Abstract
Recent clinical trials have shown that the presence of a robust human immunodeficiency virus type 1 (HIV-1)-specific T-cell response may not be sufficient to prevent or control HIV-1 infection. Studies of antigen processing in the context of infectious HIV-1 are therefore warranted. Envelope-specific, major histocompatibility complex class II-restricted murine T-cell hybridomas were tested for responsiveness to splenic antigen-presenting cells exposed to HIV-1-infected GHOST cells. Interleukin-2 assays showed that the presence of a peptide within HIV-1 did not ensure the reactivation of peptide-specific T cells. Further experiments defined the impact of gamma interferon-induced thiol reductase and cysteine proteases on the processing of HIV-1 peptides. The results highlight potential influences of peptide context on T-cell reactivation by HIV-1 and encourage the continued study of antigen processing as support for improved vaccine design.
Most current human immunodeficiency virus type 1 (HIV-1) vaccines are designed to activate HIV-1-specific T cells, with or without accompanying B cells. There have been numerous T-cell-based vaccine strategies, including the following: (i) the linkage of prominent major histocompatibility complex (MHC) class I- and class II-restricted T-cell targets (26), (ii) the use of viral proteins or fusion proteins carrying viral peptides (24), (iii) the insertion of peptides or proteins into DNA or viral vectors (9), (iv) the synthesis of ancestral or consensus sequences (22), and (v) the scrambling of peptides from more than one viral protein (31). The preferred outcome is that T cells will reactivate in the context of a sequence-similar infectious HIV-1 challenge. However, the presence of robust HIV-1-specific T cells need not translate to the prevention or control of HIV-1 infection (21). In fact, a recent phase IIb study of a T-cell-based vaccine revealed no protective effect (among 741 vaccinated and 762 unvaccinated study participants, 24 and 21 participants were infected, respectively).
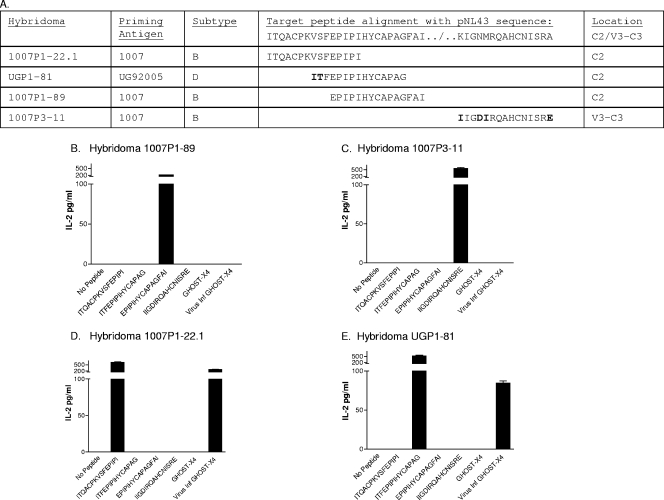
Disappointments in the field of HIV-1 vaccine development have highlighted the need for a better understanding of lymphocyte activation requirements. We therefore initiated studies of T-cell responses to HIV-1-infected cells. Specifically, murine T-cell hybridomas with known HIV-1 envelope peptide specificities (Fig. 1A) were tested for interleukin-2 (IL-2) production following stimulation with autologous splenocytes exposed to HIV-1-infected CXCR4 GHOST cells. These hybridomas originally derived from C57BL/6 mice immunized with a prime-boost regimen using either 1007 (clade B virus; GenBank accession number AF321563)- or UG92005 (clade D virus; GenBank accession number AF338704)-derived HIV-1 gp140 recombinant constructs (6, 29). The murine system afforded us access to the immortalized T cells as well as to the antigen-presenting cells (APCs) of both wild-type and knockout (KO) phenotypes (18). The fact that mouse cells are resistant to HIV-1 infection (32) also facilitated a focused study of exogenous antigen processing, a situation that may typify a fraction of human APCs (dendritic cells often resist HIV-1 infection despite internalization of the infectious particle [12, 15, 17, 34]). Our goal was to identify antigen-processing potentials in the mouse system to inform follow-up studies with human T cells.
