Abstract
Expression profiling analysis revealed ectopic high expression of mouse TFF3 in non-tumor liver tissues from the hepatocellular carcinoma (HCC) susceptible PWK/Rbrc strain. TFF3 is a member of the trefoil factor family peptides, which are small secreted proteins regulating mucosal regeneration and repair, and which are overexpressed during inflammatory processes and cancer progression. We, therefore, analyzed the TFF3 expression extensively in mouse and human HCCs. Expression of the mouse TFF3 gene was significantly increased in 6 out of 7 HCCs from a PWK spontaneous tumor model and in all 7 HCCs from an SV40T antigen-induced transgenic MT-D2C57BL/6 model. In humans, 8 of 20 HCCs (40%) had overexpression of TFF3 in both mRNA level and protein level. We then analyzed DNA methylation patterns of the TFF3 promoter region to evaluate expression regulation of promoter methylation. In mouse HCCs, we demonstrated that two CpGs, at positions −992 and +109, were hypomethylated in 13 of 14 mouse HCCs. In human HCCs, hypomethylation at CpG −260 was associated with TFF3 overexpression (p=0.04). These results indicate that TFF3 overexpression may be a critical process in mouse and human hepatocellular carcinogenesis, and the specific promoter CpG hypomethylation may be one of the regulation mechanisms of TFF3 overexpression in HCCs.
Keywords: hepatocellular carcinoma, trefoil factor 3 (TFF3), DNA methylation, expression regulation, epigenetic regulation
Introduction
Hepatocellular carcinoma (HCC) is the fifth-most common malignancy and the third-most frequent cause of cancer mortality in the world (1). Somatic mutations in more than 20 genes and abnormal genetic alterations such as frequent gain of chromosomes 1q, 6p, 8q, 17q and 20q and frequent loss of chromosomes 1p, 4q, 6q, 8p, 13, 16 and 17p have been identified in HCC (2,3). Nevertheless, crucial genetic and epigenetic changes leading to malignant transformation of liver cells have remained obscure because of considerable variance of background causal factors and heterogeneous geographical distribution of HCC.
Trefoil peptides, TFF1 (pS2), TFF2 (SP) and TFF3 (ITF), are small, 7–12 kDa secreted proteins. They are stable molecules (4) that exhibit a predominant region-specific expression pattern throughout the gastrointestinal (GI) tract (5). These three genes are evolutionarily highly conserved and are clustered on the same chromosomal region in mouse chromosome 17 and human chromosome 21. They act to maintain mucosal integrity in the GI tract by epithelium restitution with rapid cell migration (6) and resistance to apoptosis (7), and they also commit to differentiation during embryonic development of the GI tract (8). It has been suggested that TFF3, originally known as intestinal trefoil factor, enhances cell migration through modulation of E-cadherin/catenin complex function (9). Tyrosine phosphorylation of β-catenin, phosphorylation of EGF-R and activation of mitogen-activated protein kinases (MAPK) seem to be involved in this process (10). TFF3 protein has also been shown to promote invasion of transformed cells into collagen matrices (11). TFF3 overexpression is frequently observed in human cancers, including human gastric cancer (12), skin carcinoma (13), papillary-mucinous tumor in the pancreas (14) and breast cancer (15–17) and it is thought to induce cancer cell growth.
In this study, we initially identified TFF3 overexpression in PWK/Rbrc normal liver from expression profiling studies and spontaneous HCC development in the PWK strain. We also observed frequent TFF3 overexpression and promoter CpG hypomethylation in mouse HCCs. We further analyzed whether TFF3 overexpression and promoter CpG hypomethylation occurred frequently in human HCCs. Frequent TFF3 overexpression and hypomethylation of its promoter in HCCs suggest that TFF3 may be a crucial gene involved in mouse and human hepatocellular carcinogenesis.
Materials and methods
Mouse breeding and tissue preparation
The following 11 strains were bred at the Roswell Park Cancer Institute Animal Facility: C57BL/6Ros (B6), C3Hf/HesRos (C3H), NIH/Ola (NIH), A/stRos (A), C57BL/10Ros-pd (B10), BALB/cHSD (BALB), DBA/2Ros (DBA), FVB/N (FVB), PWK/Rbrc (PWK), Mus spretus (spretus) and MT-D2 C57BL/6J transgenic mice (MT-D2B6) (18). C57BR/cdJ (BR) male mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Liver tissues from 12-week-old male mice were harvested and immediately frozen in liquid nitrogen. Twelve male animals each from PWK, B6, spretus and C3H were monitored to evaluate hepatocellular cancer development at approximately one year of age. Three PWK mice died before reaching one year of age due to fighting. Seven of the nine PWK mice developed spontaneous liver tumors, while no liver tumors were observed in any of the other strains. The seven spontaneous liver tumors from PWK and seven transgene-induced liver tumors from the 10-month-old MT-D2B6 mice were used for the DNA and RNA analysis. Two serology tests (4 mice total) were performed during this experimental period, with no pathogen detected, however, Helicobacter pylori was detected in a sentinel of the same mouse colony housed in the specific pathogen-free facility.
Human HCC samples
Twenty tumors and paired non-tumor liver tissues were obtained from surgically resected materials of HCC patients who underwent partial hepatectomy at the Hepato-Biliary-Pancreatic Surgery Division, Graduate School of Medicine, University of Tokyo, Tokyo, Japan. The samples were immediately frozen and stored at −70°C, and the total RNA and high molecular weight DNA were extracted using conventional methods. There were 16 men and 4 women with a mean age of 62±13 (mean ± SD) years (range, 29–79 years). Eleven patients were positive for the hepatitis C virus antibody, 5 were positive for the hepatitis B surface antigen and 4 were negative for both markers. Tumor sizes ranged from 2.5 to 19 cm with an average of 6.7±3.8 cm (mean ± SD). Sixteen were classified as stage II HCCs, 3 as stage III and 1 as stage IV according to the TNM classification system. Among 20 patients, 10 cases were histologically diagnosed as liver cirrhosis in non-cancerous regions.
