Summary
The Hippo (Hpo) signaling pathway governs cell growth, proliferation, and apoptosis by controlling key regulatory genes that execute these processes; however, the transcription factor of the pathway has remained elusive. Here we provide evidence that the TEAD/TEF family transcription factor Scalloped (Sd) acts together with the coactivator Yorkie (Yki) to regulate Hpo pathway-responsive genes. Sd and Yki form a transcriptional complex whose activity is inhibited by Hpo signaling. Sd overexpression enhances whereas its inactivation suppresses tissue overgrowth caused by Yki overexpression or tumor suppressor mutations in the Hpo pathway. Inactivation of Sd diminishes Hpo target gene expression and reduces organ size whereas a constitutively active Sd promotes tissue overgrowth. Sd promotes Yki nuclear localization whereas Hpo signaling retains Yki in the cytoplasm by phosphorylating Yki at S168. Finally, Sd recruits Yki to the enhancer of a pathway-responsive gene diap1, suggesting that diap1 is a direct transcriptional target of the Hpo pathway.
Introduction
How an organ stops growing when it reaches its appropriate size during development and regeneration has been a fascinating problem in modern biology; yet the underlying mechanisms are still poorly understood. Although tissue growth and organ size are influenced by hormonal signals and nutrients, organ-intrinsic mechanisms may exist to coordinate cell growth, cell proliferation, and cell death in response to both global and local stimuli. Classic grafting experiments carried out in Drosophila have suggested that imaginal discs, which give rise to adult structures such as wings, legs and eyes, possess intrinsic mechanisms to monitor their growth (Bryant and Simpson, 1984). Genetic screens carried out in Drosophila have identified many tumor suppressors whose loss of function results in overgrown imaginal disc derivatives (reviewed by Hariharan and Bilder, 2006). A number of tumor suppressors fall into an emerging tumor suppressor pathway, the so-called Hippo (Hpo) pathway (Edgar, 2006; Pan, 2007).
Central to the Hpo pathway is a kinase cascade consisting of an upstream kinase Hpo (also called dMST), the Drosophila homolog of mammalian Ste20 family kinases MST1 and MST2 (Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003), and a downstream kinase Wts/Lats, a member of the Nuclear Dbf-2-related (NDR) kinase family (Justice et al., 1995; Xu et al., 1995). Hpo forms a complex with the scaffolding protein Salvador (Sav) to phosphorylate Wts and a small regulatory protein of Wts called Mats (Lai et al., 2005; Tapon et al., 2002; Wei et al., 2007; Wu et al., 2003). Wts acts in a complex with Mats to restrict cell growth and proliferation and promote cell death by regulating the transcription of Hpo pathway target genes including cyclin E (cycE), diap1, and bantam (Harvey et al., 2003; Jia et al., 2003; Lai et al., 2005; Pantalacci et al., 2003; Thompson and Cohen, 2006; Udan et al., 2003; Wu et al., 2003).
Several recent studies have identified the protocadherin Fat as a candidate receptor for the Hpo pathway (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006), as well as the intracellular signaling molecules Merlin (the Drosophila homolog of neurofibromatosis 2) and its related protein Expanded (Ex) that may link the membrane receptor(s) to the Hpo kinase cascade (Cho et al., 2006; Hamaratoglu et al., 2006; Maitra et al., 2006; Pellock et al., 2007). At the other end of the pathway, the Hpo/Wts kinase cascade appears to restrict tissue growth by phosphorylating and inactivating Yorkie (Yki), the Drosophila homolog of mammalian transcriptional coactivator YAP (Huang et al., 2005). However, the transcription factor that acts together with Yki to mediate the nuclear response to the Hpo signaling pathway has remained elusive. As such, it is not clear whether any of the known Hpo pathway-responsive genes are the direct transcriptional targets.
The mammalian Yki homolog, YAP, can function as a coactivator for several transcription factors including the Runt family member PEBP2α, the P53 family member P73, and the TEAD/TEF family of transcription factors (Strano et al., 2001; Vassilev et al., 2001; Yagi et al., 1999). However, the functional relationships between these transcription factors and YAP have not been explored in vivo. Therefore, it is not clear if any of these transcription factors mediate the physiological function of YAP in embryonic development and adult tissue homeostasis. Unlike mammals that contain four closely-related TEAD/TEF family members, Drosophila contains a single TEAD/TEF family member encoded by scalloped (sd) (Campbell et al., 1992). While strong mutations in sd cause lethality, hypomorphic mutants are viable but lose wing tissue, a phenotype attributed to the loss of the transcriptional complex formed between Sd and the product of the wing selector gene vestigial (vg) (Halder et al., 1998; Williams et al., 1991; Williams et al., 1993). In additional to its expression in wing discs, sd is also expressed in other tissues and cell types including eye discs where vg is not present (Campbell et al., 1992), suggesting that Sd may play a broader role than Vg in development. The observation that the TEAD/TEF family of transcription factors can interact with and utilize YAP as a coactivator prompted us to investigate if Sd interacts with Yki to regulate Hpo pathway target genes. Here we provide evidence that Sd functions as a transcription factor in the Hpo pathway to control tissue growth and organ size. We provide evidence that Yki-Sd directly regulates the transcription of diap1. Furthermore, we show that Sd and Hpo signaling play opposing roles in regulating Yki nuclear localization.
Results
Sd forms a transcriptional complex with Yki
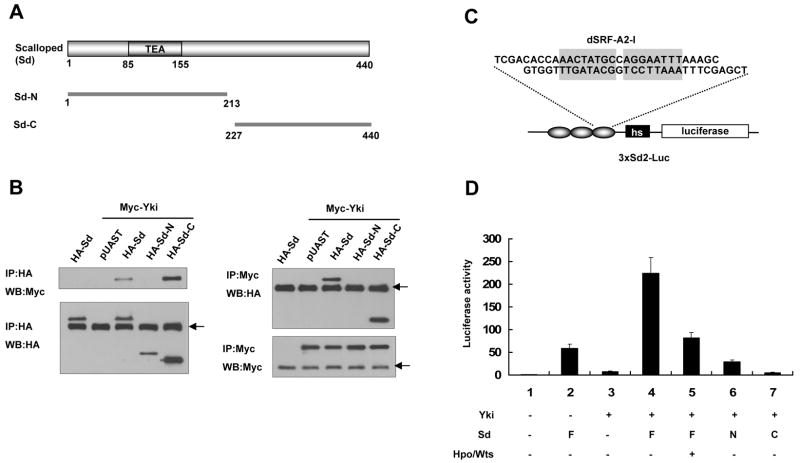
A yeast two-hybrid screen identified Sd as a binding partner for Yki (Giot et al., 2003). To determine if Sd and Yki form a complex in Drosophila cells, we transfected S2 cells with Myc-tagged Yki (Myc-Yki) and HA-tagged Sd (HA-Sd) and found they coimmunoprecipitated with each other (Fig. 1B). We also generated HA-tagged N-terminal Sd fragment (HA-SD-N) and C-terminal fragment (HA-Sd-C) (Fig. 1A), and found that HA-Sd-C but not HA-SD-N coimmunoprecipitated with Myc-Yki (Fig. 1B), suggesting that Yki interacts with the C-terminal half of Sd.
Figure 1. Sd and Yki form a transcriptional complex regulated by Hpo signaling.