FIG. 1.
The peptide sequence within HIV-1 does not ensure T-cell activation. (A) Hybridomas are listed with the names, subtypes, and target sequences of the priming antigens. The target sequences are aligned with sequences from HIV-1NL4-3. Residue differences are indicated in bold. (B to E) The IL-2 responses of hybridomas 1007P1-89, 1007P3-11, 1007P1-22.1, and UGP1-81 toward peptides (10 μM) or GHOST cells (105 cells either infected with HIV-1NL4-3 or uninfected) were measured. The HIV-1NL4-3 used for GHOST cell infections was prepared by first purifying DNA (with a Qiagen maxiprep kit) from bacteria carrying the pNL4-3 infectious clone (originally derived from HIV-1IIIB; National Institutes of Health AIDS Research and Reference Reagent Repository [catalogue no. 114 for pNL4-3]). DNA was then used to transfect 293 T cells in Dulbecco's modified Eagle's medium-l-glutamine (without fetal calf serum) with Lipofectamine plus reagent (Invitrogen) as recommended by the manufacturer. After a 3-h incubation at 37°C and 5% CO2, cells were cultured in Dulbecco's modified Eagle's medium plus 20% fetal calf serum for 2 days. The virus from 293 T-cell supernatants was expanded on GHOST-X4 cells (an osteosarcoma cell line; AIDS Research and Reference Reagent Repository) in R10 medium (RPMI 1640, 10% fetal calf serum, glutamine, and antibiotics) for 3 days and stored frozen.
The presence of a matched peptide sequence or subtype need not correlate with T-helper cell reactivation toward HIV-1-infected cells.
To conduct T-cell assays, hybridomas were washed and resuspended in complete tumor medium (13) and added to flat-bottomed microliter plates at 105 cells/well. C57BL/6 splenocytes (5 × 105 cells/well) were added to wells along with target synthetic peptides (10 μM) or HIV-1-infected cells for overnight incubation at 37°C and 10% CO2. The HIV-1-infected cells were CXCR4 GHOST cells (1 × 105) that had been infected 3 days previously with cloned HIV-1NL4-3, a derivative of HIV-1IIIB. Uninfected GHOST cells (1 × 105) served as controls. After 1 day of T-cell stimulation, supernatants were collected and IL-2 assays were performed. We note that GHOST cell supernatants were also tested in antigen presentation assays, but they did not trigger T-cell activity, perhaps because the bulk of the virus was cell associated. Results from the hybridoma assays are shown in Fig. 1B to E. Based on the comparisons of known T-cell peptide targets and the HIV-1NL4-3 sequence (Fig. 1A), one might have predicted that hybridomas 1007P1-22.1 and 1007P1-89 would respond to HIV-1NL4-3. This was not the case. Rather, hybridomas 1007P1-89 and 1007P3-11 (Fig. 1B and C) did not respond, while hybridomas 1007P1-22.1 and UGP1-81 (D and E) were both responsive. Of particular interest was the lack of responsiveness by hybridoma 1007P1-89, as its target sequence was precisely matched with that of HIV-1NL4-3. Results demonstrated that the presence of a target sequence within HIV-1 did not ensure T-cell reactivity (3, 25, 36) and illustrated the influence of the peptide context on antigen presentation (5-7, 29).
It was also clear that the subtype of the immunogen used to elicit T-cell activity did not predict T-cell responsiveness to the virus (6). For example, hybridoma UGP1-81, which was elicited by vaccination with an envelope of subtype D, was responsive to the HIV-1NL4-3 subtype B virus, while hybridoma 1007P3-11, which was elicited by vaccination with an envelope of subtype B, did not respond to another subtype B virus.
