Abstract
In Azotobacter vinelandii, deletion of the fdxA gene that encodes a well characterized seven-iron ferredoxin (FdI) is known to lead to overexpression of the FdI redox partner, NADPH:ferredoxin reductase (FPR). Previous studies have established that this is an oxidative stress response in which the fpr gene is transcriptionally activated to the same extent in response to either addition of the superoxide propagator paraquat to the cells or to fdxA deletion. In both cases, the activation occurs through a specific DNA sequence located upstream of the fpr gene. Here, we report the identification of the A. vinelandii protein that binds specifically to the paraquat activatable fpr promoter region as the E1 subunit of the pyruvate dehydrogenase complex (PDHE1), a central enzyme in aerobic respiration. Sequence analysis shows that PDHE1, which was not previously suspected to be a DNA-binding protein, has a helix–turn–helix motif. The data presented here further show that FdI binds specifically to the DNA-bound PDHE1.
One of the most exciting advances in the field of iron–sulfur ([Fe-S]) protein research has come recently with the recognition that a number of [Fe-S] proteins (e.g., iron regulatory protein, endonuclease III, fumarate nitrate reduction protein, SoxR) bind DNA or RNA and that some of them serve as sensors of iron, oxygen, superoxide, or NO and regulate gene expression (1). The study described here concerns the participation of the well characterized seven-iron ferredoxin (FdI) from Azotobacter vinelandii in an oxidative stress response. FdI is only 1 of at least 12 ferredoxins synthesized by this organism (2). Its critical role makes it likely that organisms with the same type of ferredoxin (e.g., Pseudomonas aeruginosa, Bordetella pertussis, Rhodobacter capsulatus, Afipia felis, Thiobacillus ferrooxidans, Caulobacter crescentus CB15) will have the same oxidative stress response. These organisms live in environments (e.g., soil, water, respiratory tract) where the oxygen concentration can fluctuate widely.
This story began about 10 years ago when it was reported that disruption of the gene encoding FdI (fdxA) led to a dramatic increase in the abundance of another protein (3, 4). Isolation and characterization of that small acidic protein (5, 6) and its gene (7) resulted in identification of the overexpressed protein as the specific FdI redox partner (8), NADPH:ferredoxin reductase (FPR). A. vinelandii FPR is a member of a superfamily of NADPH/NADH:ferredoxin/flavodoxin reductases and is similar to the NADPH reductase from Escherichia coli (the fpr gene product) (5, 7).
In E. coli, the fpr gene is known to be under the control of the SoxRS regulon that responds to oxidative stress caused by addition of NO or the superoxide propagator paraquat (9). Studies of FdI-mediated regulation of fpr expression in A. vinelandii have revealed both close similarities and significant differences when compared with the E. coli SoxRS system (10, 11). The similarities include the following. The DNA sequence that responds to fdxA disruption in A. vinelandii shows significant similarity to the analogous region upstream of the E. coli fpr gene, which is known to bind SoxS (11). In both organisms, the fpr genes are transcriptionally activated through that sequence when the superoxide propagator paraquat is added to cells (10). It is important to note that in A. vinelandii the level of activation is the same as that seen in the FdI deletion strain, and the level does not increase in that strain after addition of paraquat. In addition, when the A. vinelandii fpr gene is introduced into E. coli, it is activated by SoxRS (10). In spite of these similarities, a soxS homologue could not be found in A. vinelandii and a fdxA homologue could not be found in E. coli (10, 11). In A. vinelandii, a specific protein binds to the paraquat-activatable fpr promoter region (10, 11). However, that protein does not react with Abs raised against SoxS, and, after binding the specific DNA sequence, it causes a much greater gel retardation than does purified E. coli SoxS (10). The current study was aimed at identifying this A. vinelandii DNA-binding protein. Herein, this protein will be referred to as the fpr promoter region binding protein (fprBP).
Methods
Preparation of Promoter Region DNA Upstream of fpr Gene.