(A) Diagram of Sd protein and deletion constructs. The TEA DNA binding domain is between aa 85–155. (B) S2 cells were transfected with the indicated Yki and Sd constructs, followed by immunoprecipitation and western blot analyses with the indicated antibodies. Arrows indicate IgG. (C) Diagram of the 3XSd2-luc reporter gene. Three copies of a DNA fragment containing tandem Sd binding sites (shaded sequence) from the dSRF enhancer (dSRF-A2-I) were placed upstream of the heat shock basal promoter (hs) followed by the luciferase coding sequence. (D) Sd and Yki act synergistically to activate the 3XSd2-luc reporter gene. S2 cells were transfected by the indicated Yki and Sd expression constructs plus the luciferase reporter gene, followed by the dual luciferase assay. F, N, and C indicate the full-length Sd, Sd-N, and Sd-C, respectively.
To determine if Sd and Yki form a functional transcriptional complex, we generated a luciferase (luc) reporter gene (3xSd2-Luc) containing multiple copies of tandem Sd binding sites from the dSRF enhancer (Fig. 1C; Halder et al., 1998). In S2 cells, transfection of Yki did not significantly activate 3xSd2-Luc whereas expression of Sd only modestly activated the reporter gene; however, coexpression of Sd with Yki synergistically activated the reporter gene (Fig. 1D). In contrast, neither Sd-N nor Sd-C could cooperate with Yki (Fig. 1D). Cotransfection of Hpo and Wts suppressed the reporter gene expression induced by the Yki-SD complex (Fig. 1D). Taken together, these results suggest that Sd and Yki form a transcriptional complex whose activity is inhibited by the Hpo pathway.
Sd acts cooperatively with Yki to induce tissue overgrowth
To determine the functional relationship between Sd and Yki in vivo, they were expressed either alone or together in eye discs using GMR-Gal4, a Gal4 driver that induces UAS transgene expression posterior to the morphogenetic furrow. Consistent with the previous finding that Yki overexpression induces tissue outgrowth (Huang et al., 2005), expressing Yki with GMR (GMR-Yki) resulted in enlarged eyes similar to those caused by loss of function of upstream Hpo pathway components (Fig. 2B, B′). In addition, GMR-Yki induced elevated expression of cycE and diap1, and ectopic BrdU incorporation posterior to the second mitotic wave (SMW) (Fig. 2E, E′, E″). Whereas overexpression of Sd alone did not induce tissue overgrowth (Fig. 5D), coexpression of Sd with Yki enhanced the overgrowth phenotype caused by Yki alone (Fig. 2C, C′), and led to a more dramatic increase in the expression of cycE and diap1 as well as BrdU incorporation posterior to the SMW (Fig. 2F, 2F′, F″). In contrast, neither Sd-N nor Sd-C enhanced the overgrowth phenotype induced by Yki (data not shown), consistent with the observation that Sd-N or Sd-C failed to support Yki-induced activation of 3xSd2-Luc in S2 cells (Fig. 1D). Of note, coexpressing Sd with Yki resulted in severely overgrown but depigmented eyes (Fig. 2C, C′), suggesting that pigment cell differentiation might also be affected.
Figure 2. Sd synergizes with Yki to promote tissue growth and Hpo target gene expression.
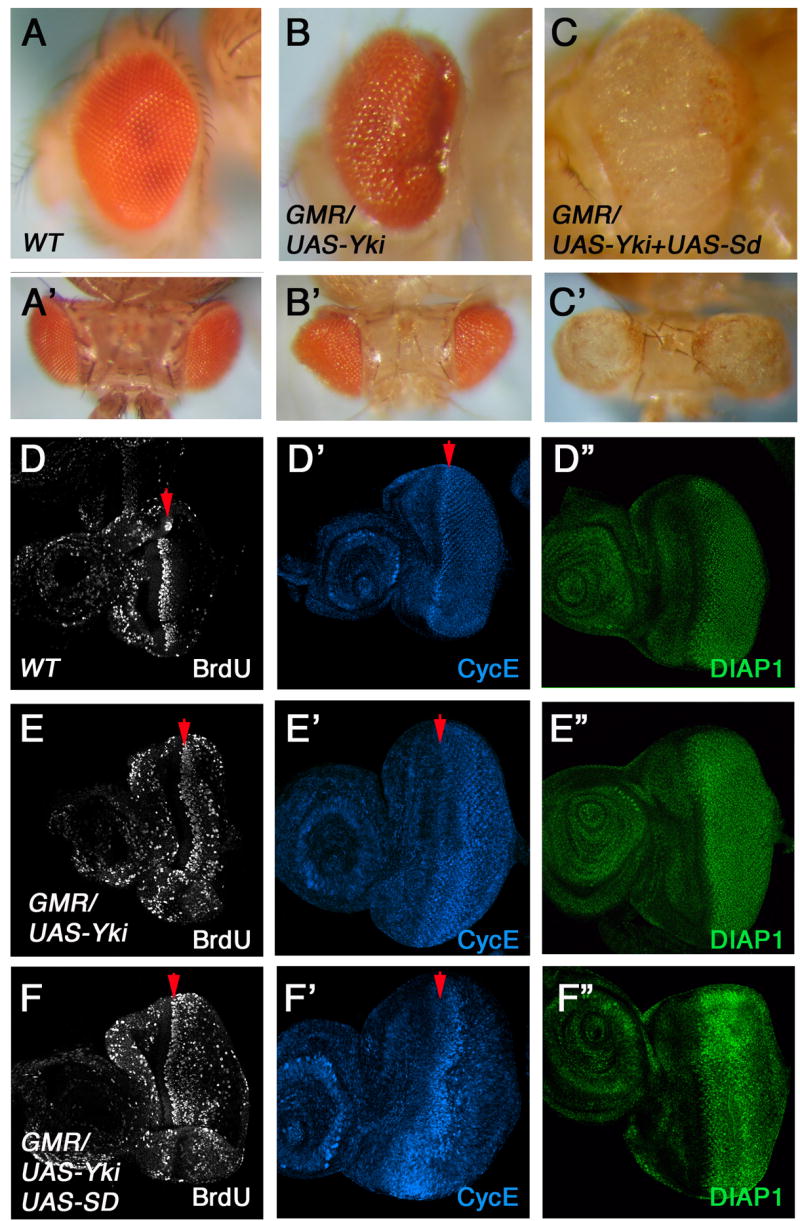
(A–C′) Side (A–C) or dorsal views (A′–C′) of wild type eye (A, A′), eyes expressing Yki (B, B′) or Yki + Sd (C, C′) with GMR-Gal4. (D–F″) BrdU incorporation (D, E, F), cycE (D′, E′, F′) and diap1 (D″, E″, F″) expression in wild type eye discs (D, D′, D″), or eye discs expressing Yki (E, E′, E″) or Yki +Sd (F, F′, F″). Arrowheads indicate the position of SMW.
Figure 5. An active form of Sd induces tissue overgrowth and activates Hpo target genes.