Quantitative real-time RT-PCR (qPCR)
Total RNA was extracted from frozen tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA). The SuperScript First Strand Synthesis System for reverse transcriptase-PCR (RT-PCR) (Invitrogen) was used to synthesize template cDNA. An identical reaction without reverse transcriptase was performed to verify the absence of genomic DNA (no-RT control). PCR was performed with Sybr-Green (Qiagen, Valencia, CA) on an iCycler iQ Real-Time Quantitative System (Bio-Rad, Hercules, CA). Cycling conditions were 30 sec at 95°C, 30 sec at 54°C and 30 sec at 72°C for 40 cycles. The specificity of each PCR reaction was tested by melting curve analysis and electrophoresis with a 2% agarose gel. In both mouse and human samples, the expression level of β-2 microglobulin was used to adjust the mRNA level of each sample (19). All qPCR experiments were performed in quadruplicate. Primer sequences used are in Table I.
Table I.
PCR and sequencing primers.
| Primer name | Sequence | |
|---|---|---|
| qPCR | mTff3F: | 5′-TAATGCTGTTGGTGGTCCTG-3′ |
| mTff3R: | 5′-CAGCCACGGTTGTTACACTG-3′ | |
| mB2MF: | 5′-CTATATCCTGGCTCACACTGA-3′ | |
| mB2MR: | 5′-GATGCTTGATCACATGTCTCGA-3′ | |
| hTFF3F: | 5′-CTCCAGCTCTGCTGAGGAGT-3′ | |
| hTFF3R: | 5′-CAGGGATCCTGGAGTCAAAG-3′ | |
| hB2MF: | 5′-ATGTCTCGCTCCGTGGCCTTA-3′ | |
| hB2MR: | 5′-ATCTTGGGCTGTGACAAAGTC-3′ | |
| Mouse sequencing | 1F: | 5′-AACAAGCCCTGGTTTTTGC-3′ |
| 1R: | 5′-TGCAGAGTCAGAAAAGACATCAG-3′ | |
| 2F: | 5′-GGCAGACTCGGTAAAGATGAAG-3′ | |
| 2R: | 5′-AGGTACTTACACAGGCCAACG-3′ | |
| Bisulfite sequencing | mCpG-1723-1569F: | 5′-TTAGATAATAGAGATTGATAATTTGAGAAA-3′ |
| mCpG-1723-1569R: | 5′-ACCTAAAATCAAAACCTAAAAAAAA-3′ | |
| mCpG-1418-1379F: | 5′-AGGTTGAATTTTTGATGTTAGAATTAGTT-3′ | |
| mCpG-1418-1379R: | 5′-ATACCCAAATAAATACCCAAAAACC-3′ | |
| mCpG-1172F: | 5′-AATGTTAGGTAGTGAGGATTAGGTAAGG-3′ | |
| mCpG-1172R: | 5′-CTACAACAACAAAAACTAACAACCATTA-3′ | |
| mCpG-992-795F: | 5′-TTTTAGTGTAGATTATTTATTGGGAGTAAG-3′ | |
| mCpG-992-795F: | 5′-AAACCTAAACAAATTTTTAAAAATTTACTT-3′ | |
| mCpG-472F: | 5′-TTTAGAAATTAGAGGTATTGAAAATAGATG-3′ | |
| mCpG-472R: | 5′-ACTTTACACTTTCTCCAAAAAAACC-3′ | |
| mCpG-370-318F: | 5′-GGTTTTTTTGGAGAAAGTGTAAAGTTA-3′ | |
| mCpG-370-318R: | 5′-CAAAACCACAAACTAAACAAAAACAT-3′ | |
| mCpG-19+109B6F: | 5′-TTATAGGAGAGAGAGAGTTTAGAGTTTATA-3′ | |
| mCpG-19+109B6R: | 5′-CAACTAAATACTTACACAAACCAAC-3′ | |
| mCpG-19+109PWKF: | 5′-TTATAGGAGAGAGAGAGTTTAGAGTTTATA-3′ | |
| mCpG-19+109PWKR: | 5′-CAACCTAAAACAAAAAAAACCAACTA-3′ | |
| hCpG-1157F: | 5′-GTTTTGGGAGTGGGTTAAATATTAGT-3′ | |
| hCpG-1157R: | 5′-AATAATCTACCTTCCTTAACCTCCC-3′ | |
| hCpG+50F: | 5′-AAATGAGGTTTTTTGGATTATGAAG-3′ | |
| hCpG+50F: | 5′-AATCAAAACAATACTCACACAAACC-3′ | |
| hCpG-90F: | 5′-AGGAGGGAGAGTTTTTTTTAAGTAAATAA-3′ | |
| hCpG-90R: | 5′-AAAACCAACCCCAACATACAA-3′ | |
| hCpG-260F: | 5′-TAAGGAATTTTTGTGTTTTAGGAGTT-3′ | |
| hCpG-260R: | 5′-ATCAATTCAACCCCCACTATTTTA-3′ |
Bisulfite sequencing
Genomic DNA from each strain was prepared from frozen tissues by the standard method of proteinase K digestion and phenol/chloroform/isoamyl alcohol extraction. Sequencing of approximately 2000 bp in the 5′-flanking region from inbred strains B6 and PWK were initially performed using the primers in Table I. Genomic DNA was bisulfite-treated, as described previously, with some modification (20). Briefly, 10 μg of genomic DNA from each mouse tissue and 4 μg of genomic DNA from human hepatocellular carcinoma tissues and corresponding non-tumor liver tissues were digested with EcoRV and then denatured. Freshly prepared sodium bisulfite (pH 5.0, 1020 μl, 3.6 M) and hydroquinone (60 μl, 10 mM) was added with a final volume of 1200 μl and then the DNA solution incubated for 16 h at 55°C. DNA was purified using the Marligen Matrix Gel Extraction System (Ijamsville, MD). After denaturation by NaOH and neutralizing, the DNA was precipitated and resuspended in 50 μl of TE. PCR primers that should amplify both methylated DNA and unmethylated DNA (Table I) were designed using the website (http://itsa.ucsf.edu/~urolab/methprimer/index1.html) of Methprimer software. PCR products were directly sequenced using an ABI PRISM 3100 DNA Sequencer (Applied Biosystems, Foster City, CA) and signals of the CpGs were estimated by comparing the peak areas of C and T. The percentage of methylation is calculated by the C/T ratio. The bisulfite sequencing was at least duplicated using independently prepared DNAs and the constancy was also confirmed.
