Abstract
Canine distemper virus (CDV) is a highly contagious virus that causes multisystemic disease in dogs. We received seven samples from dogs with CD from the United States during 2007. CDV isolates from these samples formed large, multinucleated syncytia in a Vero cell line expressing canine signaling lymphocyte activation molecule (SLAM). Based on the hemagglutinin gene sequences, the CDV isolates from three states (California, Missouri, and Oklahoma) formed two CDV genetic groups: group I (major; six of seven isolates) consisted of CDV isolates closely related to the European wildlife lineage of CDV, and group II (minor; one of seven isolates) was genetically related to the Arctic-like lineage of CDV. However, both CDV groups were genetically different from the current vaccine strains that belong to the American-1 lineage of the old (1930 to 1950) CDV isolates.
Canine distemper virus (CDV) is a Morbillivirus that affects dogs of all ages (12). In the United States, raccoons (Procyon lotor), foxes, coyotes, and wolves are susceptible to CDV infection and may play a role in the transmission of the virus. CDV is not a cause of disease in domestic cats (3); however, large cats (lions, leopards, cheetahs, and tigers) have been affected by CDV (23). CD is a multisystemic infection, frequently involving the ocular, respiratory, gastrointestinal, and nervous systems. The disease has been prevented and controlled by vaccination (25). However, there have been sporadic reports of a reemergence of CDV in the United States from 2004 to 2005 (26). Moreover, the reemergence of CDV in an area has been linked to a limited number of CDV lineages that are unique to a geographical area (5, 20, 28). Based on phylogenetic analysis of the hemagglutinin (H) gene sequences, the CDV isolates form seven lineages: American-1 (vaccines), American-2, Arctic-like, Asia-1, Asia-2, Europe, and European wildlife (22). H protein is involved in host cell binding and shows more variation (about 10% variation in amino acid sequence) among CDV isolates (30), whereas antibodies against CDV H protein provide protection against experimental infections. Thus, variation in H gene sequences has biological effects on virus-host interactions.
There is anecdotal evidence that the number of CD cases has increased as much as four- to fivefold in dogs in the last 3 years (H. van Campen, Colorado State University, Fort Collins, CO, personal communication). Thus, epidemiological studies to investigate and substantiate the apparent rise in CD clinical cases are warranted. In this study, we performed an evolutionary and genetic analysis of seven CDV isolates from the United States using the H gene sequences. The biological effects of the H gene sequence variation were investigated using an in vitro cell culture system.
(Part of this work was recently submitted as a poster presentation at the 27th Annual Meeting of the American Society for Virology, Ithaca, NY, July 2008.)
Antemortem samples included ocular swabs, nasal swabs, and peripheral blood anticoagulated with EDTA. The swabs were received in 1 to 2 ml of cold normal saline sent on ice by overnight delivery within 24 h of collection. We did not test urine samples; however, in a recent study, urine has been described as a sensitive sample for the detection of CDV in live dogs (2). Postmortem samples were from tonsils, brains, bladders, and lungs (16). Approximately 2 to 5 g of each tissue was received in tubes sent on ice by overnight delivery for virological examination. The specimens were obtained from seven suspected cases of CD from three states in the United States (Oklahoma, four; Missouri, one; California, two).
For the direct fluorescent-antibody testing, tissues were sectioned at 8-μm thickness and fixed with an acetone (75%)-methanol (25%) mixture at room temperature. Veterinary Medical Research and Development (VMRD), Pullman, WA, supplied pretitrated, lot-to-lot-certified conjugates for veterinary diagnostic applications. As part of quality control/quality assurance, we tested the conjugates before use on negative and known positive CDV controls. After the addition of ready-to-use, prediluted, fluorescein isothiocyanate-labeled, anti-CDV monoclonal antibody (VMRD, Pullman, WA) or polyclonal antibody conjugates (VMRD, Pullman, WA), the sections were incubated for 30 min at 37°C. After the unbound antibody conjugates were washed, the sections were counterstained with Evans blue for 15 min. After being mounted in buffered glycerol (pH 9.4), the sections were examined by fluorescent microscopy. Positive cells showed apple-green fluorescence in the cytoplasm, and negative cells were brick red.