For the gel shift assays shown in Figs. 1–3, several double-stranded oligonucleotides of 50 bp in length were synthesized. Each corresponds to a previously described region upstream of the fpr promoter that contains a SoxS-like sequence referred to as S and a palindromic sequence referred to as P (10, 11). As indicated in the figure legends, in some cases, one or both of these sequences were mutated. The wild-type sequence is designated by +, and the mutated sequence is designated by − (S+P+, AGTGAATCGCATATCTATATAACTGATTATCCATATCGCGATAATTTACT; S−P−, CGCAGAGTGAATCGCCGCTAGTACCGCGGTACCGATGGACGAGCGATAATTTACTA; S+P−, AGTGAATCGCATATCTATATAAGGTACCGATGGACGAGCGATAATTTACT; S−P+, AGTGACTCGGACATCTATATAACTGATTATCCATATCGCGATAATTTACT). An oligonucleotide corresponding to the mutated SoxS consensus sequence (12–14) was also synthesized and was referred to as SoxS− (ACGTAGTGACTCGGACATCTATATCGCTGATTATCCATATCGCGATAATTTAC). For determination of the apparent dissociation constant, a 317-bp DNA fragment was obtained by PCR with two primers (5′-GGGAAGCTTGCTCGAGTTCGGTCAGCG-3′ and 5′-CCCTCTAGATGAACAGGGTATCGTTCC-3′), which allowed the amplification of the DNA sequence upstream of the fpr gene. This fragment contained the sequences that we designated as P+ and S+.
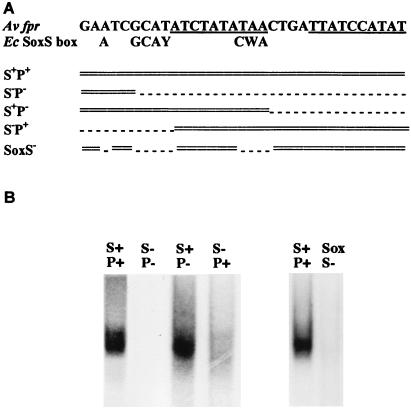
Figure 1.
A. vinelandii fprBP binds specifically to a sequence that is almost identical to the SoxS consensus sequence of E. coli. (A) The A. vinelandii fpr promoter region contains a SoxS-like sequence (referred to as S) (9) and a palindrome (underlined; referred to as P) (11). As described in Methods, a set of oligonucleotides was constructed with different regions mutated as indicated by dashed lines. (B) Gel mobility shift assays used partially purified fprBP (20 μg) and the oligonucleotides (1 fmol) indicated in A and methods described in ref. 11 and the Methods section of this paper. The gels were run for 2 hr, such that the free probe ran off the gel and only the upper shifted band is shown. For full-length gels, see refs. 10 and 11 and Fig. 4.
Figure 3.
Recombinant A. vinelandii PDHE1 is identical to fprBP. (A) Coomassie-stained 7.5% SDS/polyacrylamide gel of pure PDHE1. The molecular mass positions indicated here refer only to the gel in A (markers not shown). (B) Gel mobility shift assay with PDHE1 (50 pmol) and 1 fmol of the mutated fpr promoter region (SoxS−) (see Fig. 1) (lane 1); 1 fmol of the wild-type fpr promoter region (S+P+) (lane 2); and with addition of a 100× excess of unlabeled (S+P+) DNA to the sample (lane 3). For comparison, the position of the fprBP gel shift with (S+P+) using the partially purified fprBP is shown in lane 4, and in lane 5 with additional 100× excess of unlabeled (S+P+) DNA.
All of these DNA fragments were end-labeled with [γ-32P]ATP by using the T4 polynucleotide kinase and then were purified by Nick-column chromatography (Amersham Pharmacia).
Purification of fprBP.