Immunohistochemistry
Immunohistochemistry was done as previously described (21), with slight modifications. In brief, sections were deparaffined and rehydrated, followed by hydration. For antigen retrieval of TFF3 protein, specimens were immersed in a 10 mM citrate buffer (pH 6.0) and pressure microwaved for 30 min at 100°C. The slides were then kept at room temperature for 30 min, followed by BSA (1%) blockage for 20 min at 37°C. The treated slides were then incubated with the primary antibody (TFF3 antibody was kindly donated by Dr Werner Hoffmann) (21) in 0.5% BSA (1:750 dilution) overnight at 37°C. The Dako (Capters, CA) automated stainer system and Enhanced DAB detection kit were used for visualization. The slides then were washed, counterstained with Richard Qlien hematoxylin, mounted with Crystal Mount (Biomed, CA) and cover-slipped for micro-scopic evaluation. Sections of small intestine were used for positive control. An irrelevant rabbit IgG served as a negative control. Photographs were taken by using the Olympus Digital System (BX45, Mode DP70).
Bioinformatics and statistical analysis
TRANSFAC database (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi) was used to identify transcription factor binding sequences. Genome information was obtained from UCSC Genome Bioinformatics at http://genome.cse.ucsc.edu/and from the Celera database at http://www.celera.com. Statistical analysis was performed using Wilcoxon Signed-Ranks test, Student’s t-test or χ2 test. Differences were considered significant at p<0.05.
Results
mRNA expression of mouse TFF3
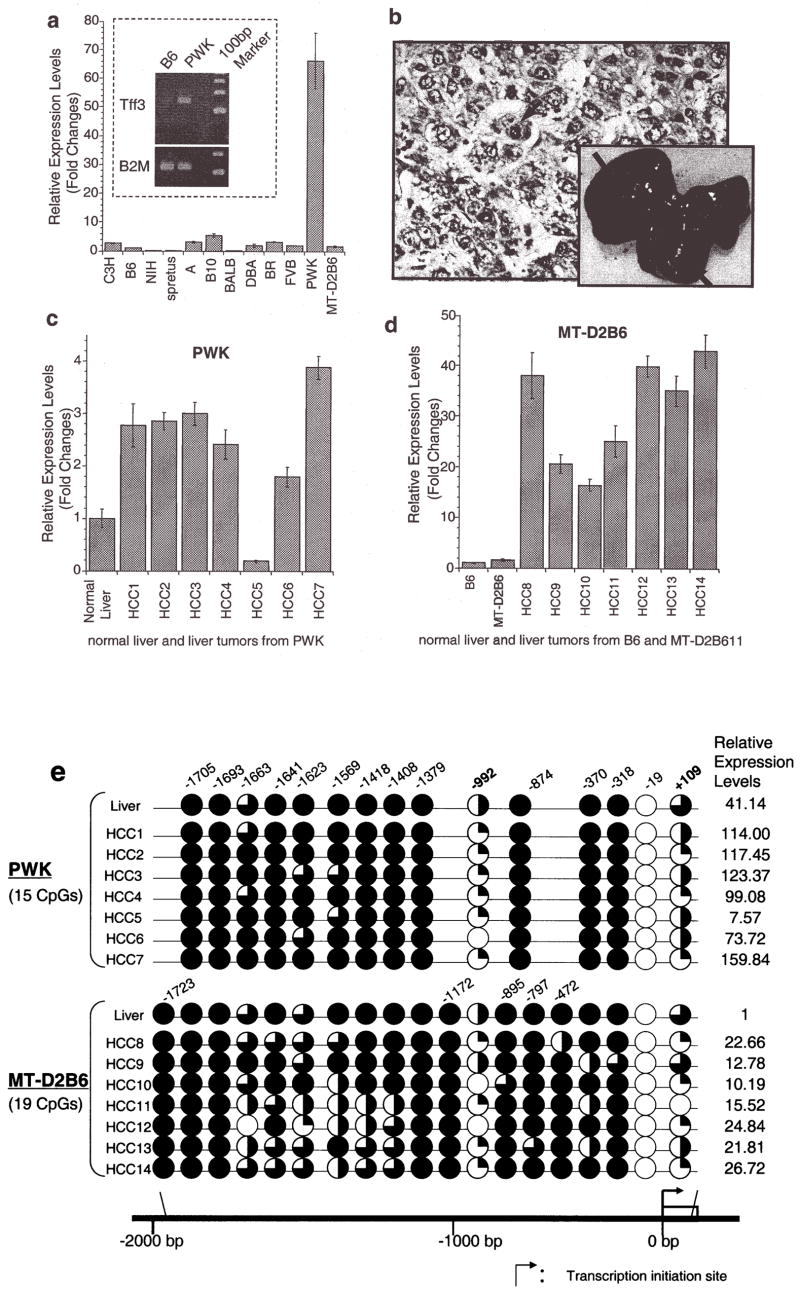
Mouse TFF3 expression levels were examined in normal liver cDNA from 12 different mouse strains (B6, NIH, C3H, spretus, A, B10, BALB, DBA, FVB, PWK, BR and MT-D2B6). Real-time quantitative RT-PCR indicated that TFF3 expression in the PWK liver was >60-fold higher than those in the B6 liver (Fig. 1a). During a 1-year observation under specific pathogen-free conditions, 7 out of 9 PWK/Rbrc males developed spontaneous liver tumors, while none of the 12 mice from B6, spretus and C3H strains developed liver tumors during the same observation periods. All seven tumors were diagnosed with well-differentiated hepatocellular carcinomas (HCCs) and all nine mice had atypical fatty degeneration in normal liver tissue (Fig. 1b). In 6 of 7 PWK spontaneous HCCs, mouse TFF3 was overexpressed relative to the corresponding normal liver (Fig. 1c). We also analyzed mouse TFF3 expression in HCCs from the SV40 T antigen-induced transgenic MT-D2B6 tumor model (18), in which 100% of mice developed HCCs. All seven tumors exhibited significantly higher levels of TFF3 expression relative to normal liver tissue (Fig. 1d), indicating that overexpression of TFF3 is common in both spontaneous and oncogenic transgene-induced HCC development.