Gamma interferon-inducible lysosomal thiol reductase (GILT) influences MHC class II-associated HIV-1 envelope processing.
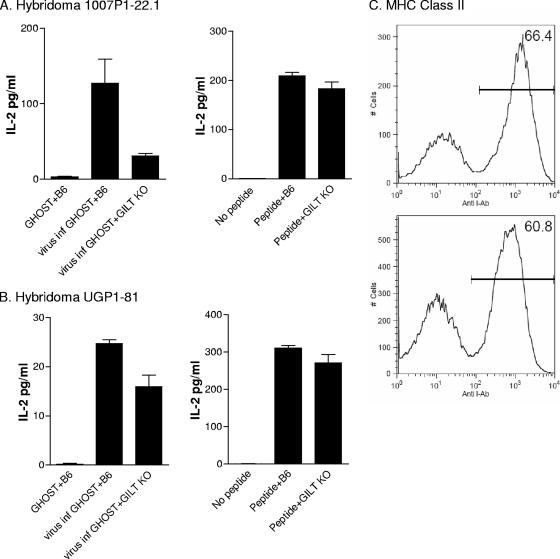
Given that hybridomas 1007P1-22.1 and UGP1-81.1 were responsive to envelope peptides presented by autologous splenocytes exposed to HIV-1-infected GHOST cells, we had an opportunity to begin the dissection of antigen-processing mechanisms necessary for T-cell reactivation. We first noted that based on the X-ray crystal structure described by Kwong et al. (16, 35), the target peptides of the 1007P1-22.1 and UGP1-81.1 hybridomas were both located in the vicinity of two antiparallel beta sheets (Fig. 2A). The target sequence of hybridoma 1007P1-22.1 straddled a disulfide-bonded cysteine residue at the base of the V1/V2 loops, while the downstream target sequence of hybridoma UGP1-81 straddled a different disulfide-bonded cysteine residue. A further mapping of hybridoma target epitopes is shown in Fig. 2C. Results confirmed that the 1007P1-22.1 and UGP1-81 hybridomas had different requirements for activation. Hybridoma 1007P1-22 responded to peptides containing the QACPKVSFEP or QACPKITFEP sequence. Hybridoma UGP1-81 responded to peptides containing the VSFEPIPIHYCAP or ITFEPIPIHYCAP sequence.
FIG. 2.
Peptide context within the three-dimensional structure of the HIV-1 envelope. (A) The positions of target peptides for hybridomas 1007P1-22.1 and UGP1-81 are shown within the predicted three-dimensional structure of the gp120 envelope protein. The crystal formation was previously described by Kwong et al. (HIV-1IIIB sequence) (16, 35). Black arrows define nonparallel beta sheets; red boxes define positions of disulfide bonds. Yellow, green, and blue regions identify positions of overlapping peptide targets for hybridomas 1007P1-22.1 and UGP1-81. (B) T-cell hybridomas were stimulated with syngeneic splenocyte APCs and an array of peptides from the C2 region of the HIV-1 envelope protein.