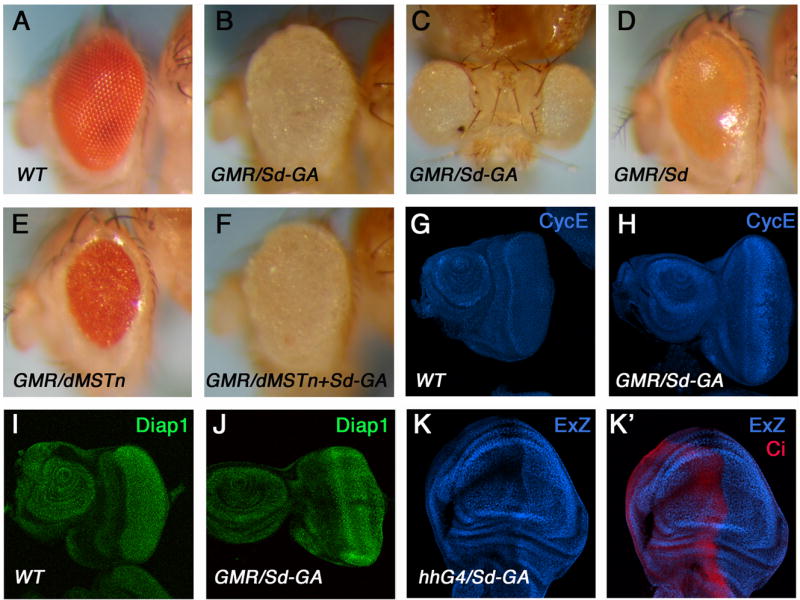
(A–D) Wild type eye (A), or eyes expressing Sd-GA (B, C) or Sd (D) with GMR-Gal4. (E, F) Adult eyes expressing dMSTn (E) or dMSTn plus Sd-GA (F) with GMR-Gal4. (G-J). Wild type eye discs (G, I) or eye discs expressing Sd-GA with GMR-Gal4 (H, J) were immunostained to show the expression of CycE (G–H) or Diap1 (I–J). (K, K′) A wing disc expressing Sd-GA with hh-Gal4 was immunostained to show the expression of ex-lacZ (blue) and Ci (red). Ci marked the A-compartment.
Sd inactivation blocks Yki-induced tissue overgrowth
If Yki regulates Hpo pathway-responsive genes through Sd, inactivation of Sd should suppress the phenotypes caused by Yki overexpression. To perturb Sd activity, we generated two RNAi transgenes that target two non-overlapping regions of Sd: Sd-RNAiN and Sd-RNAiC. Both RNAi transgenes effectively knocked down coexpressed HA-Sd (Supplementary Fig. S1) and suppressed tissue overgrowth induced by Yki (cf. Fig. 3B–D with Fig. 3A). In addition, Sd knockdown suppressed the ectopic expression of cycE and diap1 as well as ectopic incorporation of BrdU induced by Yki (cf. Fig. 3L, N, P with Fig. 3K, M, O). In wing imaginal discs, overexpression of Yki in the posterior (P) compartment resulted in enlarged P-compartment as well as elevated expression of diap1 and ex-lacZ, an enhancer trap for ex that is induced in response to loss of Hpo signaling (Supplementary Fig. S2A, C; Hamaratoglu et al., 2006). All these phenotypes were suppressed by Sd knockdown (Supplementary Fig. S2B, D). Taken together, these results suggest that Sd is essential for elevated Yki to promote overgrowth of multiple tissues.
Figure 3. Inactivation of Sd blocks tissue overgrowth induced by Yki or Hpo pathway mutations.
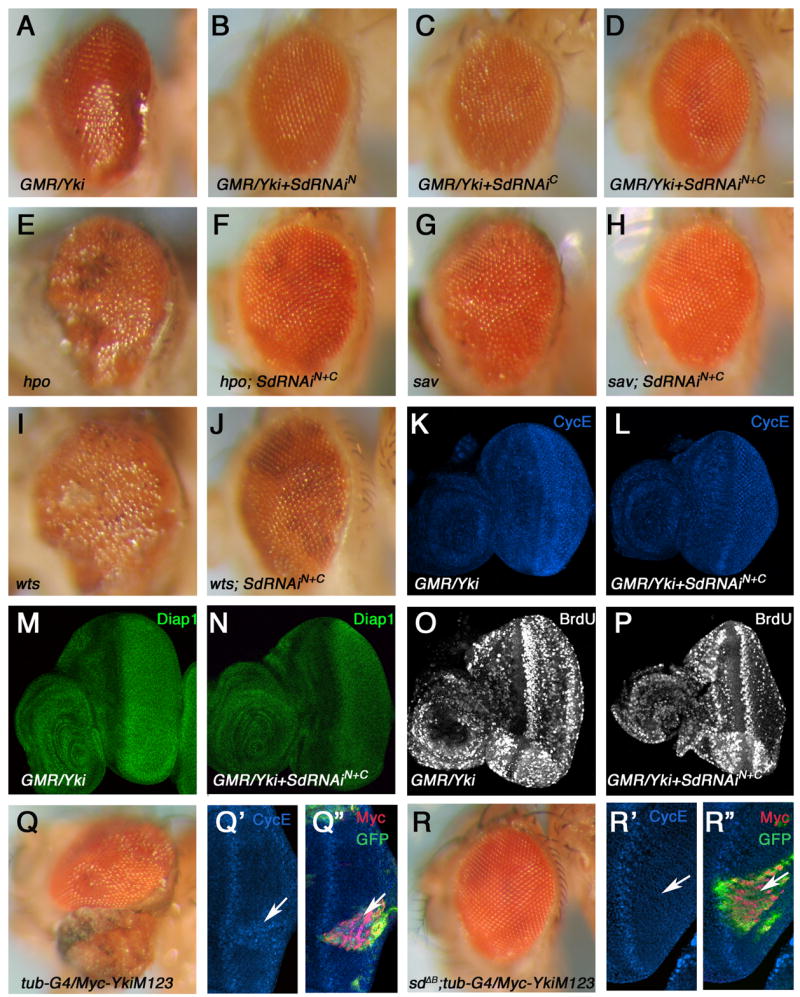
(A–D) Adult eyes of GMR-Gal4 UAS-Yki (A), GMR-Gal4 UAS-Yki; UAS-Sd-RNAiN (B), GMR-Gal4 UAS-Yki; UAS-Sd-RNAiC (C), and GMR-Gal4 UAS-Yki; UAS-Sd-RNAiN +UAS-Sd-RNAiC (D). (E–J) Adult eyes containing hpo (E), sav (G), or, wts (I) mutant clones or corresponding mutant clones that express UAS-SdRNAiN and UAS-SdRNAiC (F, H, J). (K–P) cycE (K–L) and diap1 (M–N) expression, and BrdU incorporation (O–P) in eye discs expressing Yki (K, M, O) or Yki plus Sd RNAi transgenes (L, N, P) with GMR-Gal4. (Q–Q″) Adult eye (Q) or eye disc (Q′, Q″) with YkiM123 expressing clones. YkiM123 expressing cells were recognized by Myc (red) and GFP (green) staining in discs (Q″). (R–R″) Adult eye (R) or eye disc (R′, R″) with sd B clones expressing YkiM123. sd B mutant cells expressing YkiM123 were labeled by both Myc and GFP expression (R″).
Genetic interaction between Sd and Hpo pathway components
Loss-of-function mutations in the hpo pathway components including hpo, sav, and wts resulted in similar tissue overgrowth phenotypes (Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Tapon et al., 2002; Udan et al., 2003; Wu et al., 2003). To provide further evidence that Sd acts in the Hpo pathway, we used the MARCM system to express Sd RNAi transgenes in clones mutant for hpo, sav, or wts (Lee and Luo, 2001). As shown in Fig. 3, adult eyes carrying hpo, sav, and wts mutant clones were overgrown (Fig. 3E, G, I). In contrast, adult eyes carrying hpo, sav, or wts mutant clones that express Sd RNAi transgenes exhibited nearly normal size (Fig. 3F, H, J), suggesting that tissue overgrowth caused by loss-of-function mutations in hpo, sav, and wts depends on Sd activity.