Figure 1.
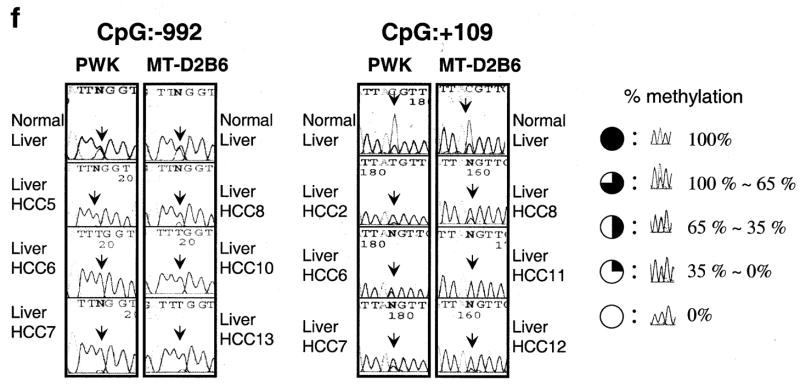
Relative expression levels of mouse TFF3 by quantitative real-time RT-PCR. (a), A bar graph of the mouse TFF3 expression levels with error bars (standard deviation) in liver among 12 mouse strains relative to B6 liver expression (B6=1) is drawn from real-time quantitative PCR. The highest expression was observed in PWK, which was 66-fold higher than that in the B6 liver. A semi-quantitative PCR result of an electrophoresis picture is also shown in the figure and confirmed the quantitative real-time PCR results of B6 and PWK livers (the top is for mouse TFF3 and the bottom is for a control of β-2 microglobulin). Each sample was loaded after 28 cycles. (b), Spontaneous liver tumors frequently developed in PWK, although no HCCs were observed in another susceptible strain of C3H during the same observation period. Pictures show a 56-week male PWK liver, where multiple spontaneous tumor nodules were observed (black arrows). An H&E staining of a spontaneous well-differentiated hepatocellular carcinoma (HCC) from PWK is shown. No normal hepatic lobular architecture was present. Instead, disorganized neoplastic hepatocytes with thickened cords were evident with marked nuclear pleomorphism and atypical mitosis (a black arrow). This tumor is indicated as HCC2 in Fig. 1c. (c), A bar graph shows the mouse TFF3 expression levels in spontaneous HCCs from PWK relative to normal liver expression and (d) HCCs from SV40 T antigen-induced transgenic MT-D2B6 relative to those in non-tumorigenic liver from MT-D2B6 at age 12 weeks. HCCs 1–7 indicate spontaneous HCCs from PWK and HCCs 8–14 indicate HCCs from MT-D2B6. A relative B6 liver expression is also shown. Six of seven tumors from PWK showed overexpression of mouse TFF3 and all tumors from MT-D2B6 showed overexpression of mouse TFF3 compared with non-tumor liver tissue from B6 and MT-D2B6. (e), Methylation status of mouse TFF3 5′-flanking regions. Approximately 2000 bp of genomic DNA in the promoter region were sequenced after bisulfite treatment. CpG identification numbers were referred to for positions from the transcription start site based on the B6 sequence. The pie chart indicates the CpG position [closed circle: 100% methylated CpG site, three-quarter circle: 99–65% methylated, half circle: moderate hypomethylation (65–35%), quarter circle: partial methylation (35–1%) and open circle indicates demethylation 0%]. Open box indicates the first exon. Red ovals indicate the differentially methylated regions in HCCs. Each expression level of TFF3 was calculated relative to normal B6 liver. (f), Bisulfite sequence results at CpGs −992 and +109 in HCCs. Black arrows indicate the cytosine of CpGs. The percentage of methylation is calculated by the C/T ratio of duplicated direct bisulfite sequencing. Both CpG position −992 and +109 are conserved between inbred B6 and PWK. HCCs from both strains were also hypomethylated at both CpG sites relative to non-tumor livers.
Extensive CpG methylation analysis of the TFF3 promoter region in mice
We performed bisulfite sequencing of approximately 2000 bp in the 5′-flanking region of mouse TFF3 to search for differentially methylated sites in the promoter (Fig. 1e). Bisulfite sequencing of seven spontaneous HCCs from PWK identified two regions, CpG −992 (7/7) and +109 (7/7), that were commonly hypomethylated in tumors compared to normal liver tissue (Fig. 1f). We also investigated methylation changes in seven individual SV40 T antigen-induced HCCs from MT-D2B6 transgenic mice. Compared to non-neoplastic liver, tumors were less methylated in CpG positions at −1663 (3/7 tumors), −1569 (5/7), −992 (6/7), −370 (3/7) and +109 (6/7) (Fig. 1e). Two CpG positions at −992 and +109 again showed the most frequent hypomethylation in HCCs from MT-D2B6.