The protein was purified from the FdI-deficient A. vinelandii strain LM100 grown under N2-fixing conditions (3). Then, 400 g of cells was suspended in 50 mM Tris⋅HCl (pH 7.5)/1 μg/ml leupeptin/10 μM PMSF/1 mM DTT/0.1% Igepal CA-630 (buffer A) and were crushed in a French press. After sedimentation at 8,000 rpm with Sorvall RC5C centrifuge for 1 hr, the supernatant was applied to a DEAE-cellulose column. The flow-through was collected and applied to a heparin column. The yellowish fprBP-containing fraction was eluted with 1 M NaCl in buffer A; it was then diluted to 0.1 M NaCl with buffer A and applied to tandem-arranged DNA affinity columns [prepared with modification of the method of Kadonaga and Tjian (15)]. The first column contained the oligonucleotide corresponding to the mutated fpr promoter region (S−P−), and the second one contained the oligonucleotide that corresponded to the wild-type fpr promoter region (S+P+) (see Fig. 1). These oligonucleotides were bound to CNBr-activated Sepharose (Amersham Pharmacia). The S+P+ column was washed with 0.1 and 0.2 M NaCl in buffer A, and fprBP was eluted with buffer A with 1 M NaCl. Then, the protein was concentrated and desalted by ultrafiltration/dialysis. The yield of partially purified fprBP was 5 mg per 400 g of cells. The protein was stored at −70°C in buffer A with 0.1 M NaCl and 20% glycerol.
Purified recombinant E1 subunit of the pyruvate dehydrogenase complex (PDHE1) was kindly provided by Aart de Kok (Agricultural University of Wageningen, The Netherlands) as an ammonium sulfate precipitate (16). Desalting and buffer exchange to buffer A with 0.1 M NaCl were done by ultrafiltration on Microcon with a molecular mass cut off of 10,000 Da (Amicon).
Gel Mobility Shift Assays.
Gel mobility shift assays were performed as described elsewhere (10), using the synthetic double-stranded oligonucleotides or the 317-bp DNA fragment. For the apparent dissociation constant determination, a titration was made with the labeled 317-bp DNA and increased quantities of recombinant PDHE1. The dried gel was analyzed by PhosphorImager densitometer (Molecular Dynamics). Band intensities of the free probe were measured with imagequant software (Molecular Dynamics). Each intensity was corrected for background differences.
Southwestern Blotting and UV Crosslinking.
Southwestern blotting was done with modification from Handen and Rosenberg (17) in 50 mM Tris⋅HCl (pH 7.5)/60 mM KCl/10% glycerol/1 mM DTT/1 mM EDTA. Samples (90 μg of total protein) were not boiled before gel electrophoresis. Incubations with 0.4 pmol of 32P-labeled wild-type fpr promoter region (S+P+) (105 cpm) were done for 2.5 hr at room temperature. UV crosslinking was done according to Cooney et al. (18). A total of 20 μg of protein was used per reaction.
Sequencing by Mass Spectrometry.
Sequencing was done at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT). Because the NH2 terminus of fprBP was blocked, the sample was digested with trypsin, and the resulting peptides were subjected to matrix-assisted laser desorption ionization mass spectrometry. Database searching was done independently by using the profound program that relies on the owl database and the peptidesearch algorithm that uses the European Molecular Biology Laboratory (EMBL)/nonredundant database. Both searches identified the same protein as PDHE1, and the percent coverage of the known protein sequence in both cases was ca. 30%.
Results and Discussion
An A. vinelandii Protein Binds to a DNA Sequence Upstream of the fpr Gene That Is Almost Identical to the E. coli SoxS Consensus Sequence.
