Abstract
The ventromedial (VMN) and arcuate (ARC) nuclei of the hypothalamus are bilateral nuclear groups at the base of the hypothalamus that are organized through the aggregation of neurons born along the third ventricle that migrate laterally. During development, GABAergic neurons and fibers surround the forming (or primordial) VMN while neurons containing GABA receptors are found within the boundaries of the emerging nucleus. To investigate the role that GABAB receptors play in establishing the VMN, Thy-1 YFP mice were utilized for live video microscopy studies. The Thy-1 promoter drives YFP expression in regions of the hypothalamus during development. Administration of the GABAB receptor antagonist saclofen and the GABAA receptor antagonist bicuculline selectively increased the rate of VMN cell movement in slices placed in vitro at embryonic day 14, when cells that form both the ARC and VMN are migrating away from the proliferative zone surrounding the third ventricle. To further test the role of GABAB receptors in VMN development, GABAB receptor knockout mice were used to examine changes in the positions of phenotypically identified cells within the VMN. Cells containing immunoreactive estrogen receptors (ER)α were located in the ventrolateral quadrant of the wild type VMN. In GABABR1 knockout mice, these ERα positive neurons were located in more dorsal positions at postnatal day (P)0 and P4. We conclude that GABA alters cell migration and its effect on final cell positioning may lead to changes in the circuitry and connections within specific nuclei of the developing hypothalamus.
The ventromedial nucleus of the hypothalamus (VMN) first appears using Nissl stains as a bilateral cell group at the base of the diencephalon around embryonic day 16/17 (E16/17) in mice (McClellan et al., 2006). The heterogeneity of the VMN contributes to the many roles it plays in neuroendocrine function. These roles include influencing female sexual behavior, feeding behavior, anxiety/ defensive behavior, and pain (Canteras et al., 1994, Dielenberg and McGregor, 2001, King, 2006). The VMN is loosely categorized into three main regions: dorsomedial, central, and ventrolateral (Saper et al., 1976, Van Houten and Brawer, 1978). The dorsomedial and central regions are characterized by the expression of the transcription factor steroidogenic factor-1 (SF-1), and the ventrolateral region can be characterized by the expression of ERα (Simerly et al., 1990, Dellovade et al., 2000). SF-1 is one critical transcription factor in VMN development as it plays a role in establishing the cytoarchitecture of the nucleus through terminal differentiation (Tran et al., 2003) and distribution of neuronal phenotypes (Dellovade et al., 2000, Davis et al., 2004a). Although SF-1 is important in VMN development, our prior work and work of others suggests that other factors, in particular, gamma-aminobutyric acid (GABA) are also likely to be involved in determining the boundaries of the nucleus by influencing the movement characteristics of migrating neurons.
The neurotransmitter GABA has an interesting relationship with the development of the VMN (Tobet et al., 1999). During early stages of development, GABA is synthesized in positions that could provide potential boundary information for the embryonic VMN. GABAergic neurons and fibers surround the embryonic VMN, and towards the end of gestation in mice, GABAergic fibers begin to infiltrate interior regions of the VMN (Tobet et al., 1999). In contrast to the late gestational in-growth of fibers, subunits for GABAA (Dellovade et al., 2001), and GABAB (Davis et al., 2002) receptors are expressed in neurons within the region of the VMN as early as E13 and throughout adulthood. In addition, physiological analyses have been performed as early as E18 and indicate the presence of functional receptors in the developing mediobasal hypothalamus of rats (Obrietan and van den Pol, 1995, Obrietan and van den Pol, 1998).
In addition to its role as the major inhibitory neurotransmitter in the adult CNS, GABA is also important in many developmental processes, including cell proliferation (LaMantia, 1995) and neuronal migration (Behar et al., 1996, Behar et al., 1998, Manent and Represa, 2007). GABA’s influence on migration can be mediated through either or both its A and B receptors. In cortical migration, GABAA and GABAB receptors play a role in the formation of cortical layers (Behar et al., 1998). Data to date indicate that the ability of GABA to influence neuronal migration within the VMN is mediated by both ionotropic GABAA (Dellovade et al., 2001) and metabotropic GABAB receptors (Davis et al., 2002). The GABAA receptor agonist muscimol caused a decrease in the percent of neurons moving within the region of the developing VMN. The addition of baclofen, a GABAB receptor agonist, to live tissue slices also caused a dose-dependent decrease in the rate of motion of cells in the region of the VMN (Davis et al., 2002). Baclofen administration did not change the probability of cells moving nor did it have an effect on the angle of cell movement. Based on the Nissl stained gross cytoarchitecture, baclofen did not influence the ability of the VMN to form.