In HCCs from PWK and MT-D2B6, two CpG sites at −992 and +109 were aberrantly hypomethylated and indicated an association between hypomethylation at these two CpGs and TFF3 overexpression (Fig. 1f). These observations suggest that promoter hypomethylation is a common event in mouse HCCs and may accelerate TFF3 overexpression.
TFF3 overexpression and hypomethylation in human HCCs
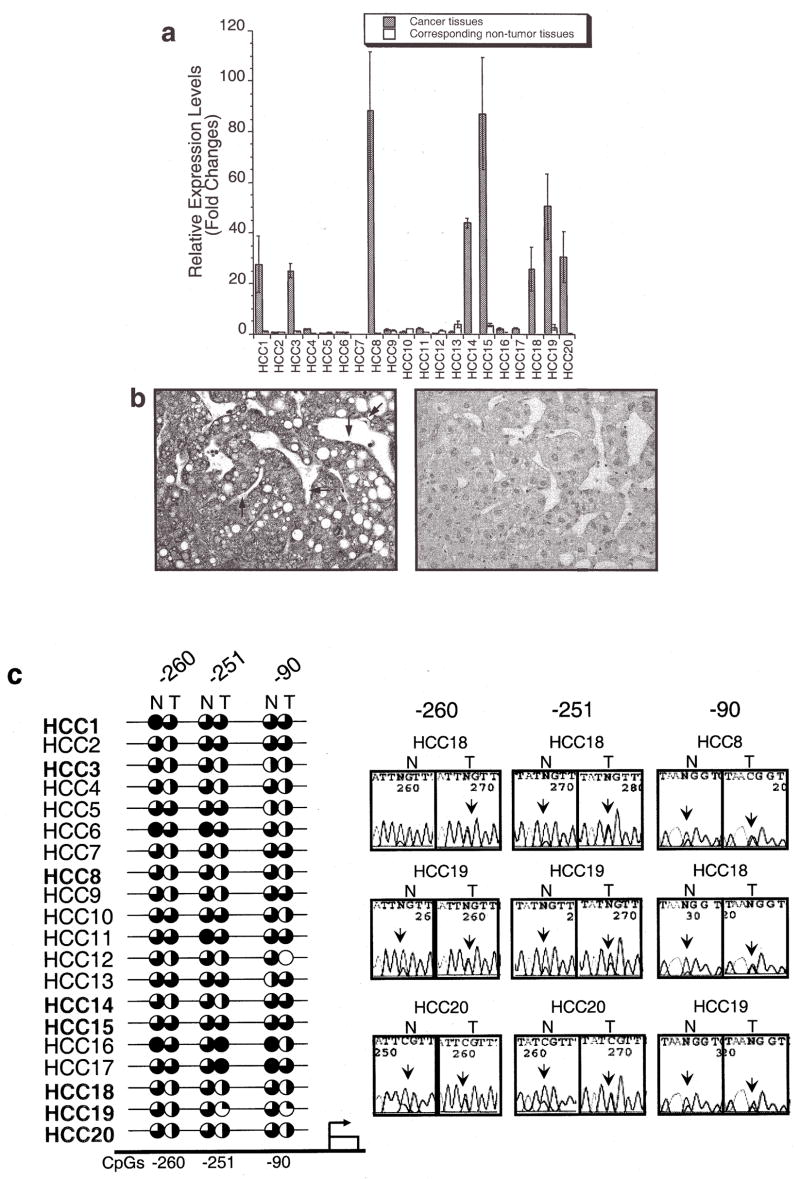
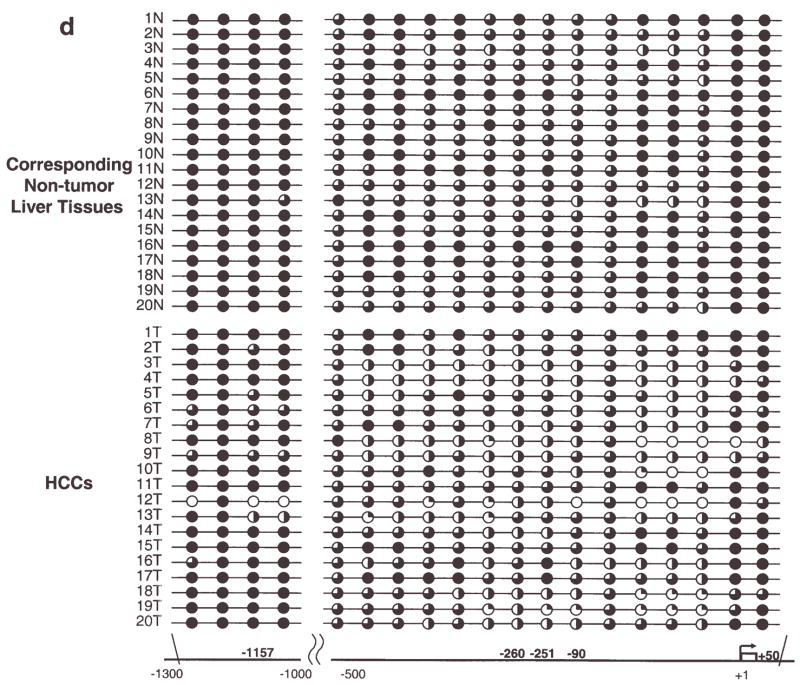
A total of 19 CpG sites, in the promoter sequence from −1300 to −1000 and −500 to +50 which were conserved between mice and humans, were examined in human tumors. HCC and paired non-tumor liver DNA were subjected to DNA methylation analysis in conjunction with expression analysis using qPCR. TFF3 was overexpressed >20-fold in 8 out of 20 human HCC samples (40%) relative to the corresponding non-tumor liver tissues (Fig. 2a). In 3 of 5 HBV, 3 of 11 HCV and 2 of 4 non-viral-infected HCC cases, TFF3 was over-expressed. The overexpression was also seen in 4 of 10 cirrhosis cases. The overexpression of TFF3 at the protein level was also confirmed by immunohistochemical analysis. TFF3 was clearly expressed in neoplastic hepatocytes, displaying fine granules within the cytoplasm (Fig. 2b), whereas liver sinus endothelial cells (Kruffer’s cells indicated by arrows) were negative. All HCC cases except 15T demonstrated hypomethylation changes in the promoter region compared to the paired non-tumor liver tissues. Hypomethylation of CpG sites −260 (7/8) and −251 (6/8) was frequently observed in the 8 overexpressed HCCs. At CpG −260, close to the HNF3/FKH binding site, CpG hypomethylation showed significant association with its expression level (p=0.04, Wilcoxon Signed-Ranks test, Fig. 2c). Although hypomethylation of CpG −1157 and CpG +50, which is equivalent to mouse −992 and +109, was seen in 35% HCCs (7/20) and 30% HCCs (6/20), respectively (Fig. 2d), there was not clear association between TFF3 overexpression and hypomethylation at promoter CpG sites in human HCCs at those sites.