For isolation, the tissues from CDV-infected samples were finely chopped, freeze-thawed twice to release the virus, and centrifuged at 8,000 × g. The clear supernatant was filtered though a 0.22-μm syringe filter. The Vero cell line was derived from the kidney of a normal, adult African green monkey (Cercopithecus) in Japan. The recombinant cell line was derived by transfection of the Vero cells with canine signaling lymphocyte activation molecule (SLAM; also known as CD150), as described before by Seki et al. (27). The inoculums (about 1 ml per 25-cm2 flask) were incubated for 1 h at 37°C with rocking every 20 min. After inoculation on a recombinant Vero cell line expressing canine SLAM, about 3.5 ml of Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA) with 5% fetal calf serum was added. The cells were examined daily for cytopathic effects (multinucleated-syncytium formation) (27). Vero cells expressing canine SLAM have been found to be useful for the primary isolation of CDV (18).
For total RNA extraction (host and viral RNAs) from specimens, QIAmp viral RNA extraction kits were used (Qiagen Inc., CA). The quality and quantity of the RNA were checked by determining the A260/A280 using a Nanodrop spectrophotometer (Nanodrop Technologies, CA).
For the detection of CDV RNA, reverse transcription (RT)-PCR based on the nucleocapsid (N) gene was targeted (15). This protocol provides high sensitivity due to the nested amplification of the target gene, the high copy number of the N gene, and the conserved sequence of the N gene among the CDV isolates. Briefly, the first-round product was amplified by the forward primer (primer 1, 5′-ATTTGGGATTGCTTAGGA-3′) and reverse primer (primer 2, 5′-GGCGCTCATCTTGGACAT-3′). The protocol was RT at 45°C for 1 h and 95°C for 3 min; 30 cycles of PCR with denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and an extension at 72°C for 1 min; and a final extension at 72°C for 7 min, with the reaction mixture held at 4°C. The small-portion (1-microliter) product of the first reaction was subjected to a second round of amplification using primer 3 (5′-GTTAGCTAGTTTCATCCT-3′) and primer 4 (5′-GGTCCTCTGTTGTCTTGG-3′). The protocol for the second round was denaturation at 95°C for 3 min; 30 cycles of denaturation at 94°C for 30 s and annealing at 54°C for 30 s; with an extension at 72°C for 1 min. The final extension was performed at 72°C for 7 min, and the reaction mixture was held at 4°C before electrophoresis. The size of the second-round amplicon was 419 base pairs, verified by including molecular size standards in agarose gel analysis.
For CDV genotyping, the H gene was used as the target (21). The forward primer (primer 204+, nucleotides 388 to 409, 5′-GAATTCGACTTCCGCGATCTCC-3′) and reverse primer (primer 232b−, nucleotides 1543 to 1519, 5′-TAGGCAACACCACTAATTTRGACTC-3′) yield an amplicon of 1,160 base pairs. The H gene RT-PCR protocol was RT at 50°C for 30 min and 94°C for 2 min. The PCR protocol was 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min and a final extension at 72°C for 10 min, with the reaction mixture held at 4°C. The positive and negative CDV controls were included in each run of both detection (N gene) and genotyping (H gene) RT-PCR protocols. For phylogenetic analysis of the H gene sequences, the amplicons were sequenced at the Oklahoma Medical Research Foundation, Oklahoma City, OK. The sequences were subjected to Basic Local Alignment Search Tool for Nucleotides (BLASTN) analysis (1) and compared to GenBank H gene sequences for CDV isolates from different species and geographic areas around the world. The percentage identities of the H gene sequences were recorded. Further, the H gene sequences were subjected to phylogenetic analysis and sequence comparison with H gene sequences of the vaccine CDV isolates (Ondersteport, Convac, Lederle, and Snyder Hill CDV isolates) deposited in GenBank. Alignments of the top 100 matches with known sequences were used to perform phylogenetic analysis by neighbor-joining using the Jukes-Cantor method (NCBI, MD).
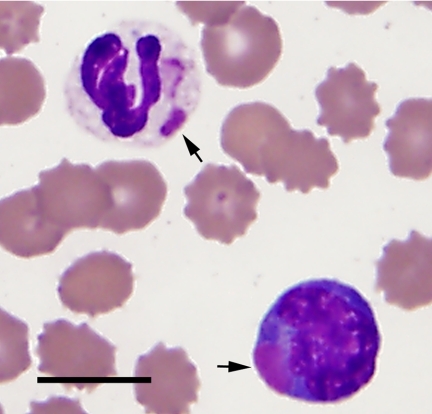
Peripheral blood films from two of the CD case samples (OADDL 07091030 and OADDL 07091031) were stained with an aqueous Romanowsky stain and examined by light microscopy. Both blood films revealed numerous eosinophilic structures within the cytoplasm of neutrophils and lymphocytes, consistent with the presence of CDV inclusions (Fig. 1). The presence of CDV inclusions was confirmed by the direct fluorescent-antibody test in both cases.