The current study further examines the role of GABAB receptors in the development of the embryonic and early postnatal murine VMN and compares the role of these receptors on the development of the neighboring arcuate nucleus (ARC). Mice in which the Thy-1 promoter drives neuron-selective yellow fluorescent protein (YFP) expression (Feng et al., 2000), were utilized for live video microscopy studies in vitro to evaluate the potential role of endogenous GABA acting at GABA receptors on the movement of one population of migrating neurons (Thy-1/YFP expressing neurons) by using GABA receptor antagonists. These mice were used as a tool to follow the movement patterns of fluorescently labeled cells in the regions of the VMN and ARC. It is unknown as to why YFP expression is found in a subset of VMN and ARC neurons at embryonic ages, but the limited pattern of expression is likely due to the insertion site of the transgene, the number of copies incorporated into each line, or the interactions of the transgene elements with flanking DNA (Feng et al., 2000). GABAB receptor knockout mice (Prosser et al., 2001) were utilized to determine the dependence of VMN formation on GABAB signaling during early development. Immunocytochemistry and in situ hybridization were used to identify cell phenotypes for ERα, BDNF (brain derived neurotrophic factor), SF-1, and specific GABAA receptor subunits to determine potential differences in cell position or expression in mice without GABAB receptor signaling.
Materials and Methods
Animals
Two lines of transgenic mice were used for the experiments in this study. GABABR1 heterozygous breeding pairs obtained from GlaxoSmithKline (Middlesex, UK) were used to generate knockout, heterozygous, and wild type mice at multiple developmental ages. Mice with disruption of GABAB receptor signaling were generated on a C57BL/6 background through the insertion of a gene encoding β-galactosidase in the coding region of the R1 subunit of the GABAB receptor (Prosser et al., 2001). Another line of transgenic mice obtained from The Jackson Laboratory (B6.Cg-Tg(Thy1-YFP)16Jrs/J; Bar Harbor, Maine) were created on a C57BL/6 background with the Thy-1 promoter driving YFP expression (Feng et al., 2000). The Thy-1 promoter in this transgenic line drives neuronal expression in the brain, including the ARC and VMN, in a spatially and temporally regulated manner during development (Tobet et al., 2003, Knoll et al., 2007). GABABR1 heterozygote mice were crossed with Thy-1/YFP mice to begin to generate a line of GABABR1 knockout and heterozygote mice that also contain fluorescently labeled YFP neurons. Animals were mated overnight and females were checked for vaginal plugs the following morning. The day of plug was designated as E0 and mice were taken at ages E15, E17, P0 (counted as day of birth - occurring at E19), and P4. Pregnant dams were anesthetized using ketamine (80mg/Kg) and xylazine (8mg/Kg) and embryos were removed by Cesarean section one at a time. Crown rump lengths were measured to verify developmental age, weights were taken for comparison, and tissue was taken for genotyping by PCR. P0 and P4 pups were anesthetized on ice and pups were perfused transcardially with either 4mL (E17) or 5mL (P0 and P4) of 4% paraformaldehyde. Heads were postfixed overnight in 4% paraformaldehyde and changed into 0.1M phosphate buffer (PB) the following day. Heads were stored in 0.1M PB at 4°C until used for immunocytochemistry or in situ hybridization. All experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the Colorado State University Animal Care and Use Committee.