In previous studies, we have established that the A. vinelandii fpr gene is transcriptionally activated in vivo, in response to oxidative stress, via an identified DNA sequence (10). We have further shown that there is an A. vinelandii protein (herein designated fprBP) that binds specifically to that sequence. If the sequence is mutated so that the protein no longer binds, then the gene is no longer activated (10, 11). In E. coli, a number of genes, including fpr, have been identified that are activated in response to oxidative stress by SoxS. This information recently led to the identification of an E. coli SoxS consensus sequence (12–14). As shown in Fig. 1A, a homologous sequence is present in the A. vinelandii fpr promoter region. That sequence is overlapping with and just upstream of the palindromic region (P) we had identified previously as being critical for A. vinelandii fprBP binding. This information therefore led us to design the constructs illustrated in Fig. 1A, where the palindromic sequence, or the SoxS-like sequence, or both, were mutated. The gel shift results, when those constructs were used, clearly show that A. vinelandii fprBP binds specifically to the defined E. coli SoxS consensus sequence (Fig. 1B). Thus, in both A. vinelandii and E. coli, where the fpr genes are activated in response to paraquat addition to cells, specific proteins bind to DNA sequences that are almost identical.
Purification and Identification of the A. vinelandii fprBP as PDHE1.
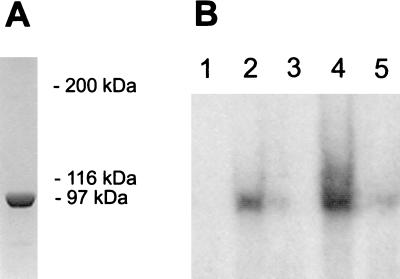
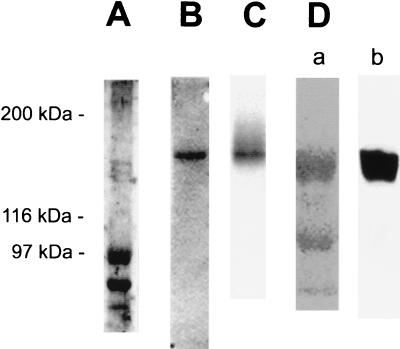
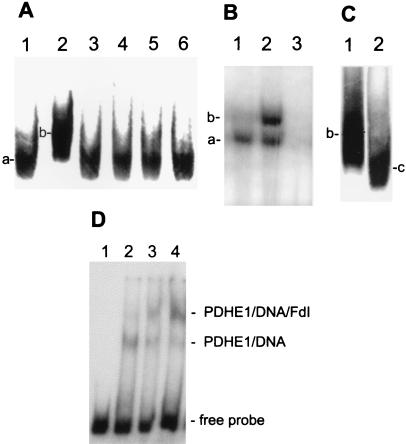
SoxS was not initially purified from wild-type E. coli cells, but rather was purified after the gene was available and the protein was overexpressed (19). Our goal then was to purify A. vinelandii fprBP to the point where a specific protein could be unambiguously identified as the DNA-binding protein for subsequent sequencing. The A. vinelandii fprBP was purified until eight bands were visible on a silver-stained SDS/polyacrylamide gel, with the highest molecular weight bands shown in Fig. 2A. Southwestern blots were used to determine which protein band bound specifically to the promoter region upstream of the fpr gene. Fig. 2B shows a typical Southwestern blot in which the gel was run for a short period of time to ensure that both large and small proteins present would be tested for DNA binding. Regardless of how long the gel was run, fprBP was always identified as a single band whose position corresponded to the major protein band in Fig. 2A that runs close to the 97,400-Da marker. UV crosslinking experiments, where the partially purified fprBP was first crosslinked to the DNA and then run on an SDS/polyacrylamide gel, also revealed a single high molecular weight band (Fig. 2C). Having thus developed a method to identify fprBP, a final Southwestern gel was run to obtain a sample for sequencing. Fig. 2D, lane a, shows a Coomassie-stained SDS/PAGE separation of partially purified fprBP that was run for a long time to ensure maximum separation of the proteins. The three bands shown in Fig. 2D, lane a, correspond to the bottom three bands from Fig. 2A that run at and below the 97,400-Da marker. Fig. 2D, lane b, shows again that it is the top band that binds DNA. That band was then excised for sequencing.
Figure 2.
