Abstract
Background
The oomycete Plasmopara viticola (Berk. and Curt.) Berl. and de Toni causes downy mildew in grapevine (Vitis vinifera L.). This pathogen is strictly biotrophic, thus completely dependent on living host cells for its survival. The molecular basis of compatibility and disease development in this system is poorly understood. We have carried out a large-scale cDNA-AFLP analysis to identify grapevine and P. viticola genes associated with the infection process.
Results
We carried out cDNA-AFLP analysis on artificially infected leaves of the susceptible cultivar Riesling at the oil spot stage, on water-treated leaves and on a sample of pure sporangia as controls. Selective amplifications with 128 primer combinations allowed the visualization of about 7000 transcript-derived fragments (TDFs) in infected leaves, 1196 of which (17%) were differentially expressed. We sequenced 984 fragments, 804 of which were identified as grapevine transcripts after homology searching, while 96 were homologous to sequences in Phytophthora spp. databases and were attributed to P. viticola. There were 82 orphan TDFs. Many grapevine genes spanning almost all functional categories were downregulated during infection, especially genes involved in photosynthesis. Grapevine genes homologous to known resistance genes also tended to be repressed, as were several resistance gene analogs and carbonic anhydrase (recently implicated in pathogen resistance). In contrast, genes encoding cytoskeletal components, enzymes of the phenylpropanoid and beta-oxidation pathways, and pathogenesis related proteins were primarily upregulated during infection. The majority of P. viticola transcripts expressed in planta showed homology to genes of unknown function or to genomic Phytophthora sequences, but genes related to metabolism, energy production, transport and signal transduction were also identified.
Conclusion
This study provides the first global catalogue of grapevine and P. viticola genes expressed during infection, together with their functional annotations. This will help to elucidate the molecular basis of the infection process and identify genes and chemicals that could help to inhibit the pathogen.
Background
Plasmopara viticola (Berk. and Curt.) Berl. and de Toni is an obligate biotrophic plant pathogen [1] that causes downy mildew, a devastating disease resulting in significant economic losses as well as environmental damage through the repetitive applications of fungicides.
Primary infection begins with over-wintering oospores, which germinate into motile zoospores that can actively locate stomata [2,3] and start the infection process. Colonization involves intercellular mycelial growth and the differentiation of haustoria, which penetrate parenchyma cells by invaginating but not breaking the plasma membrane [4]. This highly specialized nutritional strategy, which typifies biotrophic plant pathogens such as powdery mildews, downy mildews and rusts, probably involves the strict control of host cell metabolism which is diverted to maintain pathogen survival and compatibility [5]. Further infection cycles are initiated through the release of zoosporangia emerging from stomata. The cycles end with the sexual production of over-wintering oospores.
While the epidemiology of the pathogen is understood well enough to generate computer models of epidemics, the molecular aspects of the infection process are largely unknown. The main recognized role of haustoria is to obtain nutritional resources from the plant cell, but the synthesis of additional gene products and metabolites [6,7] suggests that signals are exchanged between the pathogen and host to establish and maintain compatibility and possibly to suppress defense responses [7]. Secreted virulence factors may be involved in this process [8] and four such gene products have recently been identified in other oomycetes [9-12]. Following the recent completion of Phytophthora spp. sequencing projects [13], about 700 avr homologues have been predicted based on the presence of a signal peptide and a RXRL-EER motif, typical of known cytoplasmic effectors of oomycete pathogens [9,14].
The plant's response to infection has been characterized predominantly through the study of incompatibility in the resistant species Vitis riparia, V. rupestris and Muscadinia rotundifolia. Attempts have been made to introgress resistance into cultivated V. vinifera genotypes [15-18], although the quality and the specific organolectic characteristics of wines are not easy to reproduce in interspecific hybrids. Some of resistance mechanisms have been elucidated [19-22] and they include physical barriers such as hairs and stomatal closure, the accumulation of phenolic antimicrobial compounds, increased peroxidase activity, the accumulation of pathogenesis related proteins and the hypersensitive response [23].
Molecular data from the direct investigation of compatible interactions in cultivated grapevine genotypes is scarce, and indeed downy mildew has received little attention compared to diseases carried by other biotrophic pathogens, such as powdery mildews and rusts. Understanding the basis of susceptibility would greatly assist in the development of new control strategies and the identification of pathogen and host factors required for disease progression.
One useful approach to the molecular analysis of plant-pathogen interactions is the determination of changes in steady state mRNA levels occurring in both the host and the pathogen during infection. Such transcriptomic approaches have been undertaken for different plant-oomycete interactions either by microarray analysis or alternative, open-architecture technologies, thus revealing novel information about pathogen genes [24-29]. A few studies have also included proteomic analysis [30].
The expression of selected grapevine genes during P. viticola infection was reported recently [21], but there has been no large-scale analysis and pathogen genomic information is also very scarce (fewer than 30 sequences in GenBank). In this article, we report the results of cDNA-AFLP analysis to identify infection-related transcripts in P. viticola and grapevine. Our data show that downy mildew infection of grapevine at the oil spot stage involves the downregulation of many grapevine genes with diverse functions, and the induction of pathogen genes representing important metabolic pathways such as protein synthesis, transport and energy metabolism in infected leaves.
Results
cDNA-AFLP analysis
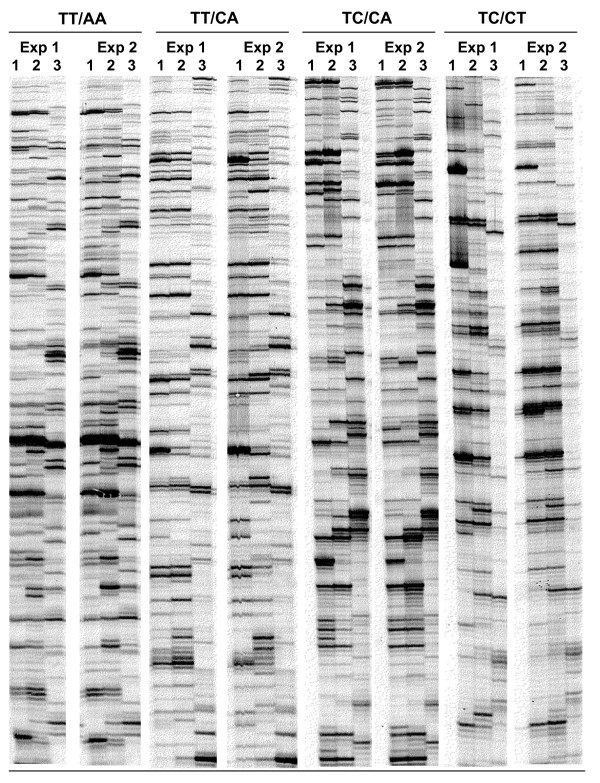
We carried out a cDNA-AFLP analysis on RNA samples of infected leaves at the oil spot stage, and on healthy control leaves and pure sporangia, as described [31,32]. The oil spot stage was chosen because the compatible interaction is well established and the mycelia produced at this stage are abundant enough to allow the detection of pathogen transcripts, even though the plant cell is still active, since various plant functions are needed to maintain pathogen survival. For each of the 128 primer combinations, 55–75 transcript derived fragments (TDFs) were visualized as bands, 25–760 bp in size, representing approximately 7000 transcripts overall. The same average number of bands per lane was obtained both from grape and from P. viticola sporangia samples. To determine the reproducibility of these profiles, the experiments were repeated using additional samples of a biological replicate (Figure 1).
Figure 1.
Expression of grapevine and P. viticola transcripts displayed by cDNA-AFLP. An example showing selective amplification with four different primer combinations, repeated in two biological replicates (Exp. 1 and Exp 2). 1: control water-treated leaves; 2: P. viticola-infected leaves; 3: P. viticola sporangia.
Detection of differentially expressed transcripts
The infection of grapevine with P. viticola resulted in the widespread modulation of steady state mRNA levels. We detected 1196 differentially expressed TDFs, corresponding to about 17% of all visualized transcripts. Each band was excised from the gel, eluted, re-amplified and purified for direct sequencing, yielding 982 cDNA fragments that gave rise to useable sequence data. Among these sequences, 599 were homologous to known expressed grapevine sequences, either as tentative consensus sequences or expressed sequence tags (EST), while 205 were homologous to genomic contigs in the newly released 8.4× Vitis Genome database [33] but were not represented in any EST databases. There were also putative annotations in the UNIPROT database for 72 TDFs, and these were assigned functional categories accordingly. Another 96 TDFs were homologous to known Phytophthora spp. sequences derived from the recently completed Phytophthora genome sequencing projects [13] and are therefore likely to be P. viticola genes expressed during infection. Finally, 82 sequences had no database matches, 65 from TDFs with similar sized bands in the sporangia sample, and 17 expressed uniquely during the infection. Because the grapevine genome is fully sequenced, the 82 additional sequences are likely to represent additional P. viticola transcripts with insufficient similarity to known genes in other oomycetes.