Genotyping
Tail DNA was extracted using a base extraction method - incubation at 95°C with 0.05M NaOH for 20 minutes, 10 minutes at room temperature, followed by addition of a Tris NaOH solution (pH 8.0) and stored at -20°C. Mice carrying the GABABR1 (Genbank accession number NW001470; (Kaupmann et al., 1997)) knockout allele were genotyped using standard Taq polymerase PCR kit (Qiagen; Valencia, CA). The PCR cycling conditions consisted of 20 seconds of denaturation at 94°C followed by 20 seconds of annealing at 56°C and 1 minute of extension at 72°C. This was repeated for 35 cycles. GABABR1 primer sets used consisted of a KO insert forward primer (CGCCTTCTTGACGAGTTCT), a WT-specific forward primer (CTCCCGAGAGTGACTGGTT) and a common reverse primer (GCCCTCTTGCCTCTCTAAA). Animals were designated as wild type, heterozygous, or knockouts. Wild type and heterozygous animals had no detectable differences on any measure and were grouped together as controls. For video microscopy studies, mice were either heterozygous or homozygous for the Thy1-YFP allele. PCR cycling conditions for YFP genotyping consisted of 30 seconds of denaturation at 94°C followed by 1 minute of annealing at 56°C and 1 minute of extension at 72°C. This was repeated for 35 cycles. YFP primer sets included a forward primer (AAGTTCATCTGCACCACCG) and a reverse primer (TCCTTGAAGAAGATGGTGCG). Sex determination was done using PCR for the Y chromosome encoded Sry gene as described previously (Luo et al., 1994).
Immunocytochemistry and In situ hybridization
For immunocytochemical experiments brains were dissected out of the head, embedded in 5% agarose, and cut into 60μm thick sections using a vibrating microtome (VT1000S; Leica Microsystems, Wetzlar, Germany). Alternating sections were collected in 0.05M phosphate buffered saline (PBS) and incubated with primary antisera. Rabbit polyclonal antisera directed against ERα was obtained from Upstate Biotechnology (C1355; Charlottesville, VA) and was diluted to 1/5000 for brightfield detection and 1/1000 for epifluorescence. Other rabbit polyclonal antibodies obtained include SF-1 (graciously provided by Dr. Ken Morohashi; diluted 1/1000), GABAA receptor subunit β3 (graciously provided by W. Sieghart; (Sperk et al., 1997); 0.045μg/mL), GABAA receptor subunit α1 (Phosphosolutions, Aurora, CO; diluted 1/200), and green fluorescent protein (GFP; Molecular Probes/Invitrogen, Carlsbad, CA; diluted 1/50,000). A chicken polyclonal GFP antibody was also used for dual label epifluorescence (ckGFP; Chemicon International, Temecula, CA; diluted 1/1000). Guinea Pig polyclonal antisera directed against GABABR1 (GP311) was generously provided by Dr. Marta Margeta-Mitrovic (Margeta-Mitrovic et al., 1999) and was used for dual label with GFP and ERα. Radial Cell (RC)2 antibody (diluted 1/3) that is selective for a protein in radial glia (Chanas-Sacre et al., 2000) and islet-1 antibody (diluted 1/30) was obtained from the developmental studies hybridoma tissue bank (DSHB; Iowa City, IA). Methods for immunocytochemical procedures using floating tissue sections were reported previously (Tobet et al., 1996, Tobet et al., 1999, Dellovade et al., 2000, Davis et al., 2002). Briefly, sections were washed in 0.05M PBS with 0.1M glycine for 30 minutes followed by a series of rinses with PBS. The sections were then placed in a 0.5% solution of sodium borohydride in PBS, rinsed in PBS, and incubated in a PBS blocking solution containing 5% normal goat serum, 0.3% Triton-X100 (Tx)/PBS and 1%H2O2. Primary antibodies were diluted in a PBS buffer containing 1% bovine serum albumin and 0.3%Tx. The sections were then placed in primary antibody for 2-3 nights at 4°C. Following the incubation in primary antibody, all steps were done at room temperature. Sections were rinsed 4 times in a PBS/ 1% normal goat serum solution containing 0.02%Tx, and then incubated in secondary antibody for 2 hours. The secondary antibodies were obtained from Jackson ImmunoResearch Labs (West Grove, PA) and were diluted in a PBS solution containing 1% normal goat serum and 0.32%Tx. Following this incubation the sections were rinsed 4 times in a PBS/ 0.02%Tx solution. For brightfield detection a 1:2500 dilution of peroxidase conjugated streptavidin in 0.32% Tx/ PBS was placed on the tissue sections for 1 hour. The sections were rinsed 4 times in a Tris buffered solution (TBS; pH 7.5) and incubated in 0.025% diaminobenzidine/ 0.02% Nickel/ 0.02% H2O2 solution diluted in TBS. This reaction was run for 5 minutes in each immunocytochemistry experiment and the sections were washed in TBS and stored at 4°C until the tissue was mounted the following day. The controls included looking at an additional GABAA receptor α1 antibody with corroborating results and antisera preabsorption, as well as incubation of the tissue without primary antibody.