Based on the locations of the peptide targets, we questioned whether disulfide bond reduction was a requirement for viral antigen processing. Therefore, the virus-processing capacities of splenocytes from C56BL/6 and GILT KO mice were compared (18). Hybridoma experiments were performed as described above, using the splenocytes from two different sources as the APCs. Again, the antigen source was CXCR4 GHOST cells (1 × 105) that had been infected 3 days previously with HIV-1. We found that GHOST cell cultures infected with either HIV-1NL4-3 or HIV-1IIIB could be used interchangeably, yielding similar results (despite possible differences between the absolute envelope content in the two HIV-1-infected cell populations). As seen in Fig. 3A, the processing of viral antigen was significantly dampened in the absence of GILT for hybridoma 1007P1-22.1 (P < 0.05, t test). Control experiments with synthetic peptide showed little difference between the C57BL/6 wild-type and KO APCs. As shown in Fig. 3B, GILT KO APCs were more comparable to C57BL/6 cells in their capacities to present viral antigen to hybridoma UGP1-81 than to hybridoma 1007P1-22.1, suggesting a lesser requirement for GILT activity. As with hybridoma 1007P1-22.1, the synthetic peptide for UGP1-81 was well presented by both types of APCs, suggesting that MHC class II expression was normal for GILT KO animals. This was confirmed by fluorescence-activated cell sorter (FACS) analyses using monoclonal antibodies specific for MHC class II molecules; the MHC expression patterns for C57BL/6 and GILT KO splenocytes were equivalent (Fig. 3C). Results are consistent with the theory that GILT assists in the unfolding of the heavily disulfide-bonded HIV-1 envelope protein and supports peptide release for MHC binding and T-cell activation. The differential requirements of the 1007P1-22.1 and UGP1-81 hybridomas for GILT activities may simply relate to the greater proximity of 1007P1-22.1 target peptides to the two disulfide bonds at the base of V1/V2. A differential requirement for thiol reductase activity has been similarly described for the presentation of two influenza virus hemagglutinin peptides to T-helper hybridomas (30).
FIG. 3.
GILT influences peptide processing for T-helper cell activation. T cells were stimulated with APCs (splenocytes from C57BL/6J or GILT KO mice) and either peptide or 105 GHOST cells (HIV-1IIIB virus infected or uninfected; results with HIV-1IIIB- and HIV-1NL4-3-infected GHOST cells were similar [HIV-1IIIB was provided by R. V. Srinivas and the NIH AIDS Research and Reference Reagent Repository]). Results are shown for hybridoma 1007P1-22.1 (A) and hybridoma UGP1-81 (B). (C) FACS profiles of splenocytes from C57BL/6 (top panel) and GILT KO (bottom panel) mice after cell staining with anti-MHC class II antibodies (Pharmingen, catalogue no. 553552) and analysis on a FACScan (Becton Dickinson).
Antigen processing in the presence of leupeptin enhances hybridoma 1007P1-22.1 activity.
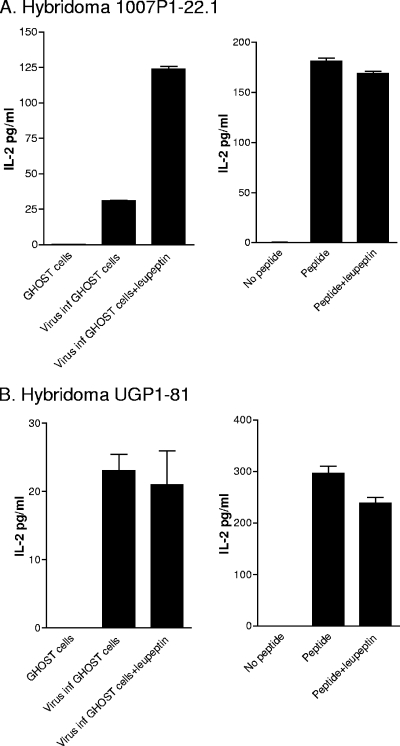
To examine the influence of an additional endolysosomal enzyme on T-cell activation, we tested the effects of leupeptin (an inhibitor of cysteine proteases) on HIV-1 viral antigen processing. Expectations were that the lack of cysteine protease might dampen antigen-processing capacities. Leupeptin experiments involved the preincubation of wild-type C57BL/6 mouse spleen cells with a drug (500 μM) for 1 hour and then incubation with GHOST cells (HIV-1 infected or uninfected) for 4 to 5 h in the continued presence of the drug. After the fixation and washing of APCs, hybridomas 1007P1-22.1 and UGP1-81 were added for overnight incubation and IL-2 assay. Synthetic peptides were used as controls. Of interest is the fact that leupeptin significantly enhanced the responsiveness of 1007P1-22.1 toward infectious HIV-1 (P < 0.05, t test) (Fig. 4A). There are a number of possible explanations for this unexpected result. First, it is possible that the cysteine protease activity yielded a target peptide fragment of suboptimal size or content for interaction with the I-Ab MHC class II molecule. Alternatively, the cysteine protease may have released an unrelated peptide with a superior affinity for the MHC peptide binding groove compared to that of the 1007P1-22.1 peptide target. Competitive inhibition may have then reduced target peptide-MHC interactions and subsequent 1007P1-22.1 T-cell activity. Hybridoma UGP1-81.1 was relatively unaffected by the presence of leupeptin during antigen processing (Fig. 4B).