Functional categories of grapevine transcripts modulated by downy mildew infection
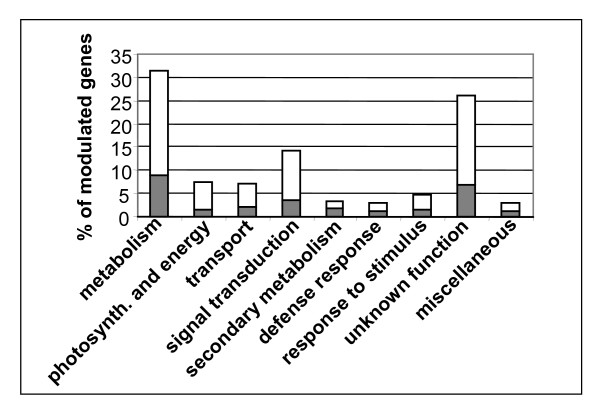
A complete list of TDFs isolated from infected grapevine is available in Additional File 1, while a selection of the most interesting TDFs is shown in Table 1. Each transcript was functionally annotated through careful analysis of the scientific literature and the Gene Ontology Database [34]. Figure 2 shows the percentages of grapevine genes assigned to different functional categories. Approximately 31% of the annotated sequences have primary metabolic roles (particularly in protein and carbohydrate metabolism), 14% are involved in signal transduction, and a further 8% in photosynthesis/energy mobilization. Other relevant groups, each accounting for 3–7% of TDFs, include secondary metabolism, cellular transport, defense, and responses to external stimuli. Approximately 26% of the modulated grapevine TDFs corresponded to tentative consensus sequences or ESTs with no known function. Most of the differentially-expressed grapevine transcripts were downregulated during the oil spot stage, and this applied across all but one of the functional categories and was especially prevalent in the photosynthesis/energy mobilization category (~82% down-regulated). The single exception was the secondary metabolism category, where 57% of the differentially-expressed genes were upregulated.
Table 1.
Selected list of modulated grape transcripts.
| TDF | Primer comb. | Accession | Lenght (bp) | I/R | Annotation | Blast score Blastn/Blastx* |
| Metabolism | ||||||
| 192 | TC – CA | TC52910 | 301 | + | Sucrose synthase (Q9SLS2) | 3.12E-63 |
| 1627 | TT – CT | TC57852 | 168 | + | UDP-glucosyltransferase HRA25 (Q9FUJ6) | 2.07E-30 |
| 245 | TA – AC | TC53221 | 169 | + | Transketolase, chloroplast precursor (O20250) | 1.92E-29 |
| 18 | TT – AA | TC67193 | 127 | + | Raffinose synthase, partial (Q8VWN6) | 9.26E-21 |
| 1279 | CT – TC | TC52362 | 175 | + | Fatty acid multifunctional protein (Q9ZPI5) | 1.12E-16 |
| 134 | TC – TT | TC67104 | 390 | + | 12-oxophytodienoate-10,11-reductase 1 (Q8LAH7) | 6.19E-24 |
| 993 | CA – CA | TC60564 | 50 | + | Gibberellin 2-oxidase (Q6TN17) | 3.63E-105 |
| 215 | TT – TA | TC55722 | 216 | + | Fatty acid hydroperoxide lyase (Q9AUD8) | 5.62E-72 |
| 152 | TC – TC | Q6X5R6 | 342 | + | (Lox2) Lipoxygenase | 4.00E-39 * |
| 1036 | CC – AT | TC58112 | 349 | + | Long-chain acyl-CoA oxidase (O64894) | 5.62E-72 |
| 1225 | CT – AC | TC53311 | 359 | + | 3-ketoacyl-CoA thiolase (Q6TXD0) | 2.10E-09 |
| 893 | CA – AC | TC67959 | 104 | + | 3-beta-hydroxy-delta5-steroid dehydrogenase | 9.77E-16 |
| 347 | TA – CG | TC54708 | 181 | + | 40S ribosomal protein S16 (Q9M5L1) | 2.91E-18 |
| 1382 | CG – TA | TC58494 | 279 | + | Ribosomal L10 protein (Q874B2) | 2.62E-56 |
| 1345 | CG – AA | TC51894 | 289 | + | Ubiquitin conjugating enzyme E2 (Q42897) | 6.56E-55 |
| 594 | TG – GA | TC60588 | 186 | + | Proteasome subunit beta type 5-B precursor (Q9LIP2) | 5.35E-37 |
| 1493 | CT – GG | TC56558 | 399 | + | Gamma-glutamylcysteine synthetase (Q6F4I8) | 4.64E-35 |
| 1293 | CT – TG | TC51806 | 244 | + | Cysteine synthase (Q43317) | 3.08E-25 |
| 79 | TC – AC | TC68684 | 457 | + | Cellulose synthase-like protein D4 (Q8GUZ9) | 2.09E-96 |
| 1594 | TT – TC | TC65238 | 179 | + | Tubulin alpha chain, partial (P33629) | 4.78E-34 |
| 232 | TA – AT | TC57434 | 161 | - | Nucleotide sugar epimerase-like protein | 8.83E-12 |
| 1630 | TT – CT | Q9ZTP5 | 105 | - | Pentose-5-phosphate 3-epimerase | 5.85E-10 * |
| 1610 | TT – CA | TC54570 | 167 | - | Sedoheptulose-bisphosphatase (Q940F8) | 1.37E-27 |
| 1668 | TT – GT | TC54602 | 412 | - | Fructose-bisphosphate aldolase (Q6RUF6) | 1.47E-50 |
| 100 | TC – AG | TC54851 | 224 | - | Alpha-mannosidase (Q2R3E0) | 9.53E-43 |
| 1567 | CG – GT | TC57827 | 211 | - | Galactose dehydrogenase, (Q84LI1) | 4.13E-33 |
| 255 | TA – AG | TC52686 | 210 | - | Carbonic anhydrase (Q5NE20) | 2.18E-39 |
| 1472 | CT – GC | TC60916 | 581 | - | Glyceraldehyde-3-phosphate dehydr. B subunit | 1.61E-126 |
| 1205 | CT – AT | TC62475 | 182 | - | Lipase class 3-like (Q6K2K7) | 6.71E-33 |
| 1689 | TT – GG | TC53435 | 141 | - | B-keto acyl reductase (O24479) | 4.93E-24 |
| 670 | CA – GA | TC62496 | 145 | - | 3-hydroxy-3-methylglutaryl coenzyme A (Q8W2E3) | 6.52E-12 |
| 151 | TC – TC | Q8H539 | 358 | - | Steroid 5alpha-reductase-like protein | 1.00E-32* |
| 1239 | CT – AG | TC69679 | 98 | - | 60S ribosomal protein L19 (Q6RYC4) | 1.65E-13 |
| 138 | TC – TT | TC59193 | 99 | - | Protein translation factor SUI1 homolog 2 (Q94JV4) | 1.49E-12 |
| 308 | TA – CA | TC51783 | 621 | - | Elongation factor 1-beta 1 (Q84WM9) | 1.99E-64 |
| 823 | TC – GG | TC54220 | 542 | - | Phytocalpain (Q6SSJ2) | 3.95E-39 |
| 1174 | CC – CC | TC63107 | 115 | - | Chaperone protein dnaJ-like (Q6H3Y3) | 9.58E-18 |
| 1530 | CC – GC | CB348741 | 62 | - | F-box protein family AtFBL5 | 1.37E-12 |
| 1476 | CT – GC | CA816379 | 345 | - | Protein At3g07360 (U-box domain-containing prot.) | 2.37E-69 |
| 1389 | CG – TT | TC65574 | 203 | - | Glycin-rich protein (Q43688) | 3.13E-35 |
| 1546 | CG – GA | TC68519 | 541 | - | Expansin, complete (Q84US7) | 8.40E-112 |
| 722 | TC – CT | TC69230 | 350 | - | Cyclin D1, partial (Q8GVE0) | 1.73E-69 |
| 664 | CA – GA | TC53870 | 228 | - | Integral membrane protein-like (Q5VRH) | 7.89E-07 |
| Photosyntesis and Energy | ||||||
| 1330 | CT – CC | TC61438 | 646 | + | Cytochrome P450 monooxygenase (Q2MJ14) | 1.38E-67 |
| 123 | TC – TA | TC68921 | 64 | + | C-type cytochrome biogenesis protein (Q7XY14) | 1.08E-06 |
| 227 | TA – AT | TC62259 | 353 | - | ATP synthase B' chain, chloroplast precursor | 1.03E-66 |
| 749 | TC – CG | TC63430 | 230 | - | NADPH-cytochrome P450 oxydoreductase | 1.20E-29 |
| 520 | TG – TG | TC65998 | 102 | - | Plastocyanin, chloroplast precursor (P17340) | 6.20E-06 |
| 1247 | CT – TA | TC66316 | 365 | - | Chloroplast photosystem II 10 kDa protein (Q2PXN6) | 5.90E-58 |
| 1156 | CC – CT | TC68056 | 154 | - | NADH-plastoq. oxidoreductase subunit 7 (Q2L953) | 2.39E-26 |
| 1049 | CC – AC | TC53584 | 561 | - | Magnesium chelatase subunit, partial | 1.28E-121 |
