 correspond to sequences within the CDSs of both locations, and of the slightly larger PCR products amplified with flanking primers specific for either MURD152 or MURD4 (Table 1).
Unique deletions in MURD152 in strains 5143 from Mexico and 9549 from Australia.
While MURD152 esxB-esxA is deleted from Mexican strain 5143 (Fig. 2), no PCR product specific for the MURD152 constellation of the strains belonging to the classical lineage was obtained with primers CH3 and CH4 (Fig. 1B), giving evidence for a larger deletion. A PCR analysis with primers corresponding to different positions of the genomic sequences flanking MURD152 demonstrated that the Mexico 5143 strain has a deletion (Fig. 3, ΔRD13A) that is replaced by an IS2404 element. This insertion-deletion (indel) event can have occurred either from an M. marinum M-like genome constellation or from an M. ulcerans Agy99-like constellation (loss of 41.8 kb or of 8 kb, respectively). The DNA sequences flanking ΔRD13A in the Mexican strain have a slightly higher identity to the corresponding sequence stretches of M. ulcerans Agy99 than to those of M. marinum M (98% versus 94% over 986 bp).
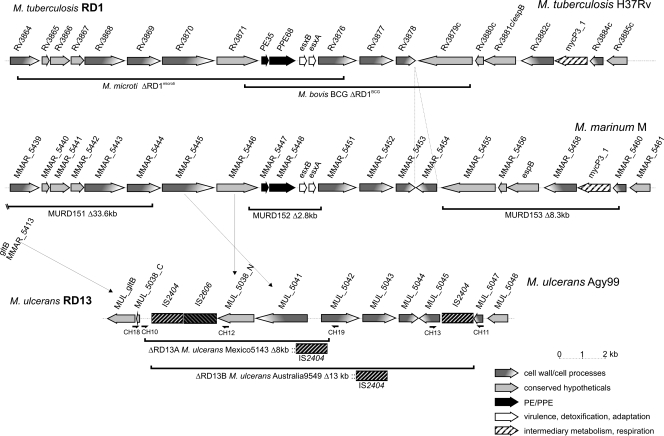
FIG. 3.
Chromosomal organization of CDSs in RD13 including deletional variations between M. ulcerans and other mycobacteria. Gene names are indicated for M. tuberculosis (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=224), M. marinum (http://www.sanger.ac.uk/Projects/M_marinum/), and M. ulcerans (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=16230), and orthologous genes are aligned. RD13 of M. ulcerans corresponds to RD1 in M. tuberculosis. Deletions in M. bovis BCG, M. microti, and various M. ulcerans strains are indicated by solid bars as marked.
Failure to obtain a PCR product with both the CH1/CH2 and the CH3/CH4 PCR primers for the Australian strain 9549 (Fig. 1) provided evidence for yet another deletion type within the MURD152 region. PCR analysis using primers located in the sequences flanking the corresponding region in the M. ulcerans Agy99 genome led to the characterization of a deletion of 13,662 bp including an IS2404 element on each of the ends of the deleted DNA segment (Fig. 3, ΔRD13B). The deleted DNA stretch was, in strain Australia 9549, replaced by an IS2404 element that, according to sequence analysis, differed from both versions of IS2404 in Agy99 that were deleted in the ΔRD13B deletions.
Sequence variation in ESAT-6 and CFP-10.
PCR products obtained with primers corresponding to MURD locus-specific flanking regions and comprising the respective esxB-esxA clusters (Fig. 2) were sequenced. Deduced amino acid sequences of all versions of M. ulcerans ESAT-6 and CFP-10 encoded in MURD4 (Mexico 5143) or MURD152 (South American and Asian strains) were compared with the M. marinum M sequences in the two loci (Fig. 4; see also the supplemental material). As expected, the translated ESAT-6 amino acid sequence of the Mexican strain clustered to and was identical with the MURD4-associated M. marinum M sequence (Fig. 4B). While the four MURD152-associated M. ulcerans ESAT-6 sequences of the Asian and the South American strains were identical to each other, their amino acid sequences differed at six positions from the MURD152-associated M. marinum sequence but only at two positions from the MURD4-associated M. marinum sequence (Fig. 4B). At the nucleotide level, the esxA genes of the Asian and South American strains appear as hybrids composed of an M. marinum MURD4 sequence stretch at the 5′ end and a MURD152 stretch at the 3′ end.