Identification of fprBP for sequencing. (A) Silver-stained 7.5% SDS/polyacrylamide gel of partially purified fprBP where the gel was run until the 62-kDa band of the Biolabs (Northbrook, IL) protein markers reached the bottom. The molecular mass positions indicated here refer only to the gel in A (markers not shown). (B) Southwestern blot, run until the dye reached the bottom of the gel, and (S+P+) DNA. (C) UV crosslinking using 20 μg of protein and (S+P+) DNA. (D) The excised fprBP band for identification by matrix-assisted laser desorption ionization mass spectrometry. Lane a, Coomassie-stained gel; and lane b, Southwestern blot identifying fprBP as the upper band. See Methods for details and references.
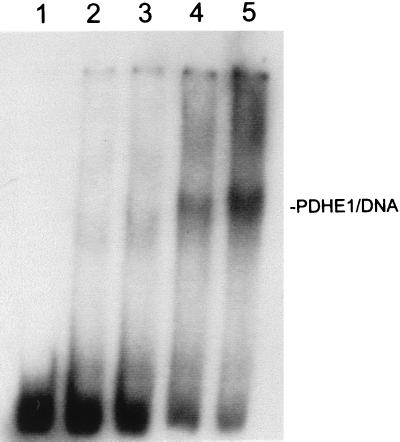
As described in Methods, sequencing data revealed that fprBP was actually an existing well known protein, PDHE1 (accession no. Y15124), an enzyme with a central role in aerobic metabolism (16) with a molecular mass of 99,800 Da per monomer. This protein had recently been overexpressed and purified by de Kok and coworkers (16), who kindly supplied us with a sample of the pure protein to confirm the identity of fprBP. As shown in Fig. 3, the purified recombinant PDHE1 runs in the same place on an SDS/polyacrylamide gel as fprBP and gives the same specific gel shift with the wild-type fpr promoter region as does fprBP, confirming its identity. Fig. 4 shows a titration with increasing amounts of recombinant PDHE1. Quantitation of these data gives an apparent dissociation constant of 180 nM under these conditions.† PDHE1 is a large protein (around 200,000 Da for the homodimer), which is a major enzyme in central metabolism and is present in large quantities in the cell (>0.6 μM),‡ so this apparent dissociation constant is well within the physiological range. As shown in Fig. 1 and elsewhere (10, 11), PDHE1 binds to a very specific DNA sequence upstream of the fpr gene, and when that sequence is mutated, PDHE1 no longer binds. Specificity was further confirmed, using recombinant PDHE1, by competition with homologous and heterologous DNA (data not shown).
Figure 4.
Titration of PDHE1. A total of 50 fmol of labeled 317-bp DNA was used per assay. Lanes: 1, DNA alone; 2–5, same as lane 1 plus increased quantities of pure PDHE1 (3.25 pmol, 6.5 pmol, 13 pmol, 19.5 pmol). Lower band is free probe; upper band is DNA/PDHE1 complex.
Although PDHE1 had previously been purified and characterized, it had not previously been suspected to be a DNA-binding protein. Analysis of the complete PDHE1 sequence by using the nnpredict program (21) or other secondary structure prediction programs (22, 23) identified the helix–turn–helix motif shown in Fig. 5 as a putative DNA-binding region. Fig. 5 also shows that the human pathogen P. aeruginosa, which has, when compared with A. vinelandii, a FdI with 97% similarity, a FPR with 91% similarity, and a PDHE1 with 86% similarity, also has the conserved helix–turn–helix motif. It thus seems likely that P. aeruginosa will have the same PDHE1/FdI-based oxidative stress response system.
Figure 5.
Sequence alignment of the putative helix–turn–helix motif region in A. vinelandii PDHE1 and P. aeruginosa PDHE1. : represents identical; . represents similar. The last line shows the predicted helix–turn–helix region in A. vinelandii PDHE1, using the nnpredict program by Kneller et al. (21) H, helix; –, turn.
FdI Binds Specifically to DNA-Bound PDHE1.