FIG. 1.
Blood film from a CDV-infected dog (OADDL 07091030). CDV inclusions (arrows) are visible within a neutrophil and a lymphocyte (aqueous Romanowsky stain; bar, 10 μm).
Six of the seven CDV-positive samples were successfully isolated in the Vero cell line with canine SLAM/CD150 (Table 1). The cytopathic effects of the CDV isolates were characterized by multinucleated syncytia that formed 1 to 2 days after inoculation. The presence of CDV was further detected by RT-PCR for the hemagglutinin gene. One CDV sample (OADDL 07061535) was tested only by RT-PCR and sequencing of the H gene; there was insufficient sample for virus isolation.
TABLE 1.
Data for 2007 OADDL CDV isolates
| Isolate | Designation in the present study | State of origin | Dogb
|
% H gene identity with:
|
Virus isolationf | CDV lineage | |||
|---|---|---|---|---|---|---|---|---|---|
| Vaccination statusa | Age (wk) | Breed | Current vaccine isolated | Missouri CDV isolate 19876e | |||||
| OADDL 07061535 | OK-1 | OK | I | 12 | Mixed | 89 | 98 | ND | European wildlife |
| OADDL 07091030 | OK-2 | OK | NV | 44 | Siberian husky | 89 | 98 | + | European wildlife |
| OADDL 07091031 | OK-3 | OK | NV | NA | NA | 89 | 99 | + | European wildlife |
| OADDL 07091032 | OK-4 | OK | NV | NA | NA | 84 | 96 | + | European wildlife |
| OADDL 07101508 | CA-1 | CA | V | 10 | American bulldog | 89 | 98 | + | European wildlife |
| OADDL 07110098 | MO-1 | MO | V | 10 | Weimaraner | 89 | 90 | + | Arctic-like |
| OADDL 07111080 | CA-2 | CA | V | 136c | Border collie | 87 | 96 | + | European wildlife |
I, incomplete vaccination; NV, not vaccinated; V, vaccinated.
NA, not available.
Animal recovered completely after supportive therapy.
All isolates had less than 90% identity with the current vaccine isolates (Ondersteport, Lederle, and Convac strains).
All isolates had greater than 95% identity with the Missouri CDV isolate 19876, except MO-1, which belongs to the Arctic-like lineage.
ND, not done; +, successful isolation in the Vero cell line with canine SLAM/CD150.
Based on RT-PCR for the H gene followed by sequencing, the level of identity among the CDV isolates (OK-1, OK-2, OK-3, OK-4, CA-1, and CA-2; major group I) was highest with the canine CDV isolate 19876 from Missouri (Table 1) that is genetically most related to a Danish mink CDV isolate (26). Thus, it was the predominant CDV variant (six of seven isolates) in this study. These six CDV isolates belonged to the European wildlife lineage of CDV isolates. However, we found one isolate (MO-1; minor group II) that was most genetically similar to the canine CDV isolate 21260 from Missouri (26) that is closely related to a lesser-panda CDV isolate. This CDV isolate belongs to the Arctic-like lineage of CDV isolates. The information on the 2007 OADDL CDV isolates is summarized in Table 1.
CDV has a negative-stranded RNA genome that encodes several proteins: nucleocapsid, polymerase, multifunctional zinc-binding protein, membrane protein and fusion (F), hemagglutinin (H), and large polymerase proteins. The H protein is responsible for attachment of the virus to the host cell receptor SLAM (30). The only known receptor for CDV is SLAM, which is present on activated T and B lymphocytes, immature thymocytes, mature dendritic cells, and activated monocytes (7). The expression of canine SLAM/CD150 has been demonstrated in vivo (31). Antibodies against H protein are responsible for protection against CDV infection (9). The hemagglutinin glycoprotein varies approximately 10% among the CDV isolates (30), and envelope protein H determines the cytopathology and tropism of the virus (30). The level of genetic variation in the F glycoprotein gene sequence of CDV is about 4% (30). Moreover, more H gene sequences than F gene sequences are available in GenBank and thus allowed robust comparison with CDV isolates from other geographic areas, other susceptible species, and vaccine isolates. In a preliminary analysis, we compared the OADDL CDV isolates with all the H gene sequences in GenBank and found that CDV isolates cluster in geographically distinct lineages. For example, all the Argentina CDV isolates formed one distinct cluster (data not shown). The South American CDV isolates were not included in the recent analysis of CDV isolates based on geography and H gene phylogeny (22). However, they form a distinct South American cluster in our analysis. In recent papers, the terms genotype, cluster, and lineage have been used interchangeably by different investigators, but the results on CDV phylogeny were similar in all the studies (20, 22, 24), including our analysis, because all the investigators used the GenBank accession sequences. A member of a particular genotype of CDV has been proposed to have more than 95% identity in the nucleotides of the H gene sequences (24) and, thus, the intragenotypic variation is less than 5% (20).