Inactivation of Sd phenocopies loss of Yki
To determine if Sd is required for normal eye growth, Sd RNAi transgenes were expressed under the control of eyeless-Gal4 (ey-Gal4). A UAS-Dicer2 transgene was coexpressed with Sd RNAi transgenes to increase RNAi efficiency (Dietzl et al., 2007). To compare the phenotypes caused by Sd inactivation with those resulted from Yki inactivation, we constructed two Yki RNAi transgenes, Yki-RNAiN and Yki-RNAiC, which target the N- and C-terminal regions of Yki, respectively. Expression of either Yki-RNAiN or Yki-RNAiC suppressed the overgrowth phenotype induced by Yki overexpression (Supplementary Fig. S3), suggesting that both RNAi constructs effectively knocked down Yki. Expression of Sd-RNAiN, Sd-RNAiC or both resulted in the formation of small eyes (Fig. 4B–D). Expression of either Yki-RNAiN or Yki-RNAiC produced a similar small eye phenotype (Fig. 4E; data not shown). The small eye phenotype caused by Sd or Yki RNAi was much less severe in the absence of Dicer2, and expression of Dicer alone did not cause any discernible phenotypes (data not shown).
Figure 4. Inactivation of Sd reduces organ size and downregulates Hpo target genes.
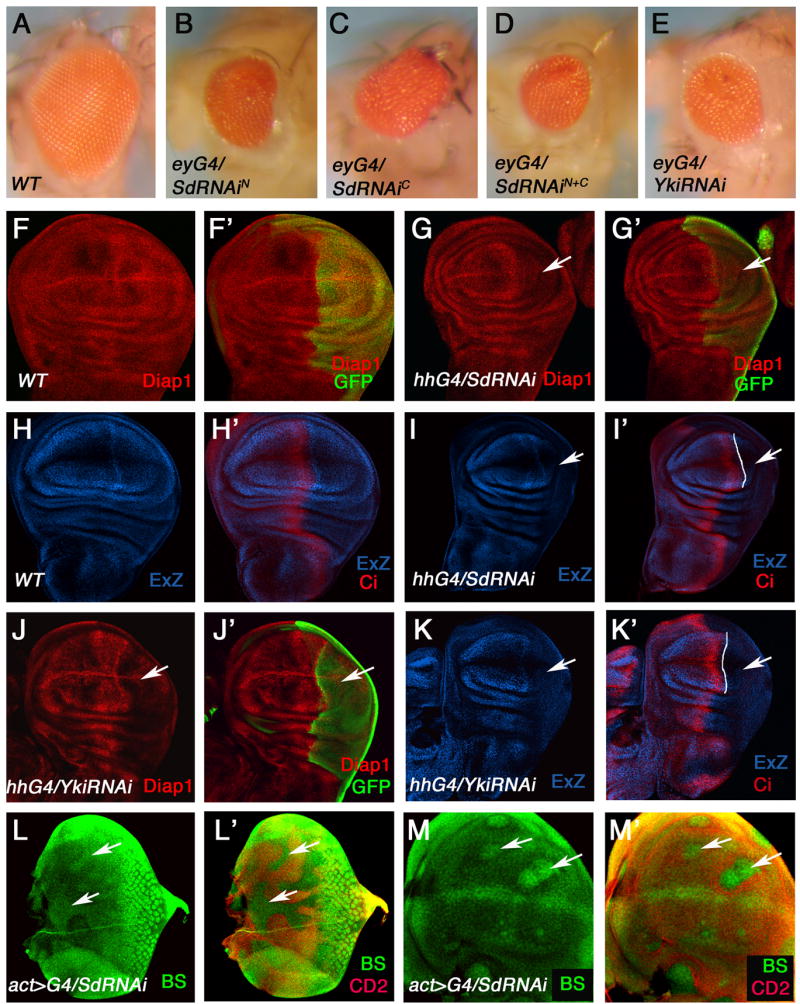
(A–E) Adult eyes of wild type (A), or expressing two copies of UAS-SdRNAiN (B), two copies of UAS-SdRNAiC (C), UAS-SdRNAiN +UAS-SdRNAiC (D), or two copies of UAS-Yki-RNAiN (E) with ey-Gal4 in the presence of UAS-Dicer2. (F–G′, J–J′) Wing discs expressing UAS-GFP (F, F′), UAS-GFP +UAS-Sd-RNAiN + UAS-Sd-RNAiC (G, G′), or UAS-GFP plus two copies of UAS-Yki-RNAi (J, J′) with hh-Gal4 were immunostained to show the expression of Diap1(red) and GFP (green). GFP marked the P-compartment (arrows). (H–I′, K, K′) Wild type wing disc (H, H′), or wing discs expressing UAS-Sd-RNAiN +UAS-Sd-RNAiC (I, I′) or two copies of UAS-Yki-RNAi (K, K′) with hh-Gal4 were immunostained to show the expression of Ci (red) and ex-lacZ (blue). The A/P boundary is indicated by white lines based on Ci expression (I′, K′). Arrows indicate the P-compartment. (L–M′) Eye (L, L′) and wing (M. M′) discs containing flip-out clones expressing UAS-Sd-RNAiN +UAS-Sd-RNAiC with act>CD2>Gal4 were immunostained to show the expression of CD2 (red) and a bantam sensor (BS; green). Cells expressing Sd-RNAi transgenes were marked by the lack of CD2 expression (arrows).
To examine the effect of Sd inactivation on the expression of Hpo pathway-responsive genes, Sd RNAi transgenes were expressed under the control of hh-Gal4, which activates UAS transgenes in the posterior (P) compartment. As shown in Fig. 4, Sd knockdown resulted in diminished expression of diap1 and ex-lacZ in P-compartment cells as well as reduced compartment size (cf. Fig. 4G, G′, I, I′ with Fig. 4F, F′, H, H′). Similar results were obtained with Yki RNAi (Fig. 4J, J′, K, K′).
The microRNA bantam is a Hpo pathway target that promotes cell growth (Nolo et al., 2006; Thompson and Cohen, 2006). To determine if Sd regulates bantam, we examined the expression of a bantam sensor (a GFP reporter inhibited by bantam) in imaginal discs that contain clones of cells expressing Sd-RNAi transgenes. Sd knockdown increased bantam sensor expression (Fig. 4L, L′, M, M′), suggesting that bantam expression was diminished when Sd is inactivated. Hence, Sd is required for bantam expression during larval development.
To complement RNAi experiments, we generated two sd deletion mutants (sdΔB and sdΔC) using the FRT/FLP-mediated genomic deletion strategy (Parks et al., 2004). The deletion in sdΔB removed sd coding sequence from aa 170 to the C-terminus, resulting in a truncated protein that lacks Yki binding domain (Supplementary Fig. S4). The deletion in sdΔC removed the entire sd coding sequence as well as a neighboring gene, CG8509 (Supplementary Fig. S4A). In transheterozygotes with a hypomorphic allele (sd1), both sdΔB and sdΔC produced wing phenotypes similar to an sd deficiency, consistent with sdΔB and sdΔC being null alleles (Supplementary Fig. S4E-G). We generated sd mutant clones at different stages during larval development and found that sdΔB and sdΔC mutant clones behaved similarly with respect to their proliferation profiles and effects on diap1 expression. Consistent with a previous study using strong sd alleles (Liu et al., 2000), early-induced clones (48 to 72 hrs AEL) for sd null alleles were barely recovered in the wing pouch region (Supplementary Fig. S5A). Late-induced sd null clones (72 to 96 hrs AEL) survived to late third instar in the wing pouch region and exhibited reduced diap1 expression (Supplementary Fig. S5C). Early-induced sd null clones (48 to 72 hrs AEL) were frequently recovered in the notal and hinge regions of wing discs as well as in eye discs and exhibited little if any change in diap1 expression (Supplementary Fig. S5B, S5D). It is possible that sufficient amounts of Sd activity persist in sd null clones to activate the basal expression of diap1. Alternatively, the basal daip1 expression depends on other transcription factor(s).