| 31 | TT – AT | TC55659 | 163 | - | Chlorophyll A/B binding protein precursor (Q32291) | 1.79E-32 |
| 427 | TG – TA | TC57132 | 209 | - | Chlorophyll A/B binding protein, precursor (P13869) | 3.62E-42 |
| 4 | TT – AA | TC55138 | 381 | - | Photosystem I assembly protein ycf3, partial | 2.10E-78 |
| 1115 | CC – TC | TC61693 | 411 | - | Photosystem I reaction center subunit II (P12372) | 2.51E-84 |
| 1322 | CT – CT | TC66994 | 275 | - | Photosystem II reaction center (Q8W536) | 2.26E-52 |
| 161 | TC – TC | TC58978 | 146 | - | Photosystem II M protein (Q6QXV8) | 3.80E-25 |
| 1393 | CG – TT | TC58567 | 103 | - | Apocytochrome f precursor (Q68RZ3) | 4.92E-10 |
| Transport | ||||||
| 1614 | TT – CA | Q3L7K6 | 185 | + | Hexose transporter | 6.26E-08 * |
| 680 | CA – GT | TC52346 | 231 | - | Transporter-like protein (Q9LSH7) | 3.88E-47 |
| 249 | TA – AC | TC66367 | 107 | - | 14-3-3 protein (Q93XW1) | 1.13E-16 |
| 1534 | CC – GG | TC57372 | 435 | - | Nitrate transporter NRT1-2 (Q9FRU5) | 8.33E-94 |
| 1438 | CG – CC | TC65826 | 270 | - | ABC transporter-like protein (Q9FT51) | 7.38E-50 |
| 133 | TC – TT | TC62785 | 219 | - | Calcium-transporting ATPase 8, plasma membr. | 4.00E-23 |
| 662 | CA – GA | TC69251 | 302 | - | ATPase alpha subunit, complete (Q3C1H4) | 7.47E-59 |
| 1679 | TT – GC | BQ798655 | 127 | - | Syntaxin {Glycine max} | 5.39E-23 |
| 792 | TC – GT | TC51721 | 180 | - | ADP-ribosylation factor 1-like protein (Q70XK1) | 2.22E-36 |
| 62 | TT – AG | TC69827 | 195 | - | Aquaporin PIP2 (Q2HZF5) | 1.45E-20 |
| Signal transduction | ||||||
| 434 | TG – TA | TC59460 | 147 | + | Protein phosphatase 2C (Q8RVG0) | 6.61E-23 |
| 240 | TA – AC | Q52QR5 | 343 | + | NAC domain protein NAC1 | 1.70E-71* |
| 1385 | CG – TA | TC55407 | 95 | + | NAC family protein (Q2Z1Y1) | 2.32E-13 |
| 687 | CA – GT | TC70344 | 687 | + | WRKY DNA-binding protein 32 (P59583) | 1.24E-05 |
| 235 | TA – AT | TC52484 | 102 | - | SOS2-like protein kinase (Q8LK24) | 1.58E-12 |
| 1298 | CT – TG | TC60214 | 164 | - | Calcium-transporting ATPase/calmodulin binding | 2.74E-28 |
| 288 | TA – TC | TC52400 | 212 | - | Calmodulin cam-207 (Q6DN29) | 1.23E-39 |
| 1538 | CC – GG | TC66781 | 267 | - | CNGC2 (Q5D6H2) | 1.22E-47 |
| 633 | TG – GC | TC69501 | 633 | - | Calcium sensor calcineurin B-like protein (Q4W3B4) | 1.02E-34 |
| 11 | TT – AA | TC61968 | 289 | - | Protein At1g01140 (CBL-interacting protein kinase) | 7.34E-54 |
| 778 | TC – GA | TC53469 | 55 | - | MAPKK (Q66MH6) | 8.34E-07 |
| 1573 | CG – GC | TC59576 | 700 | - | Receptor-like kinase with LRR repeats (Q70AH8) | 1.44E-32 |
| 1231 | CT – AG | TC52043 | 339 | - | MADS-box transcription factor FBP29 (Q9ATE2) | 2.44E-72 |
| Secondary metabolism | ||||||
| 1539 | CC – GG | TC52853 | 419 | + | Cinnamyl alcohol dehydrogenase (Q2Z1Z0) | 2.21E-08 |
| 909 | CA – AG | TC53668 | 105 | + | Stilbene synthase [Vitis vinifera] | 5.34E-17 |
| 1249 | CT – TA | TC54354 | 273 | - | Secretory laccase (Q6TDS6) | 1.11E-23 |
| 421 | TG – AG | TC51729 | 113 | - | Caffeoyl-CoA O-methyltransferase (Q43237) | 5.87E-17 |
| 362 | TG – AA | TC53331 | 208 | - | Flavanone 3-hydroxylase-like protein, (Q9FLV0) | 2.88E-34 |
| 432 | TG – TA | TC65435 | 179 | - | Limonoid UDP-glucosyltransferase (Q9MB73) | 1.29E-32 |
| Defence response | ||||||
| 1176 | CC – CG | TC63410 | 557 | + | Beta-1,3-glucanase (Q9M563) | 5.95E-120 |
| 68 | TT – AG | TC56756 | 108 | + | Pathogenesis-related protein 10.3 (Q20BD2) | 6.31E-14 |
| 724 | TC – CT | TC56512 | 253 | - | Pollen allergen-like protein, partial {A. thaliana} | 1.09E-49 |
| 1296 | CT – TG | TC62916 | 157 | - | EXECUTER1 protein (Q93YW0) | 1.72E-27 |
| 692 | CA – GC | TC57989 | 251 | - | Avr9/Cf-9 rapidly elicited protein 276 (Q84QD7) | 3.72E-23 |
| 229 | TA – AT | TC70153 | 282 | - | Flax-inducible sequence 1 (Q40255) | 6.91E-56 |
| 1173 | CC – CC | TC51855 | 262 | - | Pto-like serine/threonine kinase (Q6W0C7) | 1.69E-16 |
| 1545 | CG – GA | TC58939 | 671 | - | Ethylene-resp. element-binding factor (Q2QDF5) | 3.13E-125 |
| 584 | TA – GC | TC62111 | 83 | - | Lipid transfer protein (Q93YX9) | 4.49E-10 |
| 889 | CA – AC | TC63540 | 305 | - | DIR1 (At5g48485) {Arabidopsis thaliana} | 2.98E-58 |
| 214 | TT – TA | TC61755 | 233 | - | Enzymatic resistance protein, complete (Q3S4G9) | 1.61E-44 |
| Response to stimulus | ||||||
| 246 | TA – AC | TC63756 | 153 | + | Metallothionein-like protein (Q3HR41) | 3.69E-27 |
| 945 | CA – TT | TC53817 | 79 | + | Thioredoxin H, (Q4U0W0) | 1.68E-08 |
| 1275 | CT – TC | TC53088 | 383 | + | Glutathione S-transferase GST 24 (Q9FQD4) | 7.96E-81 |
| 748 | TC – CG | TC54876 | 240 | + | Ferritin-3, chloroplast precursor (Q948P6) | 2.59E-47 |
| 1631 | TT – CT | Q6V7W6 | 84 | + | Class III peroxidase GvPx2b | 2.76E-04 |
| 1245 | CT – TA | TC62299 | 380 | - | Auxin-induced SAUR-like protein (Q8S351) | 1.89E-79 |
| 1562 | CG – GT | TC53184 | 397 | - | Chloroplast small heat shock protein (Q6WHC0) | 1.14E-80 |
| 781 | TC – GA | TC53791 | 232 | - | Catalase (Q7XTK9) | 4.25E-45 |
| 532 | TG – CT | TC56223 | 206 | - | Peroxiredoxin Q (Q6QPJ6) | 4.38E-37 |
I/R: induced or repressed in cDNA-AFLP experiments.
Figure 2.
Grape transcripts modulated by P. viticola infection. Frequency of modulated genes in selected functional categories and percentage of upregulated (grey) or downregulated (white) transcripts, within each category.
Identification of P. viticola genes expressed in grapevine during infection
Because there is little data on P. viticola virulence factors released by the pathogen during infection, the identification of upregulated transcripts and their cross-reference to known oomycete genes was an important goal of this study. Databases containing genomic sequence information from Phytophthora sojae, P. ramorum, P. infestans and Hyaloperonospora parasitica were used for comparison. We identified 96 TDFs expressed in planta during infection that could be attributed to P. viticola based on their similarity to other oomycete sequences, 22 corresponding to non-annotated genomic contigs and 74 with functional annotations, including genes involved in protein and lipid metabolism, signal transduction, transport, response to oxidative stress and toxicity (Table 2 and Additional File 2).