FIG. 4.
Nucleotide variations (A) and amino acid sequence alignments (B) in esxB and esxA CDSs and their gene products (CFP-10 and ESAT-6, respectively). Position 1 of the nucleotide alignment reflects the start of the gene esxB. For the DNA sequences, only differing nucleotides are shown (positions as indicated). For whole-sequence alignments, see the supplemental material. Orthologous sequences of M. tuberculosis H37Rv and M. bovis AF2122/97 are included in the amino acid alignments.
The two M. marinum esxB genes differed only at three nucleotide positions at the 5′ end (Fig. 4A), encoding CFP-10 proteins with identical deduced amino acid sequences (Fig. 4B). The esxB gene of the Mexican strain differed at four positions from the M. marinum M MURD4 locus but at only one position from the MURD152 locus. While the esxB gene sequences of the South American M. ulcerans strains were identical to the MURD152-associated sequence, a frameshift mutation has converted esxB of the Asian strains to a pseudogene (Fig. 4B).
Lack of the immunodominant HspX/Acr protein in the classical lineage of M. ulcerans.
Next, we screened the worldwide M. ulcerans strain collection for the presence of the coding sequence (CDS) encoding the immunogenic protein HspX (Acr) located in MURD92 (M. marinum genome position 4271366 to 4313737) (43). Using primers (CH14 and CH15) corresponding to the hspX flanking regions, a PCR product of 791 bp comprising the complete hspX gene was obtained for all members of the ancestral lineage but for none of the strains belonging to the classical lineage (not shown). Instead, amplification of a 469-bp PCR product (primers CH16 and CH17) obtained with a complementary PCR again demonstrated the presence of the Agy99 genome constellation (related to the MURD92 deletion) in all members of the classical lineage. While strains coming from the same geographical area had identical gene sequences, Asian and South American sequences differed slightly from each other and from the M. marinum sequence (Fig. 5A; see also the supplemental material). In the case of the Mexican strain, nucleotide insertions resulted in a frameshift mutation leading to a truncated translation product (Fig. 5B).
FIG. 5.
Nucleotide variations (A) and amino acid sequence alignments (B) in the hspX CDS and its gene product. Position 1 of the nucleotide alignment reflects the start of the gene. For the DNA sequences, only differing nucleotides are shown (positions as indicated). For whole-sequence alignments see the supplemental material. Orthologous sequences of M. tuberculosis H37Rv and M. bovis AF2122/97 are included in the amino acid alignments.
DISCUSSION
The M. tuberculosis proteins ESAT-6, CFP-10, and HspX are strong T- and B-cell immunogens. This makes them suitable targets for immunodiagnostic tests (7, 8, 14, 15, 20) and potentially also for subunit vaccine development (1, 30, 35). These approaches cannot be duplicated for Buruli ulcer, since these proteins are not expressed by classical-lineage M. ulcerans strains that are found in areas of endemicity in Africa and Australia and are responsible for the vast majority of clinical cases worldwide.
The genome of the M. marinum strain M harbors two esxB-esxA gene clusters at distant chromosomal locations, one in MURD4 and the other in MURD152. Such duplications are common for proteins of the Esx protein family (46). In this report we demonstrate that all analyzed M. ulcerans strains belonging to the ancestral lineage have lost only one copy of the esxB-esxA cassette: the Asian and South American strains have lost the MURD4 copy and the Mexican strain has lost the MURD152 copy. Furthermore, a frameshift mutation has converted the remaining esxB gene of the Asian strains to a pseudogene. The basis for the high degree of identity of the N-terminal esxA nucleotide sequence located in the MURD152 locus in the South American and Asian haplotypes with the M. marinum MURD4 sequence is unclear, but the finding implies a history of homologous recombination between the two copies of the esxB and esxA genes before loss of the MURD4 region. Members of the classical lineage have lost both copies, probably in a bottleneck situation that forged this lineage.