Although the sd null mutations did not perturb diap1expression in the eye, sdΔB suppressed the overgrowth phenotype as well as ectopic cycE expression caused by excessive Yki activity (Fig. 3Q–3R″). These observations further strengthen the view that elevated Yki activity acts through Sd to promote tissue overgrowth.
A constitutively active Sd induces tissue overgrowth
To gain further evidence that Sd functions as a transcription factor in the Hpo pathway, we generated a constitutively active Sd (Sd-GA) in which the Gal4 activation domain was fused to Sd C-terminus. Strikingly, expression of Sd-GA with GMR-Gal4 resulted in overgrown eyes reminiscent to those induced by coexpression of Yki and Sd (cf. Fig. 5B–C with Fig. 2C, C′). Consistent with the overgrowth phenotype, Sd-GA induced elevated expression of Hpo target genes including cycE, diap1, and ex-lacZ (Fig. 5H, J, K, K′).
Hyperactivation of Hpo signaling by overexpressing the Hpo kinase domain (dMSTn) resulted in precocious cell death and formation of small eyes (Fig. 5E; Jia et al., 2003). Coexpression of Sd-GA with dMSTn reversed the small eye phenotype, leading to the formation of enlarged eyes similar to those caused by expressing Sd-GA alone (Fig. 5F), suggesting that Sd-GA is resistant to Hpo inhibition.
Characterization of diap1 enhancer elements
Although a number of Hpo pathway-responsive genes have been identified, it is still unclear if they are the direct transcriptional targets of the pathway. As a starting point to address this issue, we focused on the characterization of diap1. An enhancer trap line (thj5c8) inserted in the first intron of diap1 drives lacZ expression in patterns identical to the endogenous gene (Ryoo et al., 2002), suggesting that the genomic sequence near the insertion site may harbor diap1 enhancer elements. We therefore constructed several GFP reporter genes (diap1-GFP) that contain different genomic fragments flanking the thj5c8 insertion site (Fig. 6A). diap1-GFP4.3, which contains a 4.3 kb genomic fragment from +1.37 kb to +5.68 kb, drove GFP expression in patterns similar to those of thj5c8 (Fig. 6C, C′). Furthermore, diap1-GFP4.3 was activated by both Yki and Sd-GA (Fig. 6F, F′, G, G′). In contrast, diap1-GFP5.1, which carries a 5.2 kb genomic fragment from −1.31 kb to +3.86 kb, failed to activate GFP expression in either eye or wing discs (data not shown), indicating that the region between +3.86 kb and +5.68 kb is critical for diap1 expression. Consistent with this notion, diap1-GFP3.5 containing a 3.5 kb genomic fragment from +3.86 kb to +7.35 kb exhibited expression patterns similar to diap1-GFP4.3 (Fig. 6D, D′), and responded to both Yki and Sd-GA (Supplementary Fig. S6A–B). A reporter gene containing the genomic fragment from +3.86 kb and +5.68 kb (diap1-GFP1.8) drove GFP expression in both wing and eye discs; however, the GFP levels were much weaker in the wing pouch region and the antennal disc compared to diap1-GFP4.3 (Fig. 6E). In addition, diap1-GFP1.8 did not response to Yki or Sd-GA overexpression (Supplementary Fig. S6C–D). Taken together, these results suggest that the 1.8 kb genomic region from +3.86 kb and +5.68 kb contains critical diap1 enhancer elements; however, adjacent regions are required for full activation of diap1 as well as proper response to Yki-Sd.
Figure 6. Characterization of diap1 enhancers.
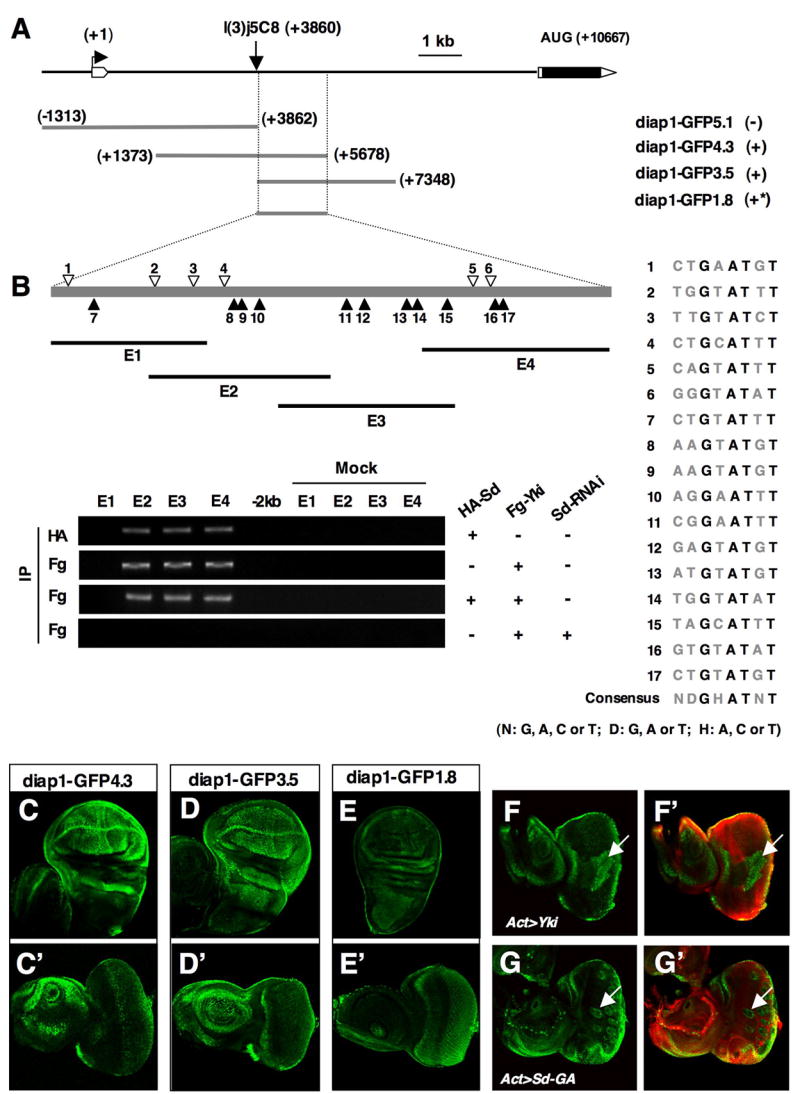
(A) Deletion analysis of diap1 enhancer elements. The diap1 genomic region containing the first intron and two neighboring exons is shown at the top. A diap1-lacZ enhancer trap line, l(3)j5C8, is inserted at +3860 bp. diap1-GFP transgenes containing indicated genomic fragments are listed. (−): no discernible expression; (*): weaker expression. (B) ChIP analysis of the 1.8 kb diap1 enhancer. Individual Sd binding consensus sites are indicated by open or filled arrowheads (filled arrowheads indicate the binding sites with reversed orientation). The sequences of individual Sd binding consensus sites are shown to the right. E1 to E4 demarcate the regions amplified by PCR in the ChIP experiments. (C–E′) GFP expression in wing (C, D, E) or eye (C′, D′, E′) discs driven by diap1-GFP4.3 (C, C′), diap1-GFP3.5 (D, D′), or diap1-GFP1.8 (E, E′). (F–G′) Eye discs that contain diap1-GFP4.3 and flip-out clones expressing Yki (F, F′) or Sd-GA (G, G′) with act>CD2>Gal4 were immunostained to the expression of GFP (green) and CD2 (red). Cells expressing Yki or Sd-GA were marked by the lack of CD2 expression.