Table 2.
List of selected putative P. viticola TDFs expressed in planta
| TDF | Primer Comb. | Lenght (bp) | Homology | Annotation | Blast score |
| 170 | TC – TG | 213 | Ps_004_22448_Jun03 | PROBABLE 50S RIBOSOMAL PROTEIN L1 [Sinorhizobium meliloti] | 6.00E-05 |
| 1272 | CT – TT | 72 | Pi_006_52843_Feb05 | 60S ribosomal protein L11 [Hyacinthus orientalis] | 2.00E-05 |
| 1214 | CT – AC | 400 | Ps_060_22857_Jun03 | 60S RIBOSOMAL PROTEIN L7A protein [Arabidopsis thaliana] | 7.00E-77 |
| 1390 | CG – TT | 114 | Pi_001_74169_Feb05 | RL35_EUPES 60S ribosomal protein L35 | 8.00E-09 |
| 1421 | CG – CA | 64 | Pi_015_57096_Feb05 | RL9_SPOFR 60S ribosomal protein L9 | 6.00E-05 |
| 1565 | CG – GT | 216 | Pi_004_46349_Feb05 | L-aspartate oxidase [Chromobacterium violaceum ATCC 12472] | 1.00E-05 |
| 1132 | CC – TG | 89 | Pi_001_82736_Feb05 | Nascent polypeptide associated complex alpha chain prot. [A. thaliana] | 6.00E-12 |
| 862 | CA – AA | 171 | Pi_030_51789_Feb05 | Polyubiquitin [Plasmodium falciparum 3D7] | 5.00E-20 |
| 1228 | CT – AC | 89 | Ps_016_22726_Jun03 | Ubiquitin-conjugating enzyme e2-16 kd [S. pombe] | 6.00E-09 |
| 1135 | CC – TG | 81 | Pi_003_44814_Feb05 | Actin depolymerizing factor – like protein [A. thaliana] | 5.00E-09 |
| 1484 | CT – GC | 127 | Pi_002_46251_Feb05 | Fructose-1 6-biphosphatase [Phytophthora infestans] | 5.00E-04 |
| 160 | TC – TC | 174 | Ps_016_22911_Jun03 | Myo-inositol-1-phosphate synthase [Nicotiana paniculata] | 2.00E-22 |
| 83 | TC – AC | 240 | Ps_005_22630_Jun03 | Putative succinate dehydrogenase flavoprotein subunit [A. thaliana] | 4.00E-12 |
| 394 | TG – AC | 277 | Ps_001_26384_Jun03 | Putative steroid binding protein [A. thaliana] | 1.00E-12 |
| 241 | TA – AC | 288 | Pi_004_52230_Feb05 | Fatty acid synthase, subunit alpha – fission yeast (S. pombe) | 1.00E-31 |
| 1604 | TT – TG | 81 | Ps_029_22780_Jun03 | 14-3-3 protein epsilon [Xenopus laevis] | 3.00E-07 |
| 875 | CA – AT | 203 | Pi_003_45566_Feb05 | ATPase, H+ transporting [Gallus gallus] | 6.00E-14 |
| 1366 | CG – AG | 129 | Ps_001_28794_Jun03 | Putative beta-subunit of K+ channels [Solanum tuberosum] | 5.00E-07 |
| 689 | CA – GC | 422 | Ps_018_22812_Jun03 | Mn superoxide dismutase [Chlamydomonas reinhardtii] | 1.00E-105 |
| 542 | TG – CG | 170 | Pi_018_51411_Feb05 | Mitochondrial Mn3+ Superoxide Dismutase | 4.00E-30 |
| 794 | TC – GT | 256 | Ps_040_22917_Jun03 | Homology to G protein beta subunit [Chlamydomonas reinhardtii] | 1.00E-21 |
| 1410 | CG – TG | 289 | Pi_005_48934_Feb05 | C3HC4-type RING zinc finger protein-like [Oryza sativa ] | 5.00E-12 |
| 270 | TA – TA | 182 | Pi_009_51114_Feb05 | GTP-binding protein ora3 – [Gallus gallus] | 5.00E-05 |
| 239 | TA – AC | 92 | Pi_001_76149_Feb05 | NADH2 dehydrogenase (ubiquinone) [Canis familiaris] | 1.00E-13 |
| 292 | TA – TC | 87 | Pi_004_51216_Feb05 | Probable atrazine chlorohydrolase [Chromobacterium violaceum ] | 3.00E-07 |
| 1274 | CT – TC | 397 | Pi_001_66405_Feb05 | Putative citrate synthase [Saccharomyces kluyveri] | 5.00E-81 |
| 552 | TA – GA | 222 | Pi_002_43637_Feb05 | Putative nuclear LIM interactor-interacting prot. [Phytophthora sojae] | 3.00E-10 |
| 1010 | CA – CC | 521 | Pi_003_48725_Feb05 | S28245 NADH2 dehydrogenase (ubiquinone) | 2.00E-34 |
| 208 | TT – TA | 64 | Pi_004_48444_Feb05 | T52339 ADP-ribosylation factor – pepper | 6.00E-11 |
| 826 | TC – GG | 291 | Pi_028_48244_Feb05 | Vacuolar H+-pyrophosphatase [Prunus persica] | 2.00E-58 |
We predicted that 87 of the P. viticola-derived fragments were of pathogen origin because similar-sized bands were present in the sporangia cDNA-AFLP lanes. The remaining nine fragments were not present in sporangia, and were thus uniquely induced by infection. These are the most likely to represent putative virulence factors. However, alignment to a database of putative avr homologs, kindly provided by Dr. J. Win, Sainsbury Laboratory (UK), failed to identify any P. viticola-derived fragments with similarity to predicted Phytophthora effectors.
Verification of representative genes by real-time RT-PCR
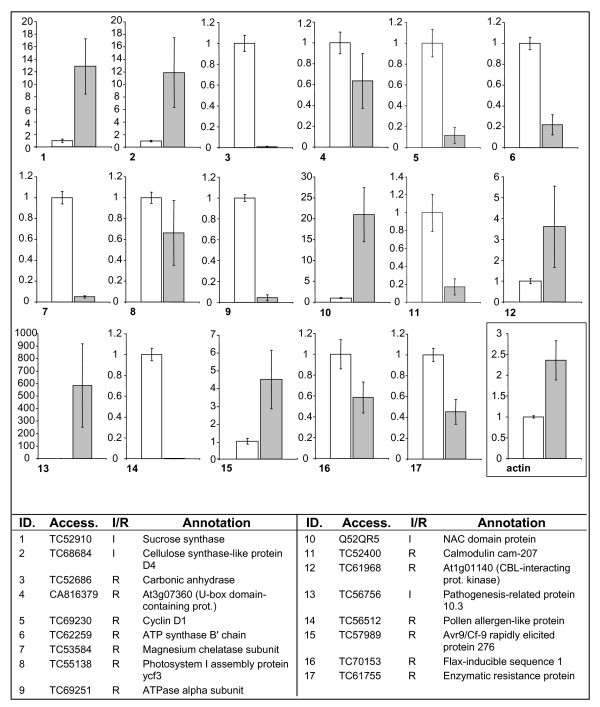
The expression level of 17 modulated grapevine transcripts was analyzed further by real-time RT-PCR, to validate cDNA-AFLP expression profiles. These genes were chosen as they represented almost all the different functional categories identified, with a preference for defense-related genes and genes possibly involved in pathogenesis. Results are shown in Figure 3. The choice of a stable reference gene for data normalization is still controversial in grapevine. Several genes that are commonly considered to be constitutively expressed, such as tubulin, ubiquitin and glyceraldehyde 3-phosphate dehydrogenase, were shown to be modulated in our experiments and in previous work [35]. Data were therefore normalized against 18S RNA expression levels, which are among the more stably-expressed genes in other oomycete infections [36]. Actin (TC60835), which was considered as a possible reference gene initially, also appeared to be modulated in the infected sample (Figure 3). The expression of the selected genes was in good agreement with profiles detected by cDNA-AFLP, except for two genes: TC57989 (avr9/Cf9 rapidly induced protein 276) and TC61968 (Protein At1g01140). These experiments allowed the detection of strong changes in gene expression (about 10-fold or more) between infected and healthy tissues for nine of the selected genes. Among defense-related transcripts, it is worth noting the ~600-fold increase in the level of mRNA for the homologue of pathogenesis-related protein 10.3 of Vitis quinquangularis (TC56756) (Figure 3, gene 13).
Figure 3.