The CDV isolate OK-1 (OADDL 07061535) was obtained from a 3-month-old, female, mixed-breed, vaccinated dog from Oklahoma with a history of conjunctivitis, nasal discharge, and weight loss. The dog had not finished the complete course of vaccination and had a history of roaming and eating garbage. This CDV isolate had maximum identity (98%) with CDV isolate 19876 (GenBank accession number AY964110.1). Based on the H gene analysis, CDV isolate 19876 belongs to the European wildlife lineage of CDV isolates along with OK-1.
The CDV isolate OK-2 (OADDL 07091030) was obtained from a tissue pool of an 11-month-old unvaccinated Siberian husky from Oklahoma. On necropsy, the conjunctival and tracheal epithelia contained intracytoplasmic, eosinophilic inclusions surrounded by clear halos. In the tonsils, there were marked lymphoid depletion and numerous inclusion bodies in the epithelium. This isolate had maximum identity (98%) with CDV isolate 19876 (canine origin, Missouri) and 94% identity with CDV isolates from Hungary (GenBank accession number EF095750.1), a Danish mink (Z47759.1), and a lesser panda (AF178039.1), CDV strain A75/17 (AF164967.1), and a morbillivirus from a German ferret isolate (X84999.1). The CDV isolate A75/17 from the United States is regarded as a virulent prototype of field CDV isolates (28). The level of identity of the H gene with the vaccine isolates (Convac, Lederle, and Ondersteport strains) was 89%.
The CDV isolate OK-3 (OADDL 07091031) was obtained from a dog in a shelter in Oklahoma. We obtained a blood tube, but no other history was available on this case. Inclusions consistent with CDV were observed in leukocytes on a peripheral blood film and further confirmed by a direct fluorescent-antibody test. The blood sample was positive for CDV by virus isolation. The H gene was sequenced and had 99% identity with CDV isolate 19876 (GenBank accession number AY964110.1) and 95% identity with CDV isolates from Hungary (EF095750.1), a Danish mink (Z47759.1), and a lesser panda (AF178039.1), CDV virus strain A75/17, CDV isolate 01-2641, and a German ferret morbillivirus strain (X84999.1).
The CDV isolate OK-4 (OADDL 07091032) was obtained from a tissue pool (bladder and lungs) from a dog adopted from an animal shelter in Oklahoma. This CDV isolate had maximum identity (96%) with CDV isolate 19876 (GenBank accession number AY964110.1). In descending order, it had 93% identity with CDV isolates from Hungary (EF095750.1), a lesser panda (AF178039.1), and a Danish mink (Z47759.1); 84% identity with the vaccine isolates; and 70% identity with the phocine distemper virus.
The CDV isolate CA-1 (OADDL 07101508) was obtained from a tissue pool from a 10-week-old, male, vaccinated American bulldog from California that died of CD. Three out of four littermates died of CD with respiratory signs, hyperkeratosis, and seizures. Of the three dead littermates, the necropsy report was available for one littermate. Its lungs were firm and congested on necropsy. The necropsy results of one of the four littermates were completely normal. The H gene sequence was 98% identical to canine origin CDV isolate 19876 (GenBank accession number AY964110.1). The CDV isolate was 94% identical to the Hungarian CDV isolate, the lesser-panda isolate (AF178039.1), and CDV strain A75/17 (AF164967). The CDV H gene sequence was 93% identical to CDV isolate 01-2641 (AY526496.1). The H gene of this CDV isolate lacked the PstI site present in all vaccine CDV isolates (10).