Since MURD152, MURD92, and MURD4 show genomic differences not only between M. marinum and M. ulcerans but also within M. ulcerans strains, we designated these RDs RD13, RD14, and RD15, respectively, in continuation of the previously assigned RDs within the species M. ulcerans (34). A detailed alignment of the chromosomal organization in RD13, which corresponds to RD1 in M. tuberculosis, is shown in Fig. 3. These RDs represent hot spots of genetic variation potentially suitable for performing genetic fingerprinting of M. ulcerans.
In addition to the previously identified five M. ulcerans indel haplotypes (27, 34), strain Australia 9549 was identified as representing a sixth indel haplotype, which is defined by ΔRD13B.
In MURD152 alone, at least three different deletion events are responsible for the indel diversity within M. ulcerans (Table 2). When this region was analyzed for variations among a collection of mycolactone-producing mycobacteria, an unclear situation was suggested for a Mexican strain (48). Here, we show that the deletion of 8 kb replaced by an IS2404 element (ΔRD13A) in the Mexican strain (or 41.8 kb with respect to the M. marinum backbone) differs from the MURD152 deletion in Agy99. This deletion is independent of yet another extended deletion of 13.7 kb (ΔRD13B) in this genomic region in the Australia 9549 strain. The latter deletion is also replaced by an IS2404 element and displays a second, large sequence polymorphism within Australian isolates in addition to the previously described RD3 (27, 34). It will be worth investigating the distribution of this indel polymorphism within a collection of Australian M. ulcerans isolates using the primer pair combination CH10/CH11, demonstrating the presence of the ΔRD13B deletion, and both CH10/CH12 and CH13/CH11, displaying positive results for strains with the sequence configuration of Agy99 (Fig. 3).
TABLE 2.
Genomic deletions and amino acid changes in CDSs of immunogenic proteins
| Strain, lineage, and haplotype | Characteristics of the indicated gene (protein) by RD and locusa
|
||||
|---|---|---|---|---|---|
| RD13 (MURD152)
|
RD14 (MURD4)
|
RD15 (MURD92), hspX (HspX) | |||
| esxA (ESAT-6) | esxB (CFP-10) | esxA-1 (ESAT-6) | esxB-1 (CFP-10) | ||
| M. marinum M | CDS | CDS | CDS | CDS | CDS |
| M. ulcerans ancestral lineage strains | |||||
| South America | CDS; A17S, Q19G, T23G, R52Q, N57K, S68A | CDS | Deletionb | Deletionb | CDS; A105S |
| Asia | CDS; A17S, Q19G, T23G, R52Q, N57K, S68A | Frameshift mutation (pseudogene) | <2.8-kb deletionc | <2.8-kb deletionc | CDS; V139F |
| Mexico | ΔRD13A | ΔRD13A | CDSd | CDSd | Frameshift mutation (pseudogene); D64A, L74R |
| M. ulcerans classical lineage strains | |||||
| Agy99, Africa, Australia | MURD152 deletion | MURD152 deletion | MURD4 deletion | MURD4 deletion | MURD92 deletion |
| Australia 9549 | ΔRD13B | ΔRD13B | MURD4 deletion | MURD4 deletion | MURD92 deletion |
Amino acid changes are in comparison to the sequence of the M. marinum protein.
The lack of PCR products as shown in Fig. 1B suggests a deletion that differs from the MURD4 deletion in the classical lineage.
The deletion in the Asian strains is less than 2.8 kb and hence differs from both the M. ulcerans Agy99 MURD4 and South American haplotype deletions.