Real-time RT-PCR analysis. Real-time RT-PCR analysis of transcript levels for 17 selected genes in healthy (white) and infected (gray) grapevine leaves. Gene annotations correspond to numbers on graphs. I/R: induced or repressed according to cDNA-AFLP experiments. The expression level of actin (TC60835) is also reported (insert). All data were normalized to the 18S rRNA expression level. Data represent fold change of gene expression in infected vs. control samples. Bars represent a 95% confidence interval, calculated on 3 technical replicates.
Discussion
Transcriptomics is a powerful approach for the global analysis of plant-pathogen interactions, and our study used this strategy to provide the first large-scale investigation of the compatible interaction between P. viticola and grapevine. We observed widespread modulation of transcriptional activity, with 17% of all transcripts showing some form of differential expression, consistent with the extensive physiological changes that affect most cells in infected tissues.
The cDNA-AFLP method for global transcriptional analysis is an open architecture technology that is appropriate for gene expression studies in non-model species. This is because prior sequence data is not required for the visual identification of differentially-expressed transcripts, in contrast to other approaches. In addition, cDNA-AFLP is particularly useful for the study of plant-pathogen interactions because the method facilitates gene discovery in both organisms simultaneously [25,37,38]. These advantages are emphasized by our discovery that 25% of our TDFs representing modulated grapevine transcripts were not previously reported in any EST database. Because of the very small number of P. viticola sequences deposited in databases thus far, all putative P. viticola transcripts expressed in planta identified in this work could be regarded as newly identified genes.
Infection with P. viticola causes widespread gene repression in grapevine
The most striking discovery in our investigation was that nearly 70% of the differentially-expressed grapevine genes we identified were downregulated during infection, possibly reflecting the exploitation of cellular resources and/or the suppression of defense responses [7]. At the oil spot stage, infection has already been established but the host cells must be kept alive to supply the pathogen with nutrients and to facilitate sporulation. This closely mirrors the early stages of natural infections, thus the activation of a non-specific senescence program seems unlikely. Additionally, most of the visualized transcripts were unaffected by infection, and 30% of the differentially-expressed genes were clearly upregulated confirming the absence of a general, global, repressive environment. Among the upregulated genes, we identified many usually considered to have "housekeeping" functions, such as a tubulin alpha chain (TC65238), a dynein light chain (TC57042) and, according to the real-time RT-PCR experiments, actin. The induction of a plant actin gene was first reported in Malva pulsilla during the biotrophic phase of interaction with Colletotrichum gloesporioides [39] and several subsequent reports supported actin's role in cytoskeleton rearrangement during incompatible interactions as well as in the maintenance of compatibility [40]. According to cytological data, it is also unlikely that the lower steady state mRNA levels could be due to the proportional increase in pathogen-derived transcripts in the mixture, as could be the case with hemibiotrophic or necrotrophic pathogens in late stages of infection [27]. In extensively colonized tissues, only apical parts of the mycelium seem to be metabolically active while older portions are totally devoid of cytoplasm [41,42]. Thus, the presence of pathogen RNA should not significantly reduce the amount of plant RNA compared to non-infected leaves. For these reasons, our data probably reflect the actual changes in mRNA levels that characterize this strictly biotrophic plant-microbe interaction. Data provided by real-time RT-PCR confirmed the original expression profiles for 15 out of 17 selected genes, further strengthening the reliability of our results.
Photosynthesis and primary carbon metabolism
The most striking transcriptional downregulation in our investigation was observed in genes related to photosynthesis, e.g. chlorophyll a-b binding proteins (TC54828, TC55242, TC56895) and photosystem components (TC53444, TC61693, TC66994), consistent with the measurable reduction in chlorophyll content during pathogenesis [43]. Transcriptional downregulation of photosynthesis-related genes has been reported previously during compatible interactions between potato and P. infestans [28] and between soybean and P. sojae [27]. Similar results for grapevine have been reported in microarray-based analyses of compatible interactions with viruses and powdery mildew [44,45]. It is well established that plants infected with biotrophic fungal pathogens, such as powdery mildews and rusts, reduce their photosynthetic rates, possibly as a result of increased invertase activity which causes carbohydrate accumulation [46]. Invertase is needed to cleave sucrose into hexose sugars, which in turn can be taken up by pathogens. In this context, the increased level of two genes with similarity to hexose transporters (Q3L7K6 and TC66367) is also worth noting. Carbohydrate accumulation may inhibit the Calvin cycle, which also limits photosynthesis [46,47]. Several genes encoding enzymes in the Calvin cycle are downregulated during infection, among them ribulose bisphosphate carboxylase/oxygenase activase (TC66665) a plastidic aldolase (TC52159), a sedoheptulose bisphosphatase (TC54570), a phosphoribulokinase (TC56646), and a plastidial glyceraldehyde-3-phosphate dehydrogenase B subunit (TC60916).
P. viticola infection also elevated mRNA levels for a sucrose synthase (TC52910), an enzyme that usually carries out sucrose degradation in plants. This reaction releases fructose and UDP-glucose residues, which are substrates for callose and cellulose synthesis. Therefore it is interesting to note that two UDP-glucosyltransferases (TC57852 and TC54299) are also among the upregulated transcripts we identified, along with a cellulose synthase-like sequence (TC68684). Moreover, since cellulose and callose are the main components of Plasmopara cell walls and septa, we speculate that the induction of these genes might reflect the supply of precursors for pathogen metabolism. Concomitantly, several genes encoding cell wall degrading enzymes are downregulated, including two pectinacetylesterases (TC54500 and TC52435) and a polygalacturonase-like protein (TC59719).
The carbonic anhydrase (CA) gene TC52686 is worth particular attention, as its downregulation during infection was established by both cDNA-AFLP and real-time RT-PCR. In C4 plants, CA catalyzes the reversible hydration of carbon dioxide to bicarbonate and provides carbon dioxide for fixation by RuBisCO. However, the role of CA in C3 plants, such as grapevine, is less clear [48,49]. The enzyme has antioxidant activity and is known to bind salicylic acid [50]. It is downregulated in tomato plants following application of the fungal toxin fusicoccin [51], in Arabidopsis following treatment with methyl jasmonate [52] and in potato infected with P. infestans [28]. Silencing of CA expression in Nicotiana benthamiana resulted in suppression of the Pto:avrPto-mediated hypersensitive response [50] and in increased susceptibility to P. infestans [28]. Taken together, these data suggest CA could be involved in the pathogen response and/or that downregulation of CA could be required for the maintenance of a compatible interaction.
Lipid metabolism
Lipid-derived molecules act as signals in plant-pathogen interactions, with jasmonic acid (JA) and related oxylipins produced from membrane-derived fatty acids through beta-oxidation, having particularly important roles [53]. Lipid accumulation is usually associated with necrogenic infections and insect infestations, but JA could also be involved in resistance against biotrophic pathogens, as suggested in grapevine for BABA-induced resistance to P. viticola [54]. During infection, low level defense responses can be activated in susceptible plants, as already reported in grapevine [21,45,55]. Therefore, it is not surprising that well-established P. viticola infections involve the upregulation of genes encoding different enzymes in the beta-oxidation pathway, such as two 3-ketoacyl-CoA thiolases (TC53311 and TC55776), an acyl-coenzyme A oxidase (TC58112) and a fatty acid multifunctional protein (TC52362), as well as a gene encoding a 12-oxophytodienoate reductase (TC67104) that could be involved in the metabolism of oxylipin signaling molecules. Fatty acid metabolism can also produce aldehydes and alcohols with antimicrobial properties, a process involving lipoxygenases and hydroperoxide lyases, examples of which were also induced by infection (CF074703 and TC55722). Other lipases were repressed during infection (DT013748, Q6K832 and TC62475). Certain genes involved in sterol biosynthesis were induced (3-beta-hydroxy-delta5-steroid dehydrogenase, TC67959) while others were repressed (steroid-5-alfa-reductase like protein, Q8H539). Because P. viticola appears fully dependent on its host for sterol biosynthesis [56,57], the modulation of transcripts involved in the sterol biosynthesis pathway needs to be investigated in more detail.
Protein metabolism
Genes related to protein metabolism were also prevalently repressed in our experiment. Among them were genes encoding ribosomal proteins, protein modification and degradation enzymes (e.g. ubiquitin-conjugating enzymes), as well as several kinases, phosphatases and peptidases, which could also be involved in intracellular and intercellular signaling. This suggested a general repression of protein synthesis and turnover. However, some genes involved in amino acid biosynthesis were induced, such as a cysteine synthase (TC51806) and a gamma-glutamylcysteine synthetase (TC56558), in agreement with previous findings [7].
Transport
About 7% of the modulated transcripts corresponded to genes involved in transport. This probably reflects the peculiar nutritional strategy of oomycetes, which rely on molecular trafficking through a modified plasma membrane with inactivated ATPases [58]. We observed the downregulation of genes encoding five different membrane ATPases (TC62785, TC53387, TC69251, TC58445, TC60214), as well as 14-3-3 proteins (TC52346, TC54584), proteins related to vesicular traffic (BQ798655) and ABC transporters (TC57412, TC65826). Genes encoding amino acid and hexose transporters were upregulated perhaps to facilitate the transfer of nutrients to the pathogen (Q1SRS8, Q3L7K6, TC66367, TC62234).