The CDV isolate MO-1 (OADDL 07110098) was obtained from nasal and conjunctival swabs of a 10-week-old, CDV-vaccinated Weimaraner dog that had clinical signs compatible with CD. The dog developed “chewing gum” seizures, thickened footpads, coughing, nasal discharge, and congested lungs. The swabs were collected before euthanasia, and CDV was isolated in a cell culture. The CDV isolate H gene had maximum identity (98%) with CDV isolates 21261 and 18133 from Missouri and 97% identity with CDV isolates from Italy (48/05 and 179/94) and Hungary (H06Bp10S, H06Bp8F, H05Bp7F, H05Bp6F, and H05BpBp5F). The H gene sequence of this CDV isolate had 95% identity with a CDV isolate from a Greenlandic dog and only 90% identity with CDV isolate 19876. It had 89% identity with the vaccine CDV isolates and 70% identity with the phocine distemper virus H gene. Moreover, this CDV isolate lacks the PstI restriction site present in all vaccine CDV isolates (10). Based on phylogenetic analysis, this isolate belongs to the Arctic-like lineage of the CDV isolates.
The CDV isolate CA-2 (OADDL 07111080) was obtained from a combination of nasal, pharyngeal, tonsil, and conjunctival swabs of a 32-month-old, neutered, male, vaccinated border collie with a history of vomiting, diarrhea, and lymphopenia. The H gene sequence had maximum identity (96%) with CDV isolate 19876. This isolate had 93% identity with the Hungarian CDV isolate, the lesser-panda CDV isolate, and the Danish mink CDV isolate; 92% identity with CDV strain A75/17 (GenBank accession number AF164967.1); and 92% identity with the German ferret CDV isolate. Based on phylogenetic analysis, this CDV isolate clusters with CDV isolates of the European wildlife lineage. This dog recovered after treatment and has been clinically healthy for the last 3 months. The survival of this dog after a natural exposure to a CDV isolate of European wildlife lineage is probably due to resistance based on age, genetic resistance, and immunity after complete vaccination with a commercial CDV vaccine. This dog had a CDV titer of 1:16 when tested by CDV serum neutralization 3 months after recovery from CDV infection.
There are recent reports of a reemergence and increased incidence of CDV on several continents: Asia (24), Australia (25), Europe (20), North America (26) and South America (5). In several reports (24), the clinical and pathological observations were not followed by molecular epidemiology findings using CDV H gene analysis. Case reports of CD in vaccinated dogs are sporadic in the United States (26). In a previous 2004 study (26), only a few Missouri CD cases (four) were analyzed. The presence of numerous CDV inclusions in peripheral blood leukocytes caught our attention (Fig. 1) and led us to investigate the H antireceptor sequences and the biological interaction of the wild-type 2007 CDV isolates with the SLAM (CD150) receptor protein in vitro. We found that five out of six OADDL 2007 CDV isolates produced multinucleated syncytia in a Vero cell line expressing canine SLAM receptor. It has been proposed that syncytial size is a correlate of the degree of virulence of the CDV isolates (8) because it will correlate with the ability of the CDV to spread from cell to cell. The aggressive spread in cell culture, the ability to produce large numbers of inclusions in canine lymphocytes that naturally express SLAM/CD150, and the ability to produce fatal infections in vaccinated dogs indicate that these canine isolates of European wildlife lineage are virulent for dogs.
Most vaccine strains of CDV were isolated from the 1930s through the 1950s (old CDV isolates from the United States; American-1 lineage) and have been used in CDV vaccines worldwide. The wild-type strains of CDV related to the vaccine strains (Ondersteport, Snyder Hill, and Lederle strains) are no longer detected in the domestic canine populations in the United States (25). Wild-type CDV isolates identified in our study are clearly genetically and phylogenetically distinct from the vaccine strains of CDV. These 2007 U.S. CDV isolates show less than 90% identity in H gene sequence with commercial vaccine CDV isolates (Table 1). The antigenic distance between the wild-type CDV isolates and vaccine CDV isolates has not been determined by cross-neutralization tests.
In a previous study (26) from Missouri, CDV isolate 19876 was detected by RT-PCR and sequencing; however, no virus isolation was performed, and thus, the type of cytopathic effect caused by Missouri CDV isolates is not described. Five of six CDV isolates described here were most identical in H gene sequence to the Missouri CDV isolate 19876. However, there were two nucleotide positions in the H gene in CDV isolate 19876 that were different in all these 2007 OADDL CDV isolates. Six out of seven of the OADDL 2007 isolates from the United States were associated with fatal outcomes due to CDV. We conclude that a large-syncytium-forming variant was the predominant CDV circulating in the United States in 2007 (Table 1). These six of seven CDV isolates belong to the European wildlife lineage (group I).