A screen using outwardly directed primers that bind in the esxB-esxA cluster and in IS2404 and IS2606 and subsequent tests with nested PCR gave evidence for the presence of both IS2404 and IS2606 in the vicinity in strain Mexico 5143 only, indicating yet further genomic changes in this region.
The described deletions also encompass CDSs surrounding the esxA, esxB, and hspX genes, indicating loss or modification of molecular apparatuses or pathways. First, PE35, essential for secretion (7), was lost in both MURD152 and ΔRD13A and is also commonly deleted in ΔRD1BCG and ΔRD1microti (Fig. 3). Second, many of the genes of the ESX-1 secretion system (the genes Rv3866/MMAR_5441 through Rv3881/MMAR_5457/espB, corresponding to the extended RD1 region) are equally affected by deletions ΔRD13A and/or MURD151 through MURD153, namely, the AAA protein family members Rv3868/MMAR_5443, Rv3871/MMAR_5446, and Rv3877/MMAR_5452 (7, 17, 19). Members of the classical lineage omit an MMAR_5457 orthologue in MURD153, which was recently described as a secreted product and renamed espB (31). Also in MURD92, hspX was jointly deleted with the coregulated Rv2032/nitroreductase gene (33).
As for ESAT-6 and CFP-10, we also found for HspX different genetic mechanisms that have led to loss of expression, comprising both deletions of genomic sequences and single-base differences (Table 2). Many of the sequence variations across the M. ulcerans haplotypes that led to the loss (of function) of these highly immunogenic proteins appear to have emerged independently of each other. This may indicate a counterselection for expression of these proteins. HspX seems to be a negative growth regulator involved in hypoxic shift-down to promote the nonreplicating persistence of M. tuberculosis (15, 20, 25). Both ESAT-6 and CFP-10 were shown to be virulence factors of M. tuberculosis, and their loss reduces infectivity due to the dysfunction of the ESX-1 secretion apparatus (5, 10, 12, 13). The mycolactone-producing and largely extracellular M. ulcerans has a profoundly different survival strategy in mammalian hosts than the intracellular M. tuberculosis; therefore, it is most likely that the pathogenicity of M. ulcerans for mammalian hosts is due to other virulence factors. Thus, our data suggest that functional disruption or complete loss of major targets of the immune response may confer a selective advantage to this emerging pathogen. Still, it is currently unclear whether pathogenicity for mammalian hosts, i.e., shedding into the environment from chronic wounds, contributes significantly to the survival of the species M. ulcerans. However, the observed loss of expression of highly immunogenic proteins caused by a variety of genomic changes may represent an indication that immune selection plays a role in the adaptation of M. ulcerans to a more stable environment.
Supplementary Material
Acknowledgments
We gratefully acknowledge F. Portaels for provision of most of the M. ulcerans strains included in this study, P. C. Small for provision of the M. marinum strain M, T. Stinear and J. Parkhill for providing the M. marinum gene annotation ahead of publication, and C. Daubenberger for stimulating discussion.
This work was partially supported by the Stanley Thomas Johnson Foundation. M. Käser was supported by a research grant from the Deutsche Forschungsgemeinschaft, KA 1842/1-1.