Figure 2.
TFF3 expression and methylation in human hepatocellular carcinomas. (a), A bar graph shows relative expression levels of TFF3 in 20 human HCCs and corresponding non-tumor liver tissues relative to non-tumor liver expression of sample HCC1 (HCC1 non-tumor liver = 1) with error bars (SD). Expression data of each sample were normalized to the β-2 microglobulin expression. Eight of 20 tumors overexpressed >20-fold higher levels compared with those in non-tumor liver tissues. Shaded bars indicates tumor tissues and white bars indicates corresponding non-tumor liver tissues. (b), Immunohistochemistry of HCC using a TFF3 polyclonal antibody. TFF3 is clearly expressed in neoplastic hepatocytes, displaying fine granules within the cytoplasma. The liver sinus endothelial cells (Kruffer’s cells indicated by arrows) are negative, serving as an internal control (left 10×20). A negative control staining of hepatocellular carcinoma section using rabbit IgG. It is characterized by disordered architecture with complex and thickened liver plates and absence of central vein and portal tracts (right 10×20). (c), Results of TFF3 methylation analysis using bisulfite sequencing. Circles show methylation status of CpG sites in tumors (T) and corresponding non-tumor liver tissues (N) with the criteria described in Fig. 1e mouse study. At CpG site −260, close to the HNF3/FKH binding site (between −251 and −90), CpG hypomethylation showed significant association with its expression level (p= 0.04, Wilcoxon Signed-Ranks test). (d), At the two conserved human CpGs (−1157 and +50) and surrounding CpG sites, non-tumor liver tissues were generally methylated, whereas several human hepatocellular carcinomas were hypomethylated. All HCC cases except 15T showed hypomethylation changes in the promoter region compared to paired non-tumor liver tissues at either site.
Discussion
The molecular mechanisms underlying HCC are complicated by its multifactorial characteristics from several risk factors, such as chronic viral infection (HBV and HCV), liver cirrhosis, α-1 antitrypsin deficiency, hemochromatosis, alcoholic liver disease and environmental factors, such as aflatoxin B1 and exposure to tobacco smoke (22). In our mouse studies, we have identified TFF3 overexpression in HCCs from a PWK spontaneous model and an MT-D2B6 transgenic model. Graveel et al also detected overexpression of TFF3 in chemical carcinogen-induced HCCs in another susceptible strain, C3H (23). In humans, overexpression of TFF3 was detected frequently in HCCs (8/20) from patients with various high risk factors (HBV, HCV, cirrhosis and alcoholism). TFF3 overexpression, therefore, appears to be a frequent event not only in spontaneous, transgene-induced and chemically-induced mouse liver tumors, but also in various risk factor-induced human HCCs. TFF3 expression in human gastric adenocarcinoma is associated with more aggressive pathological features and correlates with a reduced overall survival rate of male patients (24). TFF3 overexpression has also been reported in human breast cancer (25,26). We, therefore, speculate that ectopic expression of TFF3 may promote cancer progression during the carcinogenesis process in not only gastric and breast cancer but also in HCC.
Analysis of the TFF promoter region has identified consensus sequences for binding of HNF3/FKH transcription factor, a family of evolutionarily conserved transcription factors that are essential for the formation of gut endoderm (27). The HNF3/FKH and NF-κB binding sites have been reported as an important motif for the expression of the TFF3 gene (28,29). Hypomethylation of a HpaII site in TFF3 5′ upstream of 129 strain suggests association with tissue-specific expression of the mouse TFF3 gene (30), although this site is within 129 strain-specific insertion of a B1 repetitive element and far from the HNF3/FKH and NF-κB binding sites. We analyzed CpG sites, including this B1-inserted site up to 2000 bp from the transcription initiation site. CpG sites −260, −251 and −90 are relatively close to the HNF3/FKH and NF-κB sites (−97, −100, −226, −382, −644 and −811) and are unique in the human genome (Fig. 2c). Hypomethylation of CpG sites −260 and −251 was frequently observed in overexpressed HCCs. DNA methylation establishes a silent chromatin state by associating with methylcytosine-binding proteins that modify nucleosomes. This modification makes DNA inaccessible to transcription factors through histone deacetylation and chromatin structure changes (31). Recent studies suggest that aberrant genome-wide hypomethylation in cancer is not a random event as seen in DNA hypermethylation in promoter CpG islands in tumor suppressor genes (32). Although the TFF3 promoter region is not categorized as a CpG island in the mouse and human genomes, the TFF3 promoter hypomethylation and TFF3 overexpression was frequently observed in HCCs. Promoter CpG site hypomethylation has also been reported in a TFF3 highly expressed tissue of mouse small intestine (30) and human pancreatic ductal carcinomas (33). We also observed the partial loss of methylation in TFF3 non-expressed HCCs and corresponding non-tumor liver tissues in human, which are classified as the premalignant condition. Although further analysis is necessary, the promoter hypomethylation may be necessary but is not sufficient alone to regulate TFF3 expression mechanisms in addition to the transcription factor regulation.