Various factors such as the quality of the vaccine, poor host immune response, and genetic variability of CDV have been described as major reasons for the failure of CDV vaccines (17, 20). In a recent Australian study of CDV reemergence (25), all the infected dogs from semirural areas of Sydney, Australia, were either unvaccinated or had not completed the course of vaccination (incomplete vaccination). Moreover, changes in the H surface glycoprotein can lead to changes in virulence (20) and in tissues targeted.
In infected dogs with a history of recent vaccination with attenuated CDV, exposure to wild-type CDV before vaccination is assumed to be the source of the CDV infection. This speculation can be confirmed by a comparison of the recovered CDV with the vaccine CDV using the H gene sequences that are most variable between CDV isolates (4). In dogs with a history of recent vaccination, a thymine (T) at position 8139 on the H gene allowed us to differentiate the vaccine CDV isolates from the wild-type CDV isolates that contained cytosine (C), resulting in the H gene PCR product not being digested with the PstI restriction enzyme (5′-TATAAA-3′).
The uncontrolled movement of dogs between continents (United States and Europe) has been blamed for the recirculation of CDV variants around the globe (10). In the United States, where only one virulent CDV variant (European wildlife lineage) is predominant, there is better vaccine compliance and wider use of the CDV vaccine in dogs than in parts of Central and Eastern Europe, such as Hungary, where several different CDV variants have been reported (10). In the Hungarian study, the CDV isolates belong to two major CDV groups, those that are similar to canine CDV isolate 19876 from the United States (three Hungarian CDV isolates; group II) and those that are similar to canine CDV isolate 18133 from the United States (nine Hungarian isolates; group III). Only one Hungarian CDV isolate was in a different genetic group I (10).
In the three outbreaks in semirural areas of Australia, wildlife reservoir dingoes, feral dogs, or foxes are the most likely CDV reservoirs (25). Raccoons may serve as the reservoir of CDV viruses because of their scavenging behavior in urban areas and their spreading the virus between the domestic and wildlife populations (14, 19). CDV is endemic in raccoons, and raccoons provide a source of CDV for immunologically or genetically susceptible canine hosts that interact with them in urban and semirural areas in the United States. In rural areas, stray, unvaccinated dogs can ingest the remains of dead raccoons on the roads. We did not investigate raccoon CDV isolates in this study. In previous reports, CDV isolates from raccoons have been detected in the same genetic groups with virulent canine isolates (American-2 CDV lineage) (22). However, two of the dogs in our study were strays and had a history of eating garbage and, thus, may have been exposed to raccoons. We believe that raccoons can serve as intermediate hosts, transferring the CDV from other wildlife to dogs. The further spread of the CDV variants in the United States occurs by interstate movement of dogs associated with trade and dog shows. The circulation of the limited number of CDV variants related to the European wildlife lineage in the United States is due to the circulation between stray dogs and raccoons in urban areas and subsequent dog-to-dog transfer by the shipment of dogs from puppy-breeding areas in Oklahoma, Kansas, and Missouri to other states (e.g., California). These midwestern states have high concentrations of breeding kennels, and semirural areas have higher concentrations of raccoons and other wildlife, including foxes. Thus, there has been selection for the European wildlife CDV variants in the United States. Our speculation about raccoons as a reservoir and source of CDV in midwestern states is also suggested by histories from submitting veterinarians. Direct contact with a potential reservoir(s) is needed to transfer the fragile CDV, which does not survive well in nature.
According to the current 2006 guidelines (www.aahanet.org) by the American Animal Hospital Association, the initial puppy vaccines are administered at 6 to 8 weeks of age and the second dose is administered at 12 to 14 weeks of age. The boosters are administered at 1-year intervals; however, some newer vaccines have a USDA license claiming 3 years of protection on the label. The current CDV vaccines are providing good control of CDV infection (6), and the prevalence of clinical cases is currently low in the United States. However, the recent detection of a virulent CDV strain of the European wildlife lineage in at least three states warrants continued monitoring of CDV molecular epidemiology. There have been recent suggestions to update the CDV vaccines (5). It has been serologically demonstrated that there is about 10-fold-higher neutralization activity against the homologous novel CDV variants than against the old CDV vaccine isolates (13). Moreover, host immune responses, host genetics, age, and susceptibility are important variables in the outcome of CDV infection. Further, the issue of duration of protective immunity maintained by booster vaccination every 3 years for CDV may have to be reevaluated in light of the emergence of new variants of CDV.