FIG. 4.
The 1007P1-22.1 response is enhanced following APC drug treatment with leupeptin. T-cell hybridoma activities were measured by an IL-2 assay using syngeneic spleen cells that had been pulsed with antigen in the presence or absence of leupeptin. To prepare antigen-pulsed APCs, C56BL/6 splenocytes were first incubated with or without leupeptin (used at a final concentration of 500 μM; Calbiochem, San Diego, CA) for 1 h at 37°C and 10% CO2. Antigen was then added for a further 4-h incubation at 37°C and 10% CO2. Cells were washed twice in phosphate-buffered saline, pelleted by centrifugation, and suspended in 50 to 100 μl formaldehyde (0.1% final concentration). After 1 min, 2 to 3 ml undiluted heat-inactivated fetal calf serum (56°C, 45 min) was added and cells were washed twice with complete tumor medium. Antigens were either peptide (to yield a 10 μM final concentration), HIV-1-infected GHOST cells (HIV-1IIIB-infected GHOST cells were added to yield 105 cells/well; results with HIV-1IIIB and HIV-1NL4-3 were similar), or uninfected GHOST cells (105 cells/well).
The KO mouse and drug studies described here assist in the identification of enzymes (e.g., GILT and cysteine proteases) that may influence antigen processing in the context of HIV-1-infected cells. The studies highlight the complex nature of antigen processing, a feature that is not unique to infectious HIV-1 and which has been predicted by previous work in this and other research fields (2, 10, 20, 23, 27-29, 33). Future goals are to expand these studies to include tests of HIV-1 antigen processing in human APCs for both CD4+ and CD8+ T-cell populations. Despite obvious differences between mouse and human models, it is of interest that “hot spots” of immunodominant HIV-1 epitopes are often shared between mouse and human populations and between CD4+ and CD8+ T-cell subsets (5, 7, 29, 37). This is particularly evident for peptides in the C2 (e.g., VSFEPIPIHYCAP) and V3 regions of the HIV-1 envelope (29, 37). It is also of interest that mechanisms relevant to MHC class I and class II antigen processing, once thought to be entirely distinct, are now known to be influenced by similar intracellular enzymes and organelles (1, 19, 30). This may explain the sharing of HIV-1-specific epitopes between CD4+ and CD8+ T-cell populations, a topic worthy of further study.
A popular suggestion in the HIV-1 vaccine field has been that one or a few consensus or ancestral HIV-1 sequences may be sufficient to elicit protective T-cell responses toward diverse HIV-1 isolates (11, 14, 31). Such suggestions rely on the supposition that vaccine sequences, if matched or partially matched in the challenge virus, will be readily processed and presented to T-cell populations. However, this may not be the case if peptide contexts differ between the vaccine and the challenge virus. An improved understanding of antigen-processing mechanisms, partnered with knowledge of the HIV-1 sequence, may assist in the design of better vaccines. The formulation of antigen cocktails rather than single-antigen ancestral or consensus sequence vaccines may increase the likelihood that (at least a portion of) vaccine-induced T cells will see their targets in the event of HIV-1 exposure.
Conclusions.