At most of the CpG sites we observed the partial loss of methylation in HCCs in human study. TFFs, as secreted proteins, might not bind to specific receptors, however, this is generally at odds with the finding that TFF2 and TFF3 rapidly activate intracellular signal transduction pathways (28). TFF3 are not expressed in any cells in normal liver but involved in motogenic, antiapoptotic, scattering, proinvasive and angiogenic activities in the normal intestine, inflammatory and neoplastic diseases (34). Since TFF3 acts in a non-cell-autonomous fashion, overexpression of a partial cell population, which has the promoter CpG methylation, might be enough to contribute to cancer progression.
Although promoter hypomethylation and TFF3 over-expression were also frequent events in mouse HCCs, over-expression of TFF3 in PWK normal liver may not be explained simply by the CpG methylation status because there is no promoter methylation difference between PWK and B6 livers. It has been, however, detected that the number of CpG sites located in the transcription factor binding region of PWK was significantly smaller than that in the other m. musculus strains. In our preliminary study, four out of eight F1 hybrids between PWK and other m. musculus developed spontaneous liver tumors by the age of one year without any treatment, and intermediate mouse TFF3 expression level was observed in the liver of F1 hybrids. This suggests that PWK susceptibility is semi-dominant and that the cis-regulated element might be different between two parental strains. Susceptible BR mice, which were used to map a female-specific hepatocarcinogenesis susceptibility locus, Hcf1 (hepatocarcinogenesis in female 1) on chromosome 17 (35) had a 2-fold higher mouse TFF3 expression in the liver than that of the B6 strain, however, there was no DNA methylation difference in the promoter region. TFF3 is also located on chromosome 17 and close to Hcf1 locus. Although frequent spontaneous liver tumor development (17–100%) has been reported in C3H male mice (mouse tumor biology database: http://tumor.informatics.jax.org/FMPro?-db=TumorInstance&-format=mtdp.html&-view), none of the 12 C3H male mice aged 12 months developed HCC in the pathogen-free circumstances. C3H mice had a 2-fold higher rate of TFF3 expression than B6 mice. PWK mice had a 69-fold higher rate of TFF3 expression, and 7 out of 9 PWK mice developed HCCs. Though further analysis is necessary to explore the hypothesis that the TFF3 gene is responsible for HCC susceptibility, our data indicate that TFF3 may be an important gene involved in mouse hepatocellular tumorigenesis and their germ-line susceptibility.
In this study, we demonstrated that overexpression of TFF3 has been involved in liver tumorigenesis in mice and humans, and aberrant promoter methylation may play an important role in TFF3 expression regulation. Furthermore, ectopic TFF3 overexpression has been demonstrated in normal livers from a spontaneous HCC susceptible strain of PWK/Rbrc. This evidence indicates that TFF3 might be an important gene involved in hepatocellular carcinoma development.
Acknowledgments
The authors wish to thank Dr William Held for the MT-D2B6 mice and Drs John Cowell and Fei Song for their useful guidance throughout this study. We would also like to thank Dr Werner Hoffmann for his kind gift of the TFF3 antibody, Dr Heinz Baumann for the Hep3B cell line and Dr Norman Drinkwater for his critical comments during this study. This work was supported by a grant from the National Institute of Environmental Health Services, ES012249-01 and the Roswell Park Cancer Institute NCI Cancer Center Support Grant number CA16056.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Wong N, Lai P, Lee SW, Fan S, Pang E, Liew CT, Sheng Z, Lau JW, Johnson PJ. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size and cirrhosis. Am J Pathol. 1999;154:37–43. doi: 10.1016/S0002-9440(10)65248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59–65. [PubMed] [Google Scholar]
- 4.Thim L. Trefoil peptides: from structure to function. Cell Mol Life Sci. 1997;53:888–903. doi: 10.1007/s000180050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams GR, Wright NA. Trefoil factor family domain peptides. Virchows Arch. 1997;431:299–304. doi: 10.1007/s004280050102. [DOI] [PubMed] [Google Scholar]
- 6.Terada T, Sakagami R, Tabuchi Y, Maeda M. Characterization of the mouse TFF1 (pS2) gene promoter region. Biol Pharm Bull. 2001;24:135–139. doi: 10.1248/bpb.24.135. [DOI] [PubMed] [Google Scholar]
- 7.Taupin DR, Kinoshita K, Podolsky DK. Intestinal trefoil factor confers colonic epithelial resistance to apoptosis. Proc Natl Acad Sci USA. 2000;97:799–804. doi: 10.1073/pnas.97.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre O, Wolf C, Kedinger M, Chenard MP, Tomasetto C, Chambon P, Rio MC. The mouse one P-domain (pS2) and two P-domain (mSP) genes exhibit distinct patterns of expression. J Cell Biol. 1993;122:191–198. doi: 10.1083/jcb.122.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, El-Hariry I, Karayiannakis AJ, Wilding J, Chinery R, Kmiot W, McCrea PD, Gullick WJ, Pignatelli M. Phosphorylation of beta-catenin and epidermal growth factor receptor by intestinal trefoil factor. Lab Invest. 1997;77:557–563. [PubMed] [Google Scholar]
- 10.Kinoshita K, Taupin DR, Itoh H, Podolsky DK. Distinct pathways of cell migration and antiapoptotic response to epithelial injury: structure-function analysis of human intestinal trefoil factor. Mol Cell Biol. 2000;20:4680–4690. doi: 10.1128/mcb.20.13.4680-4690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emami S, Le Floch N, Bruyneel E, Thim L, May F, Westley B, Rio M, Mareel M, Gespach C. Induction of scattering and cellular invasion by trefoil peptides in src- and RhoA-transformed kidney and colonic epithelial cells. FASEB J. 2001;15:351–361. doi: 10.1096/fj.00-0355com. [DOI] [PubMed] [Google Scholar]