A recent study has demonstrated that SLAM-blind wild-type CDV cannot infect peripheral blood mononuclear cells (29). Moreover, the recent evolutionary studies using all CDV genes have provided compelling evidence that functional sites (amino acids 530 and 549) of H protein that interact with SLAM receptor are associated with CDV emergence in different novel host species (22). In addition to the differences in H gene sequences among CDV isolates, we predict that single-nucleotide polymorphisms in genes encoding canine SLAM may influence the outcome of CDV infection and pathogenesis. This hypothesis is supported by recent observations that specific single-nucleotide polymorphisms in genes encoding human SLAM are involved in the response of humans to vaccination with measles virus, which is related to CDV (11).
Acknowledgments
We thank Yusuke Yanagi, Department of Virology, Faculty of Medicine, Kyushu University, Fukuoka, Japan, for providing the recombinant Vero cell line expressing canine SLAM/CD150. We thank Anthony Confer for reviewing the manuscript and providing valuable scientific input based on his CDV experience.
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amude, A. M., A. A. Alfieri, and A. F. Alfieri. 2006. Antemortem diagnosis of CDV infection by RT-PCR in distemper dogs with neurological deficits without the typical clinical presentation. Vet. Res. Commun. 30:679-687. [DOI] [PubMed] [Google Scholar]
- 3.Bart, M., F. Guscetti, A. Zurbriggen, A. Pospischil, and I. Schiller. 2000. Feline infectious pneumonia: a short literature review and a retrospective immunohistological study on the involvement of Chlamydia spp. and distemper virus. Vet. J. 159:220-230. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., T. D. Jensen, E. Gottchalck, P. Arctander, M. J. Appel, R. Buckland, and M. Blixenkrone-Moller. 1997. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J. Gen. Virol. 78:367-372. [DOI] [PubMed] [Google Scholar]
- 5.Calderon, M. G., P. Remorini, O. Periolo, M. Iglesias, N. Mattion, and J. L. Torre. 2007. Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinated and nonvaccinated dogs in Argentina. Vet. Microbiol. 125:341-349. [DOI] [PubMed] [Google Scholar]
- 6.Chappuis, G. 1995. Control of canine distemper. Vet. Microbiol. 44:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocks, B. G., C. C. Chang, J. M. Carballido, H. Yssel, J. E. de Vries, and G. Aversa. 1995. A novel receptor involved in T-cell activation. Nature 376:260-263. [DOI] [PubMed] [Google Scholar]
- 8.Cosby, S. L., C. Lyons, S. P. Fitzgerald, S. J. Martin, S. Pressdee, and I. V. Allen. 1981. The isolation of large and small plaque canine distemper viruses which differ in their neurovirulence from hamsters. J. Gen. Virol. 52:345-353. [DOI] [PubMed] [Google Scholar]
- 9.Dahl, L., T. H. Jensen, E. Gottschalck, P. Karlskov-Mortensen, T. D. Jensen, L. Nielsen, M. K. Andersen, R. Buckland, T. F. Wild, and M. Blixenkrone-Moller. 2004. Immunization with plasmid DNA encoding the hemagglutinin and the nucleoprotein confers robust protection against a lethal canine distemper virus challenge. Vaccine 22:3642-3648. [DOI] [PubMed] [Google Scholar]
- 10.Demeter, Z., B. Lakatos, E. A. Palade, T. Kozma, P. Forgach, and M. Rusvai. 2007. Genetic diversity of Hungarian canine distemper virus strains. Vet. Microbiol. 122:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhiman, N., G. A. Poland, J. M. Cunningham, R. M. Jacobson, I. G. Ovsyannikova, R. A. Vierkant, Y. Wu, and V. S. Pankratz. 2007. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD 46 measles virus receptors. J. Allergy Clin. Immunol. 120:666-672. [DOI] [PubMed] [Google Scholar]
- 12.Green, C. E., and M. J. Appel. 2006. Canine distemper. p. 25-41. In C. E. Green (ed.), Infectious diseases of the dog and cat, 3rd ed. Saunders Elsevier, St. Louis, MO.