Yki and Sd bind the 1.8 kb diap1 enhancer
The 1.8 kb diap1 enhancer contains a total of 17 sites that match the TEAD/Sd binding consensus (Fig. 6B; Anbanandam et al., 2006; Guss et al., 2001; Halder et al., 1998). To determine if Yki and Sd physically interact with the diap1 enhancer, we carried out chromatin coimmunoprecipitation (ChIP) experiments. We divided the 1.8 kb diap1 enhancer into four overlapping fragments (E1 to E4) each of which contains ~600 bp amplified by the polymerase chain reaction (PCR) in the ChIP assays (Fig. 6B). HA-Sd and Fg-Yki were expressed in eye discs with GMR-Gal4 either alone or in combination, and eye discs were dissected out for ChIP analysis. We found that both HA-Sd and Flag-Yki bound to E2, E3 and E4 but not E1 (Fig. 6B). As specificity controls, we found that neither HA-Sd nor Fg-Yki were able to bind a ~600 bp DNA fragment 2 kb 5′ to the 1.8 kb diap1 enhancer (Fig. 6B). In addition, neither HA-Sd nor Fg-Yki bound to the ubx PRE element under the same condition (data not shown). Sd knockdown abolished Fg-Yki binding to the diap1 enhancer (Fig. 6B), suggesting that Yki is recruited to the diap1 enhancer through Sd.
Sd promotes whereas Hpo signaling impedes Yki nuclear localization
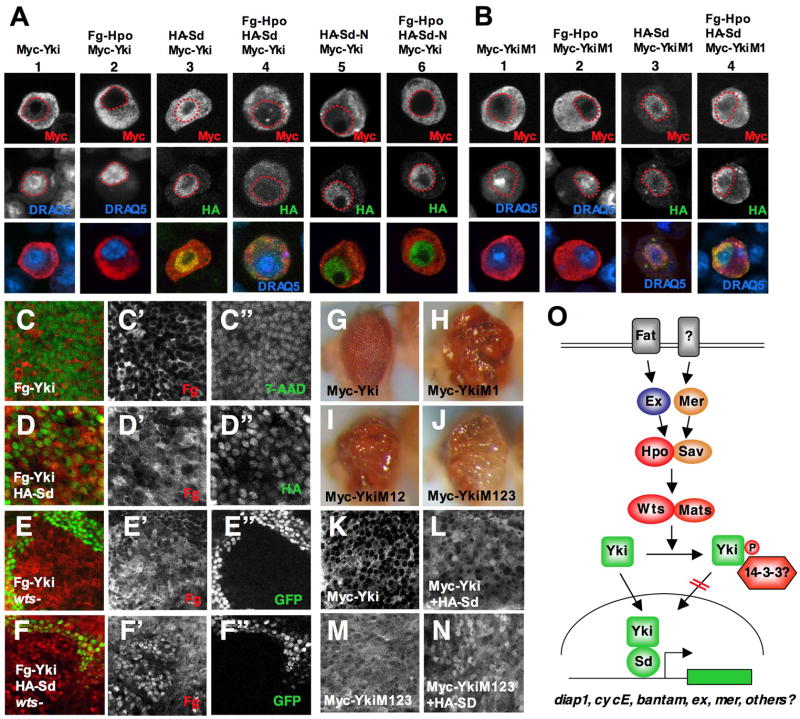
The mammalian homolog of Yki, YAP, appears to be regulated via cytoplasmic/nuclear shuttling (Basu et al., 2003), raising the possibility that Hpo signaling inhibits Yki by impeding its nuclear translocation. To test this hypothesis, we determined how the subcellular localization of epitope tagged Yki was influenced by either gain- or loss-of-Hpo signaling activity. When expressed alone in S2 cells, Myc-tagged Yki (Myc-Yki) was predominantly in the cytoplasm with low levels of nuclear staining (Fig. 7A, column 1). Coexpression of HA-Sd increased the nuclear levels of Myc-Yki (Fig. 7A, column 3). The increased nuclear localization of Myc-Yki depends on Yki-Sd interaction as coexpression of HA-Sd-N, which lacks the Yki interacting domain, failed to promote Myc-Yki nuclear translocation (Fig. 7A, column 5). On the other hand, coexpression of Flag-tagged Hpo (Fg-Hpo) abolished nuclear localization of Myc-Yki (Fig. 7A, column 2). When cotransfected with Myc-Yki in the presence of Fg-Hpo, HA-Sd but not HA-Sd-N was retained in the cytoplasm together with Myc-Yki (Fig. 7A, columns 4 and 6).
Figure 7. Hpo signaling regulates Yki nuclear localization and activity through phosphorylating S168.
(A–B) S2 cells expressing Myc-Yki (A) or Myc-YkiM1 (B) with or without Fg-Hpo and/or HA-Sd were immunostained with anti-Myc (red) and anti-HA (green) antibodies. Nuclei were marked by DRAQ5 (blue) and demarcated by red dashed lines. Images in individual channels were shown as black and white in top and middle rows and merged images in bottom rows. (C–D″) High magnification view of wing discs expressing Fg-Yki either alone (C–C″) or together with HA-Sd (D–D″) using MS1096 and immunostained with anti-Flag (red) and anti-HA (green in D) antibodies. The nuclei were labeled by 7-AAD (green in C–C″). (E–F″) High magnification view of wing discs carrying wts clones and expressing Fg-Yki either alone (E–E″) or together with HA-Sd (F–F″). wts mutant cells were marked by the lack of GFP expression (green). (G–J) Adult eyes expressing Myc-Yki (G), Myc-YkiM1 (H), Myc-YkiM12 (I), or Myc-YkiM123 (J) with GMR-Gal4. Of note, a strong transgenic line for Myc-Yki is shown here. (K–N) High magnification view of wing discs expressing Myc-Yki (K), Myc-Yki plus HA-Sd (L), Myc-YkiM123 (M), or Myc-YkiM123 plus HA-Sd (N). (O) The Drosophila Hpo pathway. See text for details.
To investigate whether Sd and Hpo signaling regulate the subcellular localization of Yki in vivo, a Flag-tagged Yki (Fg-Yki) was expressed either alone or together with HA-Sd in wing discs carrying wts mutant clones. Fg-Yki was predominantly localized in the cytoplasm of wild type wing disc cells (Fig. 7C–C″). The levels of nuclear Fg-Yki increased significantly in wts mutant clones (Fig. 7E–E″). Coexpression of HA-Sd also increased the nuclear levels of Fg-Yki (Fig. 7D–D″). Strikingly, coexpression of HA-Sd and Fg-Yki in wts mutant clones resulted in predominant nuclear localization of Fg-Yki (Fig. 7F–F″). Furthermore, the nuclear levels of endogenous Yki increased in wts or hpo mutant clones (Dong et al., 2007). Taken together, these observations suggest that Sd promotes Yki nuclear translocation by forming a complex with Yki whereas Hpo signaling impedes Yki nuclear translocation.