- 12.Leung WK, Yu J, Chan FK, To KF, Chan MW, Ebert MP, Ng EK, Chung SC, Malfertheiner P, Sung JJ. Expression of trefoil peptides (TFF1, TFF2, and TFF3) in gastric carcinomas, intestinal metaplasia, and non-neoplastic gastric tissues. J Pathol. 2002;197:582–588. doi: 10.1002/path.1147. [DOI] [PubMed] [Google Scholar]
- 13.Hanby AM, McKee P, Jeffery M, Grayson W, Dublin E, Poulsom R, Maguire B. Primary mucinous carcinomas of the skin express TFF1, TFF3, estrogen receptor and progesterone receptors. Am J Surg Pathol. 1998;22:1125–1131. doi: 10.1097/00000478-199809000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May FE, Westley BR. Expression of human intestinal trefoil factor in malignant cells and its regulation by oestrogen in breast cancer cells. J Pathol. 1997;182:404–413. doi: 10.1002/(SICI)1096-9896(199708)182:4<404::AID-PATH875>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 16.Poulsom R, Hanby AM, Lalani EN, Hauser F, Hoffmann W, Stamp GW. Intestinal trefoil factor (TFF 3) and pS2 (TFF 1), but not spasmolytic polypeptide (TFF 2) mRNAs are co-expressed in normal, hyperplastic and neoplastic human breast epithelium. J Pathol. 1997;183:30–38. doi: 10.1002/(SICI)1096-9896(199709)183:1<30::AID-PATH1085>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 17.Balcer-Kubiczek EK, Harrison GH, Xu JF, Gutierrez PL. Coordinate late expression of trefoil peptide genes (pS2/TFF1 and ITF/TFF3) in human breast, colon and gastric tumor cells exposed to X-rays. Mol Cancer Ther. 2002;1:405–415. [PubMed] [Google Scholar]
- 18.Held WA, Mullins JJ, Kuhn NJ, Gallagher JF, Gu GD, Gross KW. T antigen expression and tumorigenesis in transgenic mice containing a mouse major urinary protein/SV40 T antigen hybrid gene. EMBO J. 1989;8:183–191. doi: 10.1002/j.1460-2075.1989.tb03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekoguchi E, Sato N, Yasui A, Fukada S, Nimura Y, Aburatani H, Ikeda K, Matsuura A. A novel mitochondrial carnitine-acylcarnitine translocase induced by partial hepatectomy and fasting. J Biol Chem. 2003;278:38796–38802. doi: 10.1074/jbc.M306372200. [DOI] [PubMed] [Google Scholar]
- 20.Matsuyama T, Kimura MT, Koike K, Abe T, Nakano T, Asami T, Ebisuzaki T, Held WA, Yoshida S, Nagase H. Global methylation screening in the Arabidopsis thaliana and Mus musculus genome: applications of virtual image restriction landmark genomic scanning (Vi-RLGS) Nucleic Acids Res. 2003;31:4490–4496. doi: 10.1093/nar/gkg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiede A, Jagla W, Welte T, Kohnlein T, Busk H, Hoffmann W. Localization of TFF3, a new mucus-associated peptide of the human respiratory tract. Am J Respir Crit Care Med. 1999;159:1330–1335. doi: 10.1164/ajrccm.159.4.9804149. [DOI] [PubMed] [Google Scholar]
- 22.Nissen NN, Martin P. Hepatocellular carcinoma: the high-risk patient. J Clin Gastroenterol. 2002;35:S79–S85. doi: 10.1097/00004836-200211002-00003. [DOI] [PubMed] [Google Scholar]
- 23.Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ. Expression profiling and identification of novel genes in hepatocellular carcinomas. Oncogene. 2001;20:2704–2712. doi: 10.1038/sj.onc.1204391. [DOI] [PubMed] [Google Scholar]
- 24.Yamachika T, Werther JL, Bodian C, Babyatsky M, Tatematsu M, Yamamura Y, Chen A, Itzkowitz S. Intestinal trefoil factor: a marker of poor prognosis in gastric carcinoma. Clin Cancer Res. 2002;8:1092–1099. [PubMed] [Google Scholar]
- 25.Martin V, Ribieras S, Song-Wang XG, Lasne Y, Frappart L, Rio MC, Dante R. Involvement of DNA methylation in the control of the expression of an estrogen-induced breast-cancer-associated protein (pS2) in human breast cancers. J Cell Biochem. 1997;65:95–106. doi: 10.1002/(sici)1097-4644(199704)65:1<95::aid-jcb10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 26.Uchino H, Kataoka H, Itoh H, Koono M. Expression of intestinal trefoil factor mRNA is downregulated during progression of colorectal carcinomas. J Clin Pathol. 1997;50:932–934. doi: 10.1136/jcp.50.11.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 28.Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 29.Baus-Loncar M, Al-Azzeh ED, Romanska H, Lalani E, Stamp GW, Blin N, Kayademir T. Transcriptional control of TFF3 (intestinal trefoil factor) via promoter binding sites for the nuclear factor kappaB and C/EBPbeta. Peptides. 2004;25:849–854. doi: 10.1016/j.peptides.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Ribieras S, Lefebvre O, Tomasetto C, Rio MC. Mouse Trefoil factor genes: genomic organization, sequences and methylation analyses. Gene. 2001;266:67–75. doi: 10.1016/s0378-1119(01)00380-8. [DOI] [PubMed] [Google Scholar]
- 31.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Supp l):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 32.Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- 33.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 34.Emami S, Rodrigues S, Rodrigue CM, Le Floch N, Rivat C, Attoub S, Bruyneel E, Gespach C. Trefoil factor family (TFF) peptides and cancer progression. Peptides. 2004;25:885–898. doi: 10.1016/j.peptides.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Poole TM, Drinkwater NR. Two genes abrogate the inhibition of murine hepatocarcinogenesis by ovarian hormones. Proc Natl Acad Sci USA. 1996;93:5848–5853. doi: 10.1073/pnas.93.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]