Signal transduction
About 14% of the modulated genes had signal transduction and/or gene regulation functions, including two WRKY DNA-binding proteins (TC70344, Q1T4J9) [59], two NAC transcription factors (TC55407, Q52QR5) [60] and a phosphatase 2C (TC59460) which were induced by infection. However, the majority of genes in this category were downregulated. Several genes encoding components of the calcium signaling network were among them, including calmodulin (TC52400), calmodulin-binding proteins (TC59357, TC68333), a calcium sensor calcineurin B-like protein ('TC69501) and a calcium-dependent protein kinase (CF211026). Calcium signaling is known to be essential in some plant defense mechanisms [61,62]. Many other signaling components and transcription factors were repressed, suggesting that the suppression of endogenous signaling pathways is required to establish compatible interactions.
Secondary metabolism, defense and responses to external stimuli
Many plant defense responses involve the production of secondary metabolites [63]. In the secondary metabolism category, we found that about the same number of genes were upregulated and downregulated, in contrast to all other functional categories. For example, phenylpropanoid pathway enzymes are necessary for the biosynthesis of antimicrobial phenolic derivatives, lignanes and phytoalexins. Several genes encoding enzymes in this pathway were upregulated in infected leaves, including a caffeoyl-CoA O-methyltransferase (TC51729), a stilbene synthase (TC53668), a secretory laccase (TC54354), as well as two glucanases (TC63410, TC60651) and a pathogenesis-related protein 10.3 (TC56756). This indicates the presence of a general although weak defense response in susceptible plants. In contrast we identified homologs of a Pto-like serine/threonine kinase (TC51855) [64], the enzymatic eR protein (TC61755) [65] and the resistance protein KR4 (BQ800016) [66] all of which were downregulated. This was also the case for lipid transfer proteins, such as two homologues of the DIR1 gene (TC63540 and TC61952) [67], a homolog of the Avr9/Cf-9 rapidly elicited protein 276 [68], and a homolog of the Arabidopsis EXECUTER-1 gene (TC62916) [69]. All these genes have been assigned a function related to resistance in other pathosystems, and will be subject to further investigations. Additional genes, that respond to a variety of external stimuli and are often involved in the control of redox balance in the cell, were prevalently downregulated during infection, such as a catalase (TC53791) and a peroxiredoxin Q (TC56223).
P. viticola genes expressed in planta
The exclusively biotrophic lifestyle of P. viticola and other oomycetes complicates the identification of pathogenicity factors. Even so, we identified 96 TDFs corresponding to P. viticola transcripts expressed in planta and nine of these sequences appeared to be expressed at detectable levels only in infected tissues, not in sporangia, even though RNA from the pathogen is much more abundant in the sporangia. These nine transcripts therefore represent important candidate genes specific for the infection process.
Several P. viticola transcripts were homologous of genes involved in protein metabolism. Shan and colleagues [70] showed that several 60S ribosomal protein subunits are expressed at the onset of infection with Phythopthora nicotianae, indicating a requirement for protein synthesis in the pathogen. Several Plasmopara transcripts are homologous to enzymes involved in carbohydrate and fatty acid metabolism, in energy production, and in cellular transport. Genes encoding anti-oxidant enzymes, such as a homolog of P. nicotianae manganese superoxide dismutase [71], proteins involved in signal transduction such as a homolog of Chlamydomonas beta-subunit-like polypeptide CBLP [72] and a steroid binding protein [73] are also expressed in the interaction. All these aspects deserve further investigation in the light of their importance in fungal pathogenesis.
A large repertoire of virulence effectors is thought to be secreted by oomycete pathogens in order to manipulate their host cells [9]. Several approaches have been used to identify such factors in different plant-oomycete interactions [7,9,26,27,29,30]. Following the discovery of a conserved motif (RXRL-EER) necessary for translocation to the plant in all known oomycete avirulence proteins [12,14], bioinformatic tools have been applied to search for putative effector proteins in the different Phytophthora sequenced genomes [13]. This led to the identification of about 700 putative avirulence genes, but none of the identified P. viticola TDFs showed any similarity to predicted Phytophthora effectors.
Conclusion
This report describes the first large-scale investigation into the molecular basis of compatibility between Vitis vinifera and the strictly biotrophic pathogen Plasmopara viticola. The cDNA-AFLP technique allowed the discovery of novel genes both in grapevine and in P. viticola, as a significant proportion of TDFs are not currently represented in Vitis or in oomycete EST databases.
Our data show that infection results primarily in the downregulation of grapevine transcripts in all major functional categories, especially photosynthesis. However, certain genes required for plant-pathogen interactions are positively modulated during infection at the oil spot stage. Actin was also upregulated in infected leaves, reflecting the occurrence of important cytoskeleton modifications during downy mildew infection, and further indicating that assumption of constitutive expression for "housekeeping" genes must always be considered with caution in specific physiopathological conditions. This work also provides the largest available repertoire of P. viticola genes expressed in planta. A large amount of information concerning mRNA levels in infected grapevine is now available, which will hopefully serve as a basis to address new questions and design new experiments to elucidate further the biology of plant-oomycete interactions and the associated re-programming of host metabolism.
Methods
Plant material, inoculum and pathogen infection
Grapevine plants (cv Riesling) were grown in greenhouse at 19°C with 70–80% relative humidity. P. viticola inoculum was collected from sporulated field leaves and used for the artificial inoculations of surface-sterilized leaves. The inoculum was stored as sporangia at -20°C. Infections were initiated by spraying the third and fourth grapevine leaves with a suspension of 10,000 sporangia per ml in cold pure water. The leaves were covered for one night with plastic bags to increase humidity and the plants were kept in the same greenhouse at 19°C with a 16-h photoperiod. Within 2–3 weeks, infected leaves developed the typical oil spot symptoms. The oil spot lesions were sampled with a cork-borer and used for RNA extraction. As a control, RNA was extracted from water-treated leaves incubated under the same conditions. RNA extraction from leaves has been described [74]. Total RNA from sporangia was extracted from frozen samples (50–100 mg) with the RNeasy Mini Kit (Qiagen) following the manufacturer's protocol for plant tissues.
cDNA-AFLP analysis
The cDNA-AFLP protocol applied [31] is a modification of the original technique [32] which permits the visualization of one single cDNA fragment for each messenger originally present in the sample, thus reducing the redundancy of sequences obtained. Briefly, double-stranded cDNA was synthesized from 2 μg total RNA using the Superscript II reverse transcription kit (Invitrogen) and a biotinylated oligo-dT primer (Promega). The cDNA was digested with BstYI (restriction site RGATCY), and the 3' ends of the fragments were captured on streptavidin magnetic beads (Dynal). Digestion with MseI released yielded fragments that were ligated to adapters for amplification (BstYI-Forw: 5'-CTC GTA GAC TGC GTA GT-3'; BstYI_Rev: 5'-GAT CAC TAC GCA GTC TAC-3'; MseI-Forw: 5'-GAC GAT GAG TCC TGA G-3'; MseI-Rev: 5'-TAC ATC AGG ACT CAT-3'). Pre-amplification was performed with an MseI primer (Mse0: 5'-GAT GAG TCC TGA GTA A-3'), combined with a BstYI primer carrying either a T or a C at the 3' end (BstT0: 5'-GAC TGC GTA GTG ATC T-3'; BstC0: 5'-GAC TGC GTA GTG ATC C-3'). Pre-amplification PCR conditions were as follows: 5 min denaturation at 94°C and then 30 s denaturation at 94°C, 60 s annealing at 56°C, 60 s extension at 72°C (25 cycles), followed by 5 min at 72°C.
After preamplification, the mixture was diluted 600 fold and 5 μl was used for selective amplification with 128 primer combinations, carried out with one selective nucleotide added on the 33P-labeled BstYI primer and two selective nucleotides on the MseI primer. Touch-down PCR conditions for selective amplifications were as follows: 5 min denaturation at 94°C, followed by 30 s denaturation at 94°C, 30 s annealing at 65°C, 60 s extension at 72°C (13 cycles, scaledown of 0.7°C per cycle); 30 s denaturation at 94°C, 30 s annealing at 56°C, 60 s extension at 72°C (23 cycles) and 5 min at 72°C. Selective amplification products were separated on a 6% polyacrylamide gel in a Sequi-Gen GT Sequencing Cell (38 × 50 cm) (Bio-Rad) running for 2.5 h at 105 W and 50°C. Gels were dried onto 3 MM Whatman paper on a Gel Dryer Model 583 (Bio-Rad) and marked with Glogos II Autorad Markers (Stratagene) before exposing to Kodak Biomax MR films, for 24 h. The bands of interest were cut from the gels with a surgical blade and eluted in 100 μl of sterile distilled water. An aliquot of 5 μl was used as a template for reamplification using non-labeled primers identical to those employed for selective AFLP amplification. PCR products were purified with MultiScreen PCR μ96 plates (Millipore) and sequenced directly (BMR Genomics).