Phosphorylation of S168 inhibits Yki activity by preventing its nuclear localization
Phosphorylation of YAP at Ser127 has been implicated in mediating cytoplasmic sequestration (Basu et al., 2003). The corresponding residue in Yki is Ser168. We noticed that the sequence surrounding YAP S127/Yki S168 is highly conserved between YAP and Yki and includes two additional potential phosphorylation sites: S169 and S171 in Yki (Supplementary Fig. S7A). To determine the role of these Ser residues in regulating Yki subcellular localization and activity, we generated several Yki variants bearing Ser to Ala (SA) mutation in individual or combination of Ser residues (Supplementary Fig. S7A). When expressed in eye discs, Myc-tagged wild type Yki (Myc-Yki) induced little if any overgrowth of adult eyes (Fig. 7G; Supplementary Fig. S7B), suggesting that the N-terminal tag compromised Yki activity. Nevertheless, Myc-tagged Yki variants carrying SA mutation in S168 (Myc-YkiM1), S168 and S169 (Myc-YkiM12), or S168, S169, and S171 (Myc-YkiM123) induced dramatic overgrowth and appeared to be equally potent (Fig. 7H–J, Supplementary Fig. S7C, F, G). In contrast, Yki variants carrying SA mutation in S169 (Myc-YkiM2) or S171 (Myc-YkiM3) failed to induce eye overgrowth and behaved indistinguishable from Myc-Yki (Supplementary Fig. S7D–E). These observations suggest that phosphorylation at S168 plays a critical role in blocking Yki activity whereas S169 and S171 are not essential.
When expressed in wing discs, Myc-YkiM123 exhibited increased nuclear localization as compared with Myc-Yki (cf. Fig. 7M with Fig. 7K). Coexpression of HA-Sd further enhanced the nuclear localization of Myc-YkiM123 (7N). Similar results were obtained for Myc-YkiM1 (data not shown). These observations suggest that phosphorylation at S168 restricts Yki nuclear localization.
To determine if phosphorylation of S168 mediates the inhibition of Yki by Hpo signaling, Myc-YkiM1 and Myc-YkiM123 were transfected into S2 cells with Fg-Hpo. In contrast to Myc-Yki, which was completely excluded from the nucleus by Fg-Hpo (Fig. 7A, column 2), low levels of nuclear Myc-YkiM1 and Myc-YkiM123 were observed in the majority of transfected cells (Fig. 7B, column 2; data not shown). When HA-Sd was coexpressed, the majority of HA-Sd was retained in the nucleus with Myc-YkiM1 but sequestered in the cytoplasm with Myc-Yki in response to Hpo signaling (Fig. 7B column 4; Fig. 7A column 4). Thus, blocking S168 phosphorylation renders Yki partially resistant to Hpo-mediated inhibition.
Discussion
The Hpo pathway has emerged as a conserved signaling pathway that plays a critical role in controlling tissue growth and organ size. Despite the growing recognition of the importance of this pathway in development and cancer, the transcription factor that links the cytoplasmic components to the nuclear events has remained elusive and thus represents a major gap in the pathway. Our study demonstrates Sd is the missing transcription factor of the Hpo pathway based on several lines of genetic and biochemical evidence. First, Sd and Yki formed a transcriptional complex to activate a reporter gene in S2 cells and this transcriptional activity is inhibited by Hpo signaling. Furthermore, we demonstrated that Sd and Yki synergized in vivo to promote Hpo target gene expression and tissue overgrowth. Second and more importantly, loss of Sd function suppressed tissue overgrowth induced by Yki overexpression or loss-of-function mutations in hpo, sav, and wts. In addition, we found that Sd inactivation either by RNAi or a genetic mutation blocked the ectopic expression of Hpo responsive genes induced by excessive Yki activity. Third, RNAi knockdown of Sd phenocopied knockdown of Yki, which was manifested by reduced organ size and diminished expression of Hpo pathway-responsive genes. Fourth, a constitutively active form of Sd activated multiple Hpo pathway-responsive genes and promoted tissue overgrowth. Finally, Sd promoted Yki nuclear translocation and recruited Yki to the diap1 enhancer.
We generated several sd null alleles to further explore the consequence of loss of Sd. We found that sd null clones located in the wing pouch region exhibited growth deficit such that early-induced clones (48–72 hrs AEL) were eliminated by the end of late third instar. However, late-induced clones (72–96 hrs AEL) survived and exhibited diminished expression of diap1. In contrast, early-induced clones were recovered in the notal region of wing discs and in eye discs without showing discernible change in Diap1 levels. However, a previous study showed that yki mutant clones exhibited reduced diap1 expression in eye discs (Huang et al., 2005). It is possible that low levels of residual Sd activity persist in sd mutant clones, which are sufficient to support the basal expression of the Hpo target genes. Alternatively, Yki may act through another transcription factor to maintain the basal expression of Hpo target genes. Nevertheless, sd null mutation suppressed the overgrowth phenotype and ectopic cycE expression induced by excessive Yki activity, suggesting the residual Sd in sd mutant clones was insufficient to support the elevated Yki activity.
The identification of Hpo pathway transcription factor provided an opportunity to assess direct transcriptional targets of the pathway. To this end, we characterized the diap1 enhancer and identified a 1.8 kb enhancer element critical for diap1 expression. This region contains a total of seventeen predicted Sd binding sites. Using the ChIP assay, we demonstrated that both Sd and Yki physically interacted with the 1.8 kb diap1 enhancer and the association of Yki with the diap1 enhancer was mediated by Sd. Our results suggest that Sd recruits Yki to the diap1 enhancer to activate its transcription.
It has been shown previously that Sd acts in conjunction with Vg to promote wing development by directly regulating the expression of wing patterning genes (Guss et al., 2001; Halder et al., 1998). Here we have demonstrated that Sd acts in conjunction with Yki to control organ size by regulating the expression of genes involved in cell proliferation, cell growth, and apoptosis. These observations raise an important question of how Yki-Sd and Vg-Sd transcriptional complexes specifically select their targets. One possibility is that Vg-Sd and Yki-Sd prefer to interact with distinct Sd binding sites. Indeed, a previous study showed that binding of Vg to Sd modulated the DNA binding selectivity of Sd (Halder and Carroll, 2001). Another possibility is that target selectivity could be influenced by cofactors that bind in the vicinity of Sd binding sites. In support of this notion, previous studies have shown that wing specific enhancers contain both Sd binding sites and binding sites for transcription factors that mediate specific signaling pathways (Guss et al., 2001; Halder et al., 1998). It is also possible that Vg-Sd and Yki-Sd may share common targets. For example, diap1 could be activated by Vg-Sd in the wing pouch, which might explain why sd mutant clones in this region exhibited diminished diap1 expression.
In principle, the Hpo pathway could regulate the activity of Yki-Sd transcriptional complex at several levels. For example, Hpo signaling could regulate the formation Yki-Sd complex or the recruitment of other factor(s) to the Yki-Sd transcriptional complex. Alternatively, Hpo signaling could regulate the nuclear-cytoplasmic transport of Yki. In support of the latter possibility, Yki exhibited elevated nuclear localization in wts or hpo mutant clones (Fig. 7; Dong et al., 2007). In addition, coexpression of Hpo with Yki depleted nuclear Yki in S2 cells, suggesting that Hpo signaling impedes nuclear localization of Yki and thereby limits the amount of active Yki-Sd transcriptional complex.
Mutating Yki S168 to Ala increased nuclear localization and growth promoting activity of Yki (Fig. 7; Dong et al., 2007). In addition, Dong et al demonstrated that phosphorylation of Yki S168 was stimulated by Hpo (Dong et al., 2007). Phosphorylation of Yki by Hpo signaling increased their association with 14-3-3, which was abolished by mutating Yki S168 to Ala (our unpublished observation; Dong et al., 2007). As 14-3-3 often regulates nuclear-cytoplasmic shuttling of its interacting proteins (Muslin and Xing, 2000), these observations suggest that Hpo signaling inhibits Yki at least in part by phosphorylating Yki S168, which promotes 14-3-3 binding and cytoplasmic sequestration of Yki (Fig. 7O).