- 13.Harder, T. C., M. Kenter, H. Vos, K. Siebelink, W. Huisman, G. van Amerongen, C. Orvell, T. Barrett, M. J. G. Appel, and A. D. M. E. Osterhaus. 1996. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J. Gen. Virol. 77:397-405. [DOI] [PubMed] [Google Scholar]
- 14.Junge, R. E., K. Bauman, M. King, and M. E. Gompper. 2007. A serologic assessment of exposure to viral pathogens and Leptospira in an urban raccoon (Procyon lotor) population inhabiting a large zoological park. J. Zoo Wildl. Med. 38:18-26. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. H., K. W. Cho, H. Y. Youn, H. S. Yoo, and H. R. Han. 2001. Detection of canine distemper virus (CDV) through one step RT-PCR combined with nested PCR. J. Vet. Sci. 2:59-63. [PubMed] [Google Scholar]
- 16.Kubo, T., Y. Kagawa, H. Taniyama, and A. Hasegawa. 2007. Distribution of inclusion bodies in tissues from 100 dogs infected with canine distemper virus. J. Vet. Med. Sci. 69:527-529. [DOI] [PubMed] [Google Scholar]
- 17.Lan, N. T., R. Yamaguchi, A. Inomata, Y. Furuya, K. Uchida, S. Sugano, and S. Tateyama. 2006. Comparative analysis of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet. Microbiol. 115:32-42. [DOI] [PubMed] [Google Scholar]
- 18.Lan, N. T., R. Yamaguchi, K. Uchida, S. Sugano, and S. Tateyama. 2005. Growth profiles of recent canine distemper isolates on Vero cells expressing canine signaling lymphocyte activation molecule (SLAM). J. Comp. Pathol. 133:77-81. [DOI] [PubMed] [Google Scholar]
- 19.Lednicky, J. A., J. Dubach, M. J. Kinsel, T. P. Meehan, M. Bocchetta, L. L. Hungerford, N. A. Sarich, K. E. Witecki, M. D. Braid, C. Pedrak, and C. M. Houde. 2004. Genetically distinct American distemper virus lineages have recently caused epizootics with somewhat different characteristics in raccoons living around a large suburban zoo in the USA. Virol. J. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martella, V., F. Cirone, G. Elia, E. Lorusso, N. Decaro, M. Campolo, C. Desario, M. S. Lucente, A. L. Bellacicco, M. Blixenkrone-Moller, L. E. Carmichael, and C. Buonavoglia. 2006. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains in Italy. Vet. Microbiol. 116:301-309. [DOI] [PubMed] [Google Scholar]
- 21.Martella, V., G. Elia, M. S. Lucente, N. Decaro, E. Lorusso, K. Banyai, M. Blixenkrone-Moller, N. T. Lan, R. Yamaguchi, F. Cirone, L. E. Carmichael, and C. Buonavoglia. 2007. Canine distemper virus (CDV) by hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet. Microbiol. 122:32-42. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy, A. J., M. A. Shaw, and S. J. Goodman. 2007. Pathogen evolution and disease emergence in carnivores. Proc. Biol. Sci. 274:3165-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyers, D. L., A. Zurbriggen, H. Lutz, and A. Pospischil. 1997. Distemper: not a new disease in lions and tigers. Clin. Diagn. Lab. Immunol. 4:180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mochizuki, M., M. Hashimoto, S. Hagiwara, Y. Yoshida, and S. Ishiguro. 1999. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J. Clin. Microbiol. 37:2936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norris, J. M., M. B. Krockenberger, A. A. Baird, and G. Knudsen. 2006. Canine distemper: re-emergence of an old enemy. Aust. Vet. J. 84:362-363. [DOI] [PubMed] [Google Scholar]
- 26.Pardo, I. D. R., G. C. Johnson, and S. B. Kleiboeker. 2005. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J. Clin. Microbiol. 43:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki, F., N. Ono, R. Yamaguchi, and Y. Yanagi. 2003. Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine SLAM (CD150) and their adaptability to marmoset B95a cells. J. Virol. 77:9943-9950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon-Martinez, J., R. Ulloa-Arvizu, V. E. Soriano, and R. Fajardo. 2007. Identification of a genetic variant of canine distemper virus from clinical cases in two vaccinated dogs in Mexico. Vet. J. [Epub ahead of print.] doi: 10.1016/j.tvjl.2007.01.015. [DOI] [PubMed]
- 29.von Messling, V., N. Oezguen, Q. Zheng, S. Vongpunsawad, W. Braun, and R. Cattaneo. 2005. Nearby clusters of hemagglutinin residues sustain SLAM-dependent canine distemper virus entry in peripheral blood mononuclear cells. J. Virol. 79:5857-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenzlow, N., P. Plattet, R. Wittek, A. Zurbriggen, and A. Grone. 2007. Immunohistochemical demonstration of the putative canine distemper virus receptor CD10 in dogs with and without distemper. Vet. Pathol. 44:943-948. [DOI] [PubMed] [Google Scholar]