In A. vinelandii, the fpr gene is activated to the same extent by the same DNA sequence in response to paraquat addition to cells, or in response to the deletion of the fdxA gene that encodes FdI (10). We have previously shown that FdI does not bind directly to the fpr promoter region DNA (11). The experiment in Fig. 6 was done to test whether FdI might exert its inhibitory effects by binding directly to PDHE1. As shown in Fig. 6A, the 13,000-Da FdI binds very specifically to fprBP, causing a supershift that is not seen with FdI that has lost its iron atoms (apoFdI), or with other ferredoxins or flavodoxins. Fig. 6 B and D show that the supershift is also seen with recombinant A. vinelandii PDHE1. The supershift is maximized under these conditions† at about a 4:1 molar excess of FdI over PDHE1 (Fig. 4D), consistent with the relative quantities of the two proteins under our normal growth conditions, where oxidative stress does not occur. We have previously shown that FdI and FPR also form a very specific 1:1 complex (8). Fig. 6C shows that when FPR is added with DNA-bound PDHE1 and FdI, it competes with PDHE1 for FdI binding. Under conditions where there is no oxidative stress, FPR levels are very low relative to FdI (3). Under conditions of oxidative stress, FPR levels exceed FdI levels (5, 10).
Figure 6.
Gel mobility shift assays show that PDHE1 interacts specifically with FdI. (A) fprBP (20 μg) is incubated with (S+P+) as a DNA probe alone (lane 1) and in the presence of: A. vinelandii FdI (lane 2); A. vinelandii apoFdI (lane 3); A. vinelandii flavodoxin I (lane 4); A. vinelandii flavodoxin II (lane 5); and spinach FdI (lane 6) (20 μg each). A. vinelandii FdIII (lane 2) was also tested and does not give a supershift. (B) Recombinant PDHE1 (50 pmol) is incubated with 1 fmol of (S+P+) DNA alone (lane 1) and in the presence of: FdI (800 pmol) (lane 2) and after addition of a 100× excess of the unlabeled (S+P+) DNA-probe to the FdI/PDHE1 sample (lane 3). (C) Gel shift with recombinant PDHE1 (50 pmol) incubated with 1 fmol of (S+P+) DNA in the presence of: A. vinelandii FdI (lane 1) and A. vinelandii FdI (1.6 nmol) and Av FPR (0.7 nmol) (lane 2). In A–C, a, DNA plus DNA-binding protein (fprBP or recombinant PDHE1); b, same as a plus FdI; c, same as b plus FPR. (D) Titration of FdI. A total of 50 fmol of labeled 317-bp DNA was used per assay. Lanes: 1, DNA alone; 2, same as lane 1 plus 10 pmol of pure PDHE1; 3, same as lane 2 plus 40 pmol of FdI; 4, same as lane 2 plus 80 pmol of FdI.
Conclusion
The pyruvate dehydrogenase complex is a key enzyme in central metabolism that links glycolysis to the tricarboxylic acid cycle and is critically important in the decision of whether an organism will use anaerobic or aerobic metabolism. The above data show that in A. vinelandii, PDHE1 has a previously unsuspected role as a DNA-binding protein. The PDHE1 subunit binds specifically to the identified fpr promoter region (Figs. 3 and 4), a DNA sequence that is almost identical to the one that binds SoxS in E. coli (Fig. 1). In A. vinelandii, that promoter region is activated in vivo to the same extent in response to paraquat addition to the cell or in response to deletion of the fdxA gene that encodes FdI (10). If the DNA sequence is mutated so that PDHE1 no longer binds, then the gene is no longer transcriptionally activated in vivo in response to oxidative stress in wild-type cells (10) or in response to deletion of the fdxA gene (11). It remains to be determined whether PDHE1 serves a SoxS-like role as a transcriptional activator in A. vinelandii. The above data further show that FdI binds specifically to PDHE1; further experiments will be needed to determine how that interaction leads to FdI inhibition of fpr activation.
Acknowledgments
We thank Prof. B. Demple for bringing the SoxS consensus sequence to our attention. This research is supported by National Institutes of Health Grant GM-45209 (to B.K.B.). K.R. was supported in part by the Swedish Institute.