Sequence analysis
Homology searching by BLAST [75] was carried out against the following databases: NCBI [76] DFCI Grape Gene Index [77], Genoscope Grape Genome database [78], UNIPROT [79], PFGD Phytophthora Functional Genomics Database [80] and VBI Microbial Database [81]. Sequences were manually assigned to functional categories based on the analysis of scientific literature and also with the aid of the information reported for each sequence by The Gene Ontology Consortium [34], when available, or reported by the Swiss-Prot [82], KEGG [83] and TAIR [84] databases.
Real-time RT-PCR analysis
Real-time RT-PCR was carried out on RNA derived from two independent biological experiments. Each sample was a pool of identical quantities of RNA from the two experiments. All samples were examined in three technical replicates. First-strand cDNA was synthesized from DNase-treated total RNA using "High Capacity cDNA Reverse Transcription Kit" (Applied Biosystems). Specific primer pairs (20 b) were designed on 17 TDFs (Additional file 3) and tested by RT-PCR. Primers specific for Vitis vinifera 18S rRNA were used for the normalization of reactions. Experiments were carried out using Power SYBR Green PCR Master Mix (Applied Biosystems) in a Mx3000P QPCR Systems (Stratagene). The following thermal cycling profile was used: 95°C 10 min; 45 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s; 95°C for 1 min, 55°C for 30 s, 95°C for 30 s. Each real-time assay was tested in a dissociation protocol to ensure that each amplicon was a single product. Data were analyzed using MxPro QPCR software (Stratagene). The LinRegPCR software [85] was used to confirm that PCR efficiency was about 2 for each primer couple, and 18S rRNA expression was used as an internal control to normalize all data. Fold change in RNA expression was estimated using threshold cycles, by the ΔΔCT method [86].
Abbreviations
cDNA: Complementary DNA; DFCI: Dana-Farber Cancer Institute; EST: Expressed Sequence Tag; NCBI: National Center for Biotechnology Information; TAIR: The Arabidopsis Information Resource; TDF: Transcript-derived fragment; VBI: Virginia Bioinformatics Institute.
Authors' contributions
MP carried out the cDNA-AFLP experiments (including the extraction and reamplification of cDNA fragments) participated in sequence analysis, performed the real-time RT-PCR experiments, and contributed to data interpretation and manuscript writing. FD and AZ participated in reamplification of cDNA fragments, in real-time RT-PCR experiments and in the analysis and interpretation of data. AF participated in sequence analysis, in automatic Gene Ontology assignment and to table editing. MPz participated in experiment supervision, interpretation of data and critical reading of the manuscript. AK performed P. viticola infections, RNA extractions from sporangia and infected plant material, participated in experiment design and coordination, in interpretation of data, and in writing the manuscript. AP conceived the study, participated in its design and coordination, participated in interpretation of the data, in manual ontology assignments and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Complete list of grape transcripts modulated in P. viticola infected leaves. all genes are grouped in functional categories, according to the presentation in the paper.
Complete list of transcripts attributed to P. viticola and expressed in infected grape leaves.
List of the primers used for real-time RT-PCR experiments.
Acknowledgments
Acknowledgements
We are very grateful to Dr. Joe Win (Sainsbury Laboratory, UK) for providing the Phytophthora predicted avr database. The work was supported by the Project "Structural and functional characterization of the grapevine genome (VIGNA)", granted by the Italian Ministry of Agricultural and Forestry Policies (MIPAF), and by the Project: "Definizione di strumenti diagnostici avanzati e di target per molecole antifungine di origine naturale per la riduzione dell'impatto ambientale da fitofarmaci nel comprensorio viticolo veneto", granted by CARIVERONA Bank Foundation.
Contributor Information
Marianna Polesani, Email: polesani@sci.univr.it.
Filomena Desario, Email: desario@sci.univr.it.
Alberto Ferrarini, Email: alberto.ferrarini@sci.univr.it.
Anita Zamboni, Email: zamboni@sci.univr.it.
Mario Pezzotti, Email: mario.pezzotti@univr.it.
Andreas Kortekamp, Email: kortekam@uni-hohenheim.de.
Annalisa Polverari, Email: annalisa.polverari@univr.it.
References
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Kiefer B, Riemann M, Buche C, Kassemeyer HH, Nick P. The host guides morphogenesis and stomatal targeting in the grapevine pathogen Plasmopara viticola. Planta. 2002;215:387–393. doi: 10.1007/s00425-002-0760-2. [DOI] [PubMed] [Google Scholar]
- Kortekamp A. Leaf surface topography does not mediate tactic response of Plasmopara zoospores to stomata. J Appl Bot. 2003;77:41–46. [Google Scholar]
- Perfect SE, Green JR. Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol Plant Pathol. 2001;2:101–108. doi: 10.1046/j.1364-3703.2001.00055.x. [DOI] [PubMed] [Google Scholar]
- O'Connell RJ, Panstruga R. Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 2006;171:699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- Voegele RT, Mendgen K. Rust haustoria: nutrient uptake and beyond. New Phytol. 2003;159:93–100. doi: 10.1046/j.1469-8137.2003.00761.x. [DOI] [PubMed] [Google Scholar]
- Grenville-Briggs LJ, van West P. The biotrophic stages of oomycete-plant interactions. In: Laskin AI, Bennett JW, Gadd GM, editor. Advances in Applied Microbiology. 2005. pp. 217–243. [DOI] [PubMed] [Google Scholar]
- Chang JH, Goel AK, Grant SR, Dangl JL. Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr Op Microbiol. 2004;7:11–18. doi: 10.1016/j.mib.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Böhme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PR. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PR, Beynon JL. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Pl Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHJ, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CMB, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJG, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JKC, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BWS, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev IV, Rokhsar DS, Boore JL. Phytophthora Genome Sequences Uncover Evolutionary Origins and Mechanisms of Pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PR. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Merdinoglu D, Wiedeman-Merdinoglu S, Coste P, Dumas V, Haetty S, Butterlin G, Greif C. Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Proceedings of the Eighth International Conference on Grape Genetics and Breeding. Acta Horticulturae. 2003;603:451–456. [Google Scholar]
- Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Topfer R, Zyprian EM. Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet. 2004;108:501–515. doi: 10.1007/s00122-003-1445-3. [DOI] [PubMed] [Google Scholar]
- Di Gaspero G, Cipriani G. Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to disease resistance in grapevine. Mol Genet Genomics. 2003;269:612–623. doi: 10.1007/s00438-003-0884-5. [DOI] [PubMed] [Google Scholar]
- Doligez A, Adam-Blondon AF, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, Meredith CP, Riaz S, Roux C, This P. An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet. 2006;113:369–382. doi: 10.1007/s00122-006-0295-1. [DOI] [PubMed] [Google Scholar]
- Kortekamp A, Wind R, Zyprian E. Investigation of the interaction of Plasmopara viticola with susceptible and resistant genotypes. J Plant Dis Protec. 1998;105:475–488. [Google Scholar]
- Kortekamp A, Zyprian E. Characterization of Plasmopara resistance in grapevine using in vitro plants. J Plant Physiol. 2003;160:1393–1400. doi: 10.1078/0176-1617-01021. [DOI] [PubMed] [Google Scholar]
- Kortekamp A. Expression analysis of defense-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol Biochem. 2006;44:58–67. doi: 10.1016/j.plaphy.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Allegre M, Daire X, Heloir MC, Trouvelot S, Mercier L, Adrian M, Pugin A. Stomatal deregulation in Plasmopara viticola-infected grapevine leaves. New Phytol. 2007;173:832–840. doi: 10.1111/j.1469-8137.2006.01959.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmidl J, Lawton KA, Dangl JL, Dietrich RA. An Arabidopsis promoter element shared among genes co-regulated during systemic acquired disease resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Juwana H, Parker JE, Jones JD. cDNA-AFLP display for the isolation of Peronospora parasitica genes expressed during infection in Arabidopsis thaliana. Mol Plant Microbe Interact. 2000;13:895–898. doi: 10.1094/MPMI.2000.13.8.895. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy PD, Allen RL, Rehmany AP, Birch P, Beynon JL. Use of suppression subtractive hybridization to identify downy mildew gene expressed during infection of Arabidopsis thaliana. Mol Plant Pathol. 2003;4:501–507. doi: 10.1046/j.1364-3703.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M. Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant Microbe Interact. 2004;17:1051–1062. doi: 10.1094/MPMI.2004.17.10.1051. [DOI] [PubMed] [Google Scholar]
- Restrepo S, Myers KL, del Pozo O, Martin GB, Hart AL, Buell CR, Fry WE, Smart CD. Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol Plant Microbe Interact. 2005;18:913–922. doi: 10.1094/MPMI-18-0913. [DOI] [PubMed] [Google Scholar]
- Casimiro S, Tenreiro R, Monteiro AA. Identification of pathogenesis-related ESTs in the crucifer downy mildew oomycete Hyaloperonospora parasitica by high-throughput differential display analysis of distinct phenotypic interactions with Brassica oleracea. J Microbiol Methods. 2006;66:466–478. doi: 10.1016/j.mimet.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Bruce CR, Van West P, Grenville-Briggs LJ. Proteomic studies of plant-pathogenic oomycetes and fungi. Methods Biochem Anal. 2006;49:271–283. doi: 10.1002/0471973165.ch16. [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics. 2003;269:173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- Bachem CW, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RG. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313X.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P, French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27–37. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Eckey C, Korell M, Leib K, Biedenkopf D, Jansen C, Langen G, Kogel KH. Identification of powdery mildew-induced barley genes by cDNA-AFLP: functional assessment of an early expressed MAP kinase. Plant Mol Biol. 2004;55:1–15. doi: 10.1007/s11103-004-0275-2. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JD. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Xu R, Wei Y, Goodwin PH. Increased expression of a plant actin gene during a biotrophic interaction between round-leaved mallow, Malva pusilla, and Colletotrichum gloeosporioides f. sp. malvae. Planta. 1999;209:487–494. doi: 10.1007/s004250050752. [DOI] [PubMed] [Google Scholar]
- Lipka V, Panstruga R. Dynamic cellular responses in plant-microbe interactions. Curr Op Plant Biol. 2005;8:625–631. doi: 10.1016/j.pbi.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Langcake P, Lovell PA. Light and electron microscopical studies of the infection of Vitis spp. by Plasmopara viticola, the downy mildew pathogen. Vitis. 1980;19:321–337. [Google Scholar]
- Kortekamp A. Staining methods for the investigation of Plasmopara viticola and its infection structures in semi-thin sections. Vitis. 2005;44:45–47. [Google Scholar]
- Kortekamp A. Knocked out with Basta®! – Are herbicides effective against downy mildew of grapevine? J Plant Dis Protec.