When T-helper cell hybridomas were assessed for responsiveness to antigen in the context of live virus-infected cells, only a portion of cells (1007P1-22.1 and UGP1-81) were responsive. These results could not have been predicted by knowledge of the T-helper cell target sequence, as the immunogen target peptide used to elicit hybridoma UGP1-81 was nonidentical to that of HIV-1NL4-3, while the immunogen target peptide used to elicit hybridoma 1007P1-89 was identical to it. Furthermore, the responses could not have been predicted based on knowledge of the peptide subtype. The current study and studies of other antigenic systems emphasize the importance of peptide context on antigen presentation. In the case of HIV-1, one might expect that a particular peptide-specific T cell may have a different response pattern toward each different challenge virus. As one solution to the problem of various peptide contexts, the use of HIV-1 vaccine cocktails may be considered (13, 38). By increasing the diversity of vaccine-induced lymphocytes, the likelihood of lymphocyte reactivation toward any given HIV-1 challenge may be increased. The efficacy of the cocktail vaccine approach has now been demonstrated in a variety of fields (4, 8).
While the mouse studies begin to dissect influences on antigen processing (e.g., GILT and cysteine proteases both alter T-cell activities), further studies are needed. Future experiments with primate systems may unravel the complexities of antigen presentation in the context of various APCs (macrophage or dendritic cells), T-cells (CD4+ or CD8+ populations), and HIV-1 (CCR5 and CXCR4) sources. Such studies may advance our comprehension of basic biological concepts while informing the design of successful HIV-1 vaccines.
Acknowledgments
We thank R. V. Srinivas and the NIH AIDS Research and Reference Reagent Repository for HIV-1IIIB, bacteria containing the pNL4-3 infectious clone (catalogue no. 114 [contributor, Malcolm Martin]), and the UG92005 virus from which protein was obtained (contributor, World Health Organization). We thank V. KewalRamani and D. Littman for the GHOST cells (also from the NIH AIDS Repository). We thank Bart Jones and Pamela Freiden for their excellent technical assistance.
This work was funded in part by NIH NIAID grants P01 AI45142 and R37-AI023081. Additional funding was from the Howard Hughes Medical Institute, the Federated Department Stores, the Mitchell Fund, the Carl C. Anderson Sr. and Marie Jo Anderson Charitable Foundation, the Pendleton Fund, the Pioneer Fund, and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Ackerman, A. L., and P. Cresswell. 2004. Cellular mechanisms governing cross-presentation of exogenous antigens. Nat. Immunol. 5:678-684. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou, A. N., S. L. Blackwood, D. Mazzeo, and C. Watts. 2000. Control of antigen presentation by a single protease cleavage site. Immunity 12:391-398. [DOI] [PubMed] [Google Scholar]
- 3.Araneo, B. A., R. L. Yowell, and E. E. Sercarz. 1979. Ir gene defects may reflect a regulatory imbalance. I. Helper T cell activity revealed in a strain whose lack of response is controlled by suppression. J. Immunol. 123:961-967. [PubMed] [Google Scholar]