For generation of BDNF riboprobes, plasmids containing the coding sequence for BDNF (provided by Dr. Keith Parker) were linearized using the appropriate restriction enzyme (Pst1 - sense, EcoRV - antisense). The linearized DNA was extracted using a standard phenol chloroform extraction. The linearized DNA was then transcribed and DIG labeled using a Digoxigenin labeling kit (Roche Applied Science, Indianapolis, IN) and the appropriate RNA polymerase (T3 - sense, T7 - antisense). The DIG labeled RNA probes were then precipitated in a solution containing 1μl glycogen (20mg/ml Boehringer Manheim), 2.5μl 4M LiCl, and 75μl cold 100% EtOH. The solution was vortexed briefly, incubated at -80C, washed again with 70% EtOH, left to air dry, and resuspended in DEPC water. An optical density reading was obtained to estimate the amount of probe in solution. For BDNF 0.2ug of the riboprobe was added to 1mL of pre-hybridization buffer for each reaction. Sense probes were generated and used as a control.
For in situ hybridization experiments, tissue was collected using the same methods used for immunocytochemistry but was cut into 100μm sections. The in situ hybridization protocol used was reported previously (Dellovade et al., 2000) and was adapted for using free floating sections from previously published methods (Riddle et al., 1993) using polyvinyl alcohol in the reaction product step to enhance detection (De Block and Debrouwer, 1993). Briefly, the in situ hybridization protocol involved a series of steps occurring over 4 days that included hybridization, washes, and detection using a non-radioactive digoxigenin system. On day 1, agarose embedded brains were cut into cold DEPC-treated PBS with 12% NaCl and followed by a one hour bleaching step of 6% H2O2 in PBT (PBS with 0.1% Tween-20) at room temperature (RT). Sections were then washed with PBT and treated with 10μg/ml Proteinase K diluted in PBT, followed by a wash in 2mg/ml glycine diluted in PBT, rinsed in PBT, then placed in a post-fix solution (4% paraformaldehyde/ 0.2% glutaraldehyde diluted in PBT) for 20 minutes. After PBT rinses, the sections were moved to a prehybridization solution (for 40mls of prehybridization solution: 20ml formamide, 4ml 20X SSC, 0.182ml Yeast tRNA, 2mg heparin, 9.56ml of 1g/10ml solution dextran sulfate, 6.26ml DEPC H2O). The RNA probes were heated to 85°C for 5 minutes to denature, and riboprobes were then transferred to the vials containing the tissue and prehybridization solution. The sections were left overnight at 60°C to hybridize. The following day the sections were washed with a 50% formamide solution (25% 20X SSC, pH4.5 and 25% MQH2O) in a 60°C shaking water bath, and then washed in a second solution (50% formamide; 10% 20X SSC, pH4.5; 40% MQH2O) in a 60°C shaking water bath. The sections were washed at room temperature (RT) in a fresh solution of TBST (1L solution: 8g NaCL, 0.2g KCl, 25mls 1M Tris HCl, 10mls Tween-20, up to 1L MQH2O). The tissue was placed in a pre-block solution consisting of 10% sheep serum diluted in TBST for at least 1 hour at RT and anti-dig-AP antibody (1:2000) in 1% sheep serum/ TBST was put into each boat and then shaken overnight at 4°C in a humid chamber. The following day, the antibody was removed and the tissue was washed in TBST. Next, the sections were washed 5 times in TBST, rocking at RT, and then left overnight in TBST at 4°C. On the final day, the sections were washed in NTMT (2% 5M NaCl; 5% 2M Tris, pH 9.5; 5% 1M MgCl2; 1% Tween-20; diluted in MQH2O), and were incubated in 4 ml of a color detection mix in the dark and monitored every 1-2 hours for signal detection. The NBT/BCIP color detection mix recipe (25ml) consists of 12.5mls 20% polyvinyl alcohol; 12.3mls 200mM Tris, pH 9.5; 0.5mls 5M NaCl; 0.125mls 1M MgCl2; 0.225 mls Levamisol; 0.5mls NBT/BCIP. Once the reaction was determined complete based on high contrast signal over background, the sections were washed in NTMT, and then washed in a post substrate solution (5ml 1M Tris-HCl; 1ml 0.5M EDTA; 4.5g NaCl; 0.5ml MQH2O). Following final washes in PBT the sections were stored at 4°C until mounted onto glass slides and coverslipped using aqueous mounting medium.