The Hpo pathway appears to restrict cell growth and control organ size in mammals (Camargo et al., 2007; Dong et al., 2007; Zhao et al., 2007). The finding that Sd is critical for Yki-induced tissue growth has raised the interesting possibility that the effect of YAP in promoting tissue growth may rely on the TEAD/TEF family of transcription factors. Corroborating this hypothesis, TEAD-2/TEF-4 protein purified from mouse cells was associated predominantly with YAP (Vassilev et al., 2001). Furthermore, YAP can bind to and stimulate the trans-activating activity of all four TEAD/TEF family members (Vassilev et al., 2001). The TEAD/TEF family members exhibit overlapping but distinct spatiotemporal expression patterns and thus may have redundant but unique roles during development (Kaneko and DePamphilis, 1998; Kaneko et al., 2007; Yasunami et al., 1996). It will be important to determine which TEAD/TEF family members are involved in the mammalian Hpo pathway and whether YAP employs distinct sets of TEAD/TEF transcription factors in different tissues. As abnormal activation of YAP is associated with multiple types of cancer (Dong et al., 2007; Overholtzer et al., 2006; Zender et al., 2006; Zhao et al., 2007), disrupting YAP-TEAD/TEF interaction may provide a new strategy for cancer therapeutics.
Experimental procedures
Mutants and Transgenes
hpo/dMSTBF33, hpo/dMSTJM1, savSH13, and wts/latsX1 are strong or null alleles (Jia et al., 2003; Xu et al., 1995). sd1 and the sd deficiency, Df(1)Exel6251, are described in Flybase (http://flybase.bio.indiana.edu). sdΔB and sdΔC were generated by FLP-mediated deletion using corresponding pairs of P-elements from Exelixis collection (Parks et al., 2004; Supplementary Information). To construct diap1-GFP transgenes, genomic fragments flanking the thj5c8 insertion site were cloned between the Bgl II and Kpn I sites in the PH-Stinger vectors (Barolo et al., 2000). Sd and Yki constructs are described in Supplementary Information. Other transgenes used in this study include bantam sensor (Brennecke et al., 2003), ex-lacZ (Hamaratoglu et al., 2006), GMR-Gal4 (Freeman, 1996), ey-Gal4 (Newsome et al., 2000), MS1096 (Wang et al., 1999), act>CD2>Gal4 (Pignoni et al., 1997).
Generation of mutant clones
FRT/FLP mediated mitotic recombination was used to generate mutant clones as previously described (Jiang and Struhl, 1995). The MARCM system was used to express transgenes in mitotic clones (Lee and Luo, 2001). The genotypes for generating clones are as follows. hpo clones expressing Sd-RNAi: ey-FLP/+; FRT42D dMSTBF33(or dMSTJM1)/FRT42D tub-Gal80; UAS-Sd-RNAi(N+C)/tub-Gal4 UAS-GFP. wts or sav clones expressing Sd-RNAi: ey-FLP UAS-GFP; UAS-Sd-RNAi/+; FRT82B latsX1 (or savSH13)/tub-Gal4 FRT82 tub-Gal80. sd clones: sdΔB (or sdΔC) FRT19/hs-HA-GFP-HA hs-FLP FRT19. sd clones expressing YkiM123: sdΔBFRT19/tub Gal80 FRT19; ey-flp; tub-Gal4 UAS-mCD2-GFP/UAS-Myc-YkiM123. YkiM123 expressing clones: FRT19/tub Gal80 FRT19; ey-flp; tub-Gal4 UAS-mCD2-GFP/UAS-Myc-YkiM123. wts clones expressing Fg-Yki/HA-Sd: MS1096 hs-flp; UAS-Fg-Yki (+UAS-HA-Sd); FRT82 wtsX1/FRT82 hs-Myc-GFP.
Cell culture, transfection, immunoprecipitation, western blot analysis, luciferase reporter assay, and ChIP assay
S2 cells were cultured in the Schneider’s Drosophila Medium (Invitrogen) with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of Streptomycin. Transfection was carried out using Calcium Phosphate Transfection Kit (Specialty Media) according to manufacturer’s instructions. An ubiquitin-Gal4 construct was cotransfected with pUAST expression vectors for all the transfection experiments. Immunoprecipitation and western blot analysis were performed using standard protocols as previously described (Zhang et al., 2005). Luciferase reporter gene assay and ChIP assay are described in Supplementary Information.
Immunostaining
Immunostaining of imaginal discs and cultured cells was carried out as described (Jia et al., 2004; Jiang and Struhl, 1995). Antibodies or dyes used in this study: rabbit anti-βGal (Cappel), rabbit and mouse anti-Flag (Sigma), rabbit and mouse anti-HA (Santa Cruz), rabbit anti-GFP (Santa Cruz), Rat anti-Ci (Motzny and Holmgren, 1995), mouse anti-Diap1 (Yoo et al., 2002), Rat anti-CycE (Richardson et al., 1995), mouse anti-Arm (Jiang and Struhl, 1998), and mouse anti-Myc (Santa Cruz), 7-AAD (Molecular Probes ), and DRAQ5 (Biostatus Ltd).
Supplementary Material
Acknowledgments
We thank Tao Yue for technical assistance, Drs. Liqun Luo, Steve Cohen, Jessica Treisman, Konrad Basler, Helen Richardson, Bruce Hay, Hyung don Ryoo, the Bloomington stock center, and the Exelixis Collection at Harvard Medical School for reagents and fly stocks. We thank Dr. Keith Wharton for helping with photography. This work is supported by grants from NIH, Leukemia and Lymphoma Society Scholar Program, and Welch Foundation to J.J., and J.J. is a Eugene McDermott Endowed Scholar in Biomedical Science at UTSW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques . 2000;29:726–728. 730–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Molecular cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 Increases Organ Size and Expands Undifferentiated Progenitor. Cells Curr Biol. 2007 doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes & development. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Chao T, Jiang J. Using Immunoprecipitation to study Protein-Protein interaction in the Hedgehog Signaling Pathway. Methods in Molecular Biology. 2007;397:215–229. doi: 10.1007/978-1-59745-516-9_15. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nature genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science (New York, NY) 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science (New York, NY) 2001;292:1164–1167. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- Halder G, Carroll SB. Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development (Cambridge, England) 2001;128:3295–3305. doi: 10.1242/dev.128.17.3295. [DOI] [PubMed] [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes & development. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature cell biology. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annual review of genetics. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog Signalling Activity of Smoothened Requires Phosphorylation by Protein Kinase A and Casein Kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes & development. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and Hedgehog signalling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes & development. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Developmental genetics. 1998;22:43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Kohn MJ, Liu C, DePamphilis ML. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45:577–587. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Liu X, Grammont M, Irvine KD. Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Developmental biology. 2000;228:287–303. doi: 10.1006/dbio.2000.9939. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cellular signalling. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development (Cambridge, England) 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & development. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nature cell biology. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nature genetics. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Developmental biology. 2007;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development [published erratum appears in Cell 1998 Feb 20;92(4):following 585] Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Richardson H, O’Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development (Cambridge, England) 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nature cell biology. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. The Journal of biological chemistry. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature cell biology. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes & development. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes & Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. The EMBO journal. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes and Development. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development (Cambridge, England) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development (Cambridge, England) 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. The EMBO journal. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunami M, Suzuki K, Ohkubo H. A novel family of TEA domain-containing transcription factors with distinct spatiotemporal expression patterns. Biochemical and biophysical research communications. 1996;228:365–370. doi: 10.1006/bbrc.1996.1667. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nature cell biology. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Developmental cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.