- 4.Biagini, R. E., S. A. Schlottmann, D. L. Sammons, J. P. Smith, J. C. Snawder, C. A. Striley, B. A. MacKenzie, and D. N. Weissman. 2003. Method for simultaneous measurement of antibodies to 23 pneumococcal capsular polysaccharides. Clin. Diagn. Lab. Immunol. 10:744-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S. A., T. D. Lockey, C. Slaughter, K. S. Slobod, S. Surman, A. Zirkel, A. Mishra, V. R. Pagala, C. Coleclough, P. C. Doherty, and J. L. Hurwitz. 2005. T cell epitope “hotspots” on the HIV type 1 gp120 envelope protein overlap with tryptic fragments displayed by mass spectrometry. AIDS Res. Hum. Retroviruses 21:165-170. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S. A., K. S. Slobod, S. Surman, A. Zirkel, X. Zhan, and J. L. Hurwitz. 2006. Individual HIV type 1 envelope-specific T cell responses and epitopes do not segregate by virus subtype. AIDS Res. Hum. Retroviruses 22:188-194. [DOI] [PubMed] [Google Scholar]
- 7.Brown, S. A., J. Stambas, X. Zhan, K. S. Slobod, C. Coleclough, A. Zirkel, S. Surman, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2003. Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in antibody activity. J. Immunol. 171:4140-4148. [DOI] [PubMed] [Google Scholar]
- 8.Clark, H. F., P. A. Offit, S. A. Plotkin, and P. M. Heaton. 2006. The new pentavalent rotavirus vaccine composed of bovine (strain WC3)-human rotavirus reassortants. Pediatr. Infect. Dis. J. 25:577-583. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. 2004. AIDS vaccines. HIV dodges one-two punch. Science 305:1545-1547. [DOI] [PubMed] [Google Scholar]
- 10.Eisenlohr, L. C., W. Gerhard, and C. J. Hackett. 1988. Individual class II-restricted antigenic determinants of the same protein exhibit distinct kinetics of appearance and persistence on antigen-presenting cells. J. Immunol. 141:2581-2584. [PubMed] [Google Scholar]
- 11.Finnefrock, A. C., X. Liu, D. W. Opalka, J. W. Shiver, D. R. Casimiro, and J. H. Condra. 2007. HIV type 1 vaccines for worldwide use: predicting in-clade and cross-clade breadth of immune responses. AIDS Res. Hum. Retroviruses 23:1283-1292. [DOI] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B., P. C. Groot, M. A. Nolte, S. J. van Vliet, S. T. Gangaram-Panday, G. C. van Duijnhoven, G. Kraal, A. J. van Oosterhout, and Y. van Kooyk. 2002. Marginal zone macrophages express a murine homologue of DC-SIGN that captures blood-borne antigens in vivo. Blood 100:2908-2916. [DOI] [PubMed] [Google Scholar]
- 13.Hurwitz, J. L., X. Zhan, S. A. Brown, M. Bonsignori, J. Stambas, T. D. Lockey, R. Sealy, S. Surman, P. Freiden, B. Jones, L. Martin, J. Blanchard, and K. S. Slobod. 2008. HIV-1 vaccine development: tackling virus diversity with a multi-envelope cocktail. Front. Biosci. 13:609-620. [DOI] [PubMed] [Google Scholar]
- 14.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 15.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 16.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maréchal, V., F. Clavel, J. M. Heard, and O. Schwartz. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maric, M., B. Arunachalam, U. T. Phan, C. Dong, W. S. Garrett, K. S. Cannon, C. Alfonso, L. Karlsson, R. A. Flavell, and P. Cresswell. 2001. Defective antigen processing in GILT-free mice. Science 294:1361-1365. [DOI] [PubMed] [Google Scholar]
- 19.Monu, N., and E. S. Trombetta. 2007. Cross-talk between the endocytic pathway and the endoplasmic reticulum in cross-presentation by MHC class I molecules. Curr. Opin. Immunol. 19:66-72. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, T. Y., and A. Y. Rudensky. 1999. The role of lysosomal proteinases in MHC class II-mediated antigen processing and presentation. Immunol. Rev. 172:121-129. [DOI] [PubMed] [Google Scholar]
- 21.Nature. 2007. HIV vaccine failure prompts Merck to halt trial. Nature 449:390. [DOI] [PubMed] [Google Scholar]
- 22.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussbaum, A. K., C. Kuttler, K. P. Hadeler, H. G. Rammensee, and H. Schild. 2001. PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53:87-94. [DOI] [PubMed] [Google Scholar]