Footnotes
Published ahead of print on 6 February 2008.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Aagaard, C., M. Govaerts, V. Meikle, A. J. Vallecillo, J. A. Gutierrez-Pabello, F. Suarez-Guemes, J. McNair, A. Cataldi, C. Espitia, P. Andersen, and J. M. Pollock. 2006. Optimizing antigen cocktails for detection of Mycobacterium bovis in herds with different prevalences of bovine tuberculosis: ESAT6-CFP10 mixture shows optimal sensitivity and specificity. J. Clin. Microbiol. 44:4326-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablordey, A., P. A. Fonteyne, P. Stragier, P. Vandamme, and F. Portaels. 2007. Identification of a new variable number tandem repeat locus in Mycobacterium ulcerans for potential strain discrimination among African isolates. Clin. Microbiol. Infect. 13:734-736. [DOI] [PubMed] [Google Scholar]
- 3.Ablordey, A., R. Kotlowski, J. Swings, and F. Portaels. 2005. PCR amplification with primers based on IS2404 and GC-rich repeated sequence reveals polymorphism in Mycobacterium ulcerans. J. Clin. Microbiol. 43:448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adusumilli, S., A. Mve-Obiang, T. Sparer, W. Meyers, J. Hayman, and P. L. Small. 2005. Mycobacterium ulcerans toxic macrolide, mycolactone modulates the host immune response and cellular location of M. ulcerans in vitro and in vivo. Cell. Microbiol. 7:1295-1304. [DOI] [PubMed] [Google Scholar]
- 5.Brodin, P., M. I. de Jonge, L. Majlessi, C. Leclerc, M. Nilges, S. T. Cole, and R. Brosch. 2005. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J. Biol. Chem. 280:33953-33959. [DOI] [PubMed] [Google Scholar]
- 6.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodin, P., L. Majlessi, L. Marsollier, M. I. de Jonge, D. Bottai, C. Demangel, J. Hinds, O. Neyrolles, P. D. Butcher, C. Leclerc, S. T. Cole, and R. Brosch. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. [DOI] [PubMed] [Google Scholar]
- 9.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 10.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632-1636. [DOI] [PubMed] [Google Scholar]
- 11.Chemlal, K., G. Huys, P. A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong, B. C., P. C. Hill, R. H. Brookes, S. Gagneux, D. J. Jeffries, J. K. Otu, S. A. Donkor, A. Fox, K. P. McAdam, P. M. Small, and R. A. Adegbola. 2006. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J. Infect. Dis. 193:1279-1286. [DOI] [PubMed] [Google Scholar]
- 13.de Jonge, M. I., G. Pehau-Arnaudet, M. M. Fretz, F. Romain, D. Bottai, P. Brodin, N. Honore, G. Marchal, W. Jiskoot, P. England, S. T. Cole, and R. Brosch. 2007. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 189:6028-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demissie, A., E. M. Leyten, M. Abebe, L. Wassie, A. Aseffa, G. Abate, H. Fletcher, P. Owiafe, P. C. Hill, R. Brookes, G. Rook, A. Zumla, S. M. Arend, M. Klein, T. H. Ottenhoff, P. Andersen, and T. M. Doherty. 2006. Recognition of stage-specific mycobacterial antigens differentiates between acute and latent infections with Mycobacterium tuberculosis. Clin. Vaccine Immunol. 13:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DesJardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (HspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGiuseppe Champion, P. A., and J. S. Cox. 2007. Protein secretion systems in Mycobacteria. Cell. Microbiol. 9:1376-1384. [DOI] [PubMed] [Google Scholar]
- 17.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677-1693. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Pelayo, M. C., K. C. Caimi, J. K. Inwald, J. Hinds, F. Bigi, M. I. Romano, D. van Soolingen, R. G. Hewinson, A. Cataldi, and S. V. Gordon. 2004. Microarray analysis of Mycobacterium microti reveals deletion of genes encoding PE-PPE proteins and ESAT-6 family antigens. Tuberculosis (Edinburgh) 84:159-166. [DOI] [PubMed] [Google Scholar]
- 19.Guinn, K. M., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haile, Y., G. Bjune, and H. G. Wiker. 2002. Expression of the mceA, esat-6 and hspX genes in Mycobacterium tuberculosis and their responses to aerobic conditions and to restricted oxygen supply. Microbiology 148:3881-3886. [DOI] [PubMed] [Google Scholar]
- 21.Harboe, M., T. Oettinger, H. G. Wiker, I. Rosenkrands, and P. Andersen. 1996. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilty, M., M. Käser, J. Zinsstag, T. Stinear, and G. Pluschke. 2007. Analysis of the Mycobacterium ulcerans genome sequence reveals new loci for variable number tandem repeats (VNTR) typing. Microbiology 153:1483-1487. [DOI] [PubMed] [Google Scholar]