Slice Preparation and Live Video Microscopy
Mice were time mated and fetuses were taken at E14, a developmental age when GABAB receptors are expressed in the hypothalamus in the region of the developing VMN (Davis et al., 2002). Pregnant mice were deeply anesthetized with ketamine (80mg/Kg) and xylazine (8mg/Kg) and fetuses were taken by Cesarean section one at a time. The brain, with the pituitary and surrounding cartilage attached, was dissected out of the skull. Dissections were done in Krebs buffer (126mM NaCl, 2.5mM KCl, 1.2mM NaH2PO4, 1.2mM MgCl2, and 2.5mM CaCl2 and an additional 11mM glucose and 25mM NaHCO3) on ice and tissue was taken for sex determination (Sry PCR) and genotyping (GABAB PCR). Heads were processed for live slice preparations following previously published methods (Tobet et al., 2003). Briefly, dissections were limited to a maximum of two hours or eight pups to minimize cellular damage or death. Following each dissection, brains were embedded in 8% agarose and cut in a coronal plane at 250μm in Krebs buffer using a vibrating microtome (Leica VT1000S). Slices that contained regions of the hypothalamus that included the VMN were chosen for video microscopy and were transferred to media (Neurobasal; GIBCO-Invitrogen Corporation, Carlsbad, CA), with 10% L-glutamine, 2% B-27, 1.1% glucose, 2% pen-strep, 2% glutamine and incubated at 36°C with 5% CO2 for 35 minutes. Following the 35-minute incubation period, each slice was plated onto glass bottom dishes pre-coated by the manufacturer with poly-d-lysine (MatTek; P35G-0-20-C) and coated with a 1:1 dilution of Vitrogen (Cohesion Technologies, Inc., Palo Alto, CA) and water. The slices were put back into the 36°C incubator for up to 1 hour to promote adherence. To ensure that slices would move minimally during video observation, 40μL of a Vitrogen solution (1mL Vitrogen, 125μL 10XMEM, 23μL penicillin/streptomyocin, and 33μL 1M sodium bicarbonate) was placed over each slice. The slices were then placed in the 36°C incubator for 1.5 hours to allow the Vitrogen to polymerize. 1mL of Neurobasal media was pipetted into each dish. The slices were maintained at 36°C and 5% CO2 until use for video microscopy- as early as the following morning and as late as 3 days post plating.
In preparation for video microscopy, slices were washed 3 times with warm Neurobasal media (GIBCO-Invitrogen), and placed on a heated stage maintained at 37°C with 5% CO2 and with fresh Neurobasal medium in the dish. All data was collected on either a Nikon TE200 microscope or a Nikon TE2000-U (Nikon USA, Melville, NY) with a 20x plan Apo phase objective. Cells expressing yellow fluorescent protein (YFP) were imaged using Metamorph software (Molecular Devices Inc., Sunnyvale, CA). A digital camera captured a z-stack series of three images at 5μm intervals through the tissue. A set of three images was taken every 5 minutes throughout the duration of the video experiment. At least 1.5 hours of baseline video microscopy was taken before the addition of GABA receptor antagonists, saclofen (10μM-100μM, GABAB; Sigma-Aldrich, St. Louis, MO) or bicuculline (10μM, GABAA; Sigma-Aldrich, St. Louis, MO). The concentrations used were chosen based on the intent to block GABA signaling through its receptors in live brain slices (Harrison et al., 1990, Dellovade et al., 2001). Once a drug was administered to the slice, an additional 1.5 hours of video was taken to compare treatments.