Abbreviations
- FdI
ferredoxin I
- FPR
NADPH:ferredoxin reductase
- PDHE1
subunit E1 of the pyruvate dehydrogenase complex from A. vinelandii
- fprBP
fpr promoter region binding protein
Footnotes
The apparent dissociation constant calculations (see Methods) assume that the recombinant PDHE1 is 100% active and therefore likely represent an overestimation. For these experiments, a larger piece of DNA was used to try to more closely mimic the in vivo situation. PDHE1 is an allosteric enzyme that binds Mg2+, thiamin pyrophosphate, pyruvate, acetyl-CoA, and AMP, and it should be fruitful to investigate how these factors influence DNA binding and binding to FdI. The experiments reported here were also done with air-oxidized seven-iron FdI; various reduced forms and an oxygen-sensitive eight-iron form have not yet been examined.
The exact levels have not been published, but approximate levels have been calculated based on published purification yields (20). The calculations show that at a minimum, the E1 subunit represents 0.5% of the total proteins in a cell-free extract (concentration ≈0.6 μM).
References
- 1.Beinert H, Holm R H, Münck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.GaoSheridan H S, Pershad H R, Armstrong F A, Burgess B K. J Biol Chem. 1998;273:5514–5519. doi: 10.1074/jbc.273.10.5514. [DOI] [PubMed] [Google Scholar]
- 3.Morgan T V, Lundell D J, Burgess B K. J Biol Chem. 1988;263:1370–1375. [PubMed] [Google Scholar]
- 4.Martin A E, Burgess B K, Iismaa S E, Smartt C T, Jacobson M R, Dean D R. J Bacteriol. 1989;171:3162–3167. doi: 10.1128/jb.171.6.3162-3167.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isas J M, Burgess B K. J Biol Chem. 1994;269:19404–19409. [PubMed] [Google Scholar]
- 6.Prasad G S, Kresge N, Muhlberg A B, Shaw A, Jung Y-S, Burgess B K, Stout C D. Protein Sci. 1998;7:2541–2549. doi: 10.1002/pro.5560071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isas J M, Yannone S M, Burgess B K. J Biol Chem. 1995;270:21258–21263. doi: 10.1074/jbc.270.36.21258. [DOI] [PubMed] [Google Scholar]
- 8.Jung Y-S, Roberts V A, Stout C D, Burgess B K. J Biol Chem. 1999;274:2978–2987. doi: 10.1074/jbc.274.5.2978. [DOI] [PubMed] [Google Scholar]
- 9.Demple B. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 10.Yannone S M, Burgess B K. J Biol Inorg Chem. 1998;3:253–258. [Google Scholar]
- 11.Yannone S M, Burgess B K. J Biol Chem. 1997;272:14454–14458. doi: 10.1074/jbc.272.22.14454. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Demple B. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 13.Fawcett W P, Wolf R E., Jr Mol Microbiol. 1994;14:669–679. doi: 10.1111/j.1365-2958.1994.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 14.Fawcett W P, Wolf R E., Jr J Bacteriol. 1995;177:1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadonaga J T, Tjian R. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengeveld A F, Westphal A H, de Kok A. Eur J Biochem. 1997;250:260–268. doi: 10.1111/j.1432-1033.1997.0260a.x. [DOI] [PubMed] [Google Scholar]
- 17.Handen J S, Rosenberg H F. Front Biosci. 1997;2:9–11. doi: 10.2741/A166. [DOI] [PubMed] [Google Scholar]
- 18.Cooney A J, Tsai S Y, Tsai M J. In: Transcription Factors. 1st Ed. Latchman D S, editor. Oxford: IRL; 1993. p. 61. [Google Scholar]
- 19.Li Z, Demple B. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 20.Bresters T W, De Abreu R A, de Kok A, Visser J, Veeger C. Eur J Biochem. 1975;59:335–345. doi: 10.1111/j.1432-1033.1975.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 21.Kneller D G, Cohen F E, Langridge R. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 22.Geourjon C, Deleage G. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 23.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]