- 23.Hilty, M., D. Yeboah-Manu, D. Boakye, E. Mensah-Quainoo, S. Rondini, E. Schelling, D. Ofori-Adjei, F. Portaels, J. Zinsstag, and G. Pluschke. 2006. Genetic diversity in Mycobacterium ulcerans isolates from Ghana revealed by a newly identified locus containing a variable number of tandem repeats. J. Bacteriol. 188:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, Y., F. Movahedzadeh, N. G. Stoker, and A. R. Coates. 2006. Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infect. Immun. 74:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, P. D., T. Stinear, P. L. Small, G. Pluschke, R. W. Merritt, F. Portaels, K. Huygen, J. A. Hayman, and K. Asiedu. 2005. Buruli ulcer (M. ulcerans infection): new insights, new hope for disease control. PLoS. Med. 2:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaser, M., S. Rondini, M. Naegeli, T. Stinear, F. Portaels, U. Certa, and G. Pluschke. 2007. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol. Biol. 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacGurn, J. A., and J. S. Cox. 2007. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 75:2668-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial “core” genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin, B., J. S. Chon, J. A. Macgurn, F. Carlsson, T. L. Cheng, J. S. Cox, and E. J. Brown. 2007. A Mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS. Pathog. 3:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mve-Obiang, A., R. E. Lee, E. S. Umstot, K. A. Trott, T. C. Grammer, J. M. Parker, B. S. Ranger, R. Grainger, E. A. Mahrous, and P. L. Small. 2005. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 73:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purkayastha, A., L. A. McCue, and K. A. McDonough. 2002. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr gene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rondini, S., M. Käser, T. Stinear, M. Tessier, C. Mangold, G. Dernick, M. Naegeli, F. Portaels, U. Certa, and G. Pluschke. 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg. Infect. Dis. 13:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roupie, V., M. Romano, L. Zhang, H. Korf, M. Y. Lin, K. L. Franken, T. H. Ottenhoff, M. R. Klein, and K. Huygen. 2007. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect. Immun. 75:941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, S. A., J. E. Johndrow, P. Manzanillo, and J. S. Cox. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178:3143-3152. [DOI] [PubMed] [Google Scholar]
- 38.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stinear, T. P., H. Hong, W. Frigui, M. J. Pryor, R. Brosch, T. Garnier, P. F. Leadlay, and S. T. Cole. 2005. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 187:1668-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stinear, T. P., G. A. Jenkin, P. D. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stinear, T. P., A. Mve-Obiang, P. L. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stinear, T. P., M. J. Pryor, J. L. Porter, and S. T. Cole. 2005. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology 151:683-692. [DOI] [PubMed] [Google Scholar]
- 43.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stragier, P., A. Ablordey, L. M. Bayonne, Y. L. Lugor, I. S. Sindani, P. Suykerbuyk, H. Wabinga, W. M. Meyers, and F. Portaels. 2006. Heterogeneity among Mycobacterium ulcerans isolates from Africa. Emerg. Infect. Dis. 12:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, T., W. L. Lee, D. C. Alexander, S. Grinstein, and J. Liu. 2006. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell. Microbiol. 8:1417-1429. [DOI] [PubMed] [Google Scholar]
- 46.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 47.Torrado, E., S. Adusumilli, A. G. Fraga, P. L. Small, A. G. Castro, and J. Pedrosa. 2007. Mycolactone-mediated inhibition of tumor necrosis factor production by macrophages infected with Mycobacterium ulcerans has implications for the control of infection. Infect. Immun. 75:3979-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yip, M. J., J. L. Porter, J. A. Fyfe, C. J. Lavender, F. Portaels, M. Rhodes, H. Kator, A. Colorni, G. A. Jenkin, and T. Stinear. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry, III. 1998. The 16-kDa alpha-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. USA 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.