Imaging and Analysis
For immunocytochemical analysis, each section of tissue that contained a region of the VMN, easily identified based on cell and background density, was labeled as A, B, C, or D; extending from rostral to caudal, respectively, (McClellan et al., 2006). Digital images were taken on an Olympus BH2 microscope with an Insight QE digital camera using Spot Advanced software (Draper UT). Images were taken from the sections labeled B and C, those most centrally located within the nucleus, and then normalized for optimal contrast using Adobe Photoshop software (Version CS for Macintosh). For the analysis of cells containing immunoreactive ERα, images were opened in IP Lab software (Scanalytics Inc. part of BD Biosciences, Rockville, MD) and grids (50μm × 50μm) were placed over the images with the boundaries being the base of the brain and the third ventricle (Davis et al., 2004b). Rows and columns extended from the base of the brain moving dorsally and laterally from the third ventricle. Row 1 was designated as the row closest to the base of the brain with more than 50% of the boxes containing tissue. Areas were determined following standard segmentation of the most densely reactive cellular elements in each image. The area of ERα immunoreactive cells was measured in each box and designated as immunoreactive cells residing in the region of the ARC or VMN. Totals were calculated by region and by row, and statistical significance were determined by ANOVA for genotype × age and using row as a repeated measures using SPSS software (SPSS Inc.; Chicago, IL). Immunoreactive SF-1 and the in situ hybridization signal for BDNF were analyzed as total area within the VMN by one-way ANOVA for genotype. As there was no evidence for a heterozygote phenotype for these measures the data depicted is control (wild type + heterozygotes) versus homozygous knockout. For dual label fluorescence, images were taken on a Zeiss LSM510 Meta confocal microscope.
Video analysis has been described in detail previously (Bless et al., 2005, Knoll et al., 2007). Video sequences were evaluated for the presence of visibly moving Thy-1 YFP neurons. These images were aligned to normalize for slice movement using Image J software (NIH, Bethesda, MD), and movement analysis was performed using MetaMorph (Molecular Devices, Downington, PA). A fluorescent cell was “tracked” by manually following the center of each cell and recording the position for each time point. The rate, or average speed, was determined by calculating the speed between each frame and taking an average of all speeds. Data was first collected for all moving cells and later each cell was classified as being part of the VMN or ARC. Data for the current study was collected from 20 slices, from 16 litters.
Results
GABA receptor antagonists caused an increase in movement of fluorescently labeled neurons in the region of the VMN
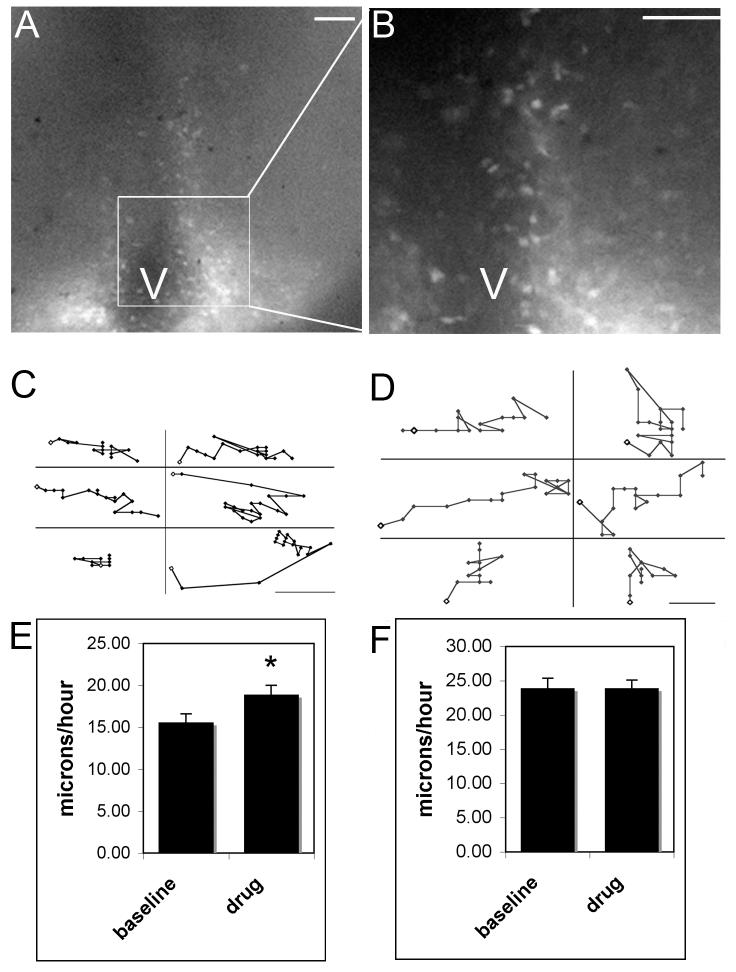
Previous experiments have shown the effect of the GABAB receptor agonist, baclofen, on the movement of neurons randomly labeled with the lipophilic carbocyanine dye DiI in the region of the VMN (Davis et al., 2002). For the current study transgenic Thy-1/YFP mice were used as a tool to visualize cell movements in the region of the VMN and ARC. Neurons expressing YFP are found in the embryonic hypothalamus in numbers that make it possible to visualize distinct cells in the VMN and ARC for the purpose of tracking and motion analysis. The subpopulation of ARC and VMN neurons that are YFP-positive was stable for number and location of cells across individuals in slices placed in vitro at E14. Transgenic Thy-1/YFP mice were used to determine the effect of GABA receptor antagonists on the movement characteristics of fluorescently labeled VMN and ARC neurons. YFP expression driven by the Thy-1 promoter was best seen in the more sub-ventricular and medial populations of VMN and ARC neurons in the 250μm thick slices