- 24.Pinter, A., W. J. Honnen, S. C. Kayman, O. Trochev, and Z. Wu. 1998. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine 16:1803-1811. [DOI] [PubMed] [Google Scholar]
- 25.Shastri, N., G. Gammon, S. Horvath, A. Miller, and E. E. Sercarz. 1986. The choice between two distinct T cell determinants within a 23-amino acid region of lysozyme depends on their structural context. J. Immunol. 137:911-915. [PubMed] [Google Scholar]
- 26.Shirai, M., C. D. Pendleton, J. Ahlers, T. Takeshita, M. Newman, and J. A. Berzofsky. 1994. Helper-cytotoxic T lymphocyte (CTL) determinant linkage required for priming of anti-HIV CD8+ CTL in vivo with peptide vaccine constructs. J. Immunol. 152:549-556. [PubMed] [Google Scholar]
- 27.Sinnathamby, G., M. Maric, P. Cresswell, and L. C. Eisenlohr. 2004. Differential requirements for endosomal reduction in the presentation of two H2-E(d)-restricted epitopes from influenza hemagglutinin. J. Immunol. 172:6607-6614. [DOI] [PubMed] [Google Scholar]
- 28.Sjolander, S., A. Bolmstedt, L. Akerblom, P. Horal, S. Olofsson, B. Morein, and A. Sjolander. 1996. N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology 215:124-133. [DOI] [PubMed] [Google Scholar]
- 29.Surman, S., T. D. Lockey, K. S. Slobod, B. Jones, J. M. Riberdy, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2001. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 98:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tewari, M. K., G. Sinnathamby, D. Rajagopal, and L. C. Eisenlohr. 2005. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat. Immunol. 6:287-294. [DOI] [PubMed] [Google Scholar]
- 31.Thomson, S. A., A. B. Jaramillo, M. Shoobridge, K. J. Dunstan, B. Everett, C. Ranasinghe, S. J. Kent, K. Gao, J. Medveckzy, R. A. Ffrench, and I. A. Ramshaw. 2005. Development of a synthetic consensus sequence scrambled antigen HIV-1 vaccine designed for global use. Vaccine 23:4647-4657. [DOI] [PubMed] [Google Scholar]
- 32.Tsurutani, N., J. Yasuda, N. Yamamoto, B.-I. Choi, M. Kadoki, and Y. Iwakura. 2007. Nuclear import of the preintegration complex is blocked upon infection by human immunodeficiency virus type 1 in mouse cells. J. Virol. 81:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unanue, E. R. 2002. Perspective on antigen processing and presentation. Immunol. Rev. 185:86-102. [DOI] [PubMed] [Google Scholar]
- 34.Wieland, C. W., E. A. Koppel, J. den Dunnen, S. Florquin, A. N. McKenzie, Y. van Kooyk, T. van der Poll, and T. B. Geijtenbeek. 2007. Mice lacking SIGNR1 have stronger T helper 1 responses to Mycobacterium tuberculosis. Microbes Infect. 9:134-141. [DOI] [PubMed] [Google Scholar]
- 35.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 36.Yowell, R. L., B. A. Araneo, A. Miller, and E. E. Sercarz. 1979. Amputation of a suppressor determinant on lysozyme reveals underlying T-cell reactivity to other determinants. Nature 279:70-71. [DOI] [PubMed] [Google Scholar]
- 37.Zhan, X., J. L. Hurwitz, S. A. Brown, and K. S. Slobod. 2007. HIV-1 envelope T cell epitope “hotspots” among mice and humans and among CD4+ and CD8+ T cell subpopulations. AIDS Res. Hum. Retroviruses 23:471-476. [DOI] [PubMed] [Google Scholar]
- 38.Zhan, X., L. N. Martin, K. S. Slobod, C. Coleclough, T. D. Lockey, S. A. Brown, J. Stambas, M. Bonsignori, R. E. Sealy, J. L. Blanchard, and J. L. Hurwitz. 2005. Multi-envelope HIV-1 vaccine devoid of SIV components controls disease in macaques challenged with heterologous pathogenic SHIV. Vaccine 23:5306-5320. [DOI] [PubMed] [Google Scholar]