- Espinoza C, Vega A, Medina C, Schlauch K, Cramer G, Arce-Johnson P. Gene expression associated with compatible viral diseases in grapevine cultivars. Funct Integr Genomics. 2007;7:95–110. doi: 10.1007/s10142-006-0031-6. [DOI] [PubMed] [Google Scholar]
- Fung RW, Gonzalo M, Fekete C, Kovacs LG, He Y, Marsh E, McIntyre LM, Schachtman DP, Qiu W. Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 2008;146:236–249. doi: 10.1104/pp.107.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes JD, Lee PJ, Horton P, Lewis DH. Invertase: Understanding Changes in the Photosynthetic and Carbohydrate Metabolism of Barley Leaves with Powdery Mildew. New Phytol. 1994;126:213–222. doi: 10.1111/j.1469-8137.1994.tb03939.x. [DOI] [Google Scholar]
- Walters DR, McRoberts N. Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci. 2006;11:581–586. doi: 10.1016/j.tplants.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Graham D, Reed ML, Patterson BD, Hockley DG, Dwyer MR. Chemical properties, distribution, and physiology of plant and algal carbonic anhydrases. Ann N Y Acad Sci. 1984;429:222–237. doi: 10.1111/j.1749-6632.1984.tb12340.x. [DOI] [PubMed] [Google Scholar]
- Williams TG, Flanagan LB, Coleman JR. Photosynthetic Gas Exchange and Discrimination against 13CO2 and C18O16O in Tobacco Plants Modified by an Antisense Construct to Have Low Chloroplastic Carbonic Anhydrase. Plant Physiol. 1996;112:319–326. doi: 10.1104/pp.112.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA. 2002;99:11640–11645. doi: 10.1073/pnas.182427699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick UB, Schaller A. cDNA microarray analysis of fusicoccin-induced changes in gene expression in tomato plants. Planta. 2002;216:83–94. doi: 10.1007/s00425-002-0887-1. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol. 2005;43:229–260. doi: 10.1146/annurev.phyto.43.040204.135951. [DOI] [PubMed] [Google Scholar]
- Hamiduzzaman MM, Jakab G, Barnavon L, Neuhaus JM, Mauch-Mani B. β-Aminobutyric Acid-Induced Resistance Against Downy Mildew in Grapevine Acts Through the Potentiation of Callose Formation and Jasmonic Acid Signaling. Mol Plant Microbe Interact. 2005;18:819–829. doi: 10.1094/MPMI-18-0819. [DOI] [PubMed] [Google Scholar]
- Lin H, Doddapaneni H, Takahashi Y, Walker MA. Comparative analysis of ESTs involved in grape responses to Xylella fastidiosa infection. BMC Plant Biol. 2007;7:8–20. doi: 10.1186/1471-2229-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD. Biosynthesis and requirement for sterols in the growth and reproduction of Oomycetes. ACS Symp Ser. 1987;325:304–328. [Google Scholar]
- Mikes V, Milat ML, Ponchet M, Ricci P, Blein JP. The fungal elicitor cryptogein is a sterol carrier protein. FEBS Letters. 1997;416:190–192. doi: 10.1016/S0014-5793(97)01193-9. [DOI] [PubMed] [Google Scholar]
- Panstruga R. Establishing compatibility between plants and obligate biotrophic pathogens. Curr Opin Plant Biol. 2003;6:320–326. doi: 10.1016/S1369-5266(03)00043-8. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, Wendehenne D, Pugin A. Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact. 2006;19:711–724. doi: 10.1094/MPMI-19-0711. [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. Calcium in plant defense-signalling pathways. New Phytol. 2006;171:249–269. doi: 10.1111/j.1469-8137.2006.01777.x. [DOI] [PubMed] [Google Scholar]
- La Camera S, Gouzerh G, Dhondt S, Hoffmann L, Fritig B, Legrand M, Heitz T. Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol Revs. 2004;198:267–284. doi: 10.1111/j.0105-2896.2004.0129.x. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–43. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Taler D, Galperin M, Benjamin I, Cohen Y, Kenigsbuch D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Pl Cell. 2004;16:172–184. doi: 10.1105/tpc.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang Y, Wang Q, Luo G, Zhang Z, He C, He SJ, Zhang J, Gai J, Chen S. Characterization of an NBS-LRR resistance gene homologue from soybean. J Plant Physiol. 2004;161:815–822. doi: 10.1016/j.jplph.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Rowland O, Ludwig A, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, Fritz-Laylin L, Nekrasov V, Sjölander K, Yoshioka H, Jones JD. Functional Analysis of Avr9/Cf-9 Rapidly Elicited Genes Identifies a Protein Kinase, ACIK1, That Is Essential for Full Cf-9-Dependent Disease Resistance in Tomato. Pl Cell. 2005;17:295–310. doi: 10.1105/tpc.104.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Marshall JS, Hardham AR. Gene expression in germinated cysts of Phytophthora nicotianae. Mol Plant Pathol. 2004;5:317–330. doi: 10.1111/j.1364-3703.2004.00231.x. [DOI] [PubMed] [Google Scholar]
- Blackman LM, Mitchell HJ, Hardham AR. Characterisation of manganese superoxide dismutase from Phytophthora nicotianae. Mycol Res. 2005;109:1171–1183. doi: 10.1017/S0953756205003771. [DOI] [PubMed] [Google Scholar]
- Schloss JA. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol Gen Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- Blein JP, Coutos-Thévenot P, Marion D, Ponchet M. From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defense mechanisms. Trends Plant Sci. 2002;7:293–296. doi: 10.1016/S1360-1385(02)02284-7. [DOI] [PubMed] [Google Scholar]
- Wielgoss A, Kortekamp A. Comparison of PR-1 expression in different grapevine culture systems after inoculation with a host- and a non-host pathogen. Vitis. 2006;45:9–13. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI http://www.ncbi.nlm.nih.gov/
- DFCI Grape Gene Index http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=grape
- Genoscope Grape Genome database http://www.genoscope.cns.fr/externe/English/Projets/Projet_ML/index.html
- UNIPROT http://www.expasy.uniprot.org/
- PFGD Phytophthora Functional Genomics Database http://www.pfgd.org/
- VBI Microbial Database http://phytophthora.vbi.vt.edu/
- Swiss-Prot http://expasy.org/sprot/
- KEGG http://www.genome.jp/kegg/
- TAIR http://www.arabidopsis.org/
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of grape transcripts modulated in P. viticola infected leaves. all genes are grouped in functional categories, according to the presentation in the paper.
Complete list of transcripts attributed to P. viticola and expressed in infected grape leaves.
List of the primers used for real-time RT-PCR experiments.