of the current study (Fig. 1A; higher magnification Fig. 1B). Numbers of YFP positive neurons seen in 20 slices from 16 litters averaged 23 +/- 2.4 neurons (VMN) and 19 +/- 4.6 neurons (ARC). The field of view for watching movement in 250μm thick slices did not include the entire VMN or ARC regions; therefore there is no total count for all fluorescently labeled cells in the regions of interest. There are also limitations in our ability to see YFP-positive neurons due to the thickness of the slices and the short exposure time (30ms) used to prevent photobleaching. Movement patterns of the neurons that could be followed were similar to patterns observed previously in a related mouse line (C57Bl/6J) with DiI labeled neurons (Dellovade et al., 2001); cells moved along dorsal/ventral and radial orientations throughout the developing VMN. Pharmacological manipulation of GABA receptors, either through administration of bicuculline (GABAA receptor antagonist), or saclofen (GABAB receptor antagonist), increased the rate of cell movement by approximately 20% (F1,48 = 4.07, p < 0.05; Fig. 1E). There were no differences seen between the two drug treatment groups or with the location of YFP cells in the nucleus. Sample tracings of individual cells illustrate the distance traveled based on the change in speed before (Fig. 1C), and after (Fig. 1D) drug treatment. For comparison, cells in the ARC region were also analyzed for average movement speeds before and after drug treatment. Moving neurons in this region did not respond to the addition of either bicuculline or saclofen (Fig. 1F). There was a significant difference in the baseline speed between neurons moving in the region of the ARC and VMN. YFP-positive cells in the ARC moved about 30% faster than those in the VMN (F1,64= 5.45, p < 0.02). The addition of saclofen following bicuculline or bicuculline following saclofen to the slices did not have any additional effects on the rate of motion of cells in the VMN or ARC (data not shown). To control for non-specificity, a cannabinoid receptor -1 agonist (WIN-55, 100μM; Sigma-Aldrich) was also administered to live slices. The CB-1 receptor mRNA is highly expressed in the VMN of the developing rat (Berrendero et al., 1998). There were no changes in the movement direction or speed of neurons exposed to this drug (data not shown). To determine if the movement of YFP neurons was affected by the direct response of GABA on these neurons, we looked for the presence of GABA receptors on YFP-positive neurons. YFP-positive neurons in E15 sections were surrounded by a network of immunoreactive GABAB (Fig. 2C) and GABAA (Fig. 2F and I) receptor subunits in the VMN. GABA receptors are expressed predominantly in non-nuclear cellular compartments while YFP primarily is a cytoplasmic marker (YFP expression is also seen in the nuclear regions of neurons likely due to leakage of the fluorescent protein). White arrows indicate examples where distinct YFP neurons were surrounded by GABA receptor expression.
Figure 1.
Brain slices 250μm thick were used for the collection of video microscopy data indicating an increase in neuronal movement in the region of the developing VMN but not the ARC in response to GABA antagonists. The entire region of the hypothalamus that includes the VMN and ARC (A) has fluorescently labeled cells located near the ventricles (V). The boxed region represents the region magnified in (B; scale bars = 100μm). Migrating neurons did not follow strictly radial or dorsal/ ventral movement patterns, rather they changed directions and speeds numerous times within any given time period. Scatter plots for VMN (C) and ARC (D) cells represent sample neurons tracked over 90 minute time periods (scale bars = 5μm) with baseline shown on the left and drug response on the right of each panel. Movement did not occur between every frame and some cells appear to have moved more or farther than others. In the VMN (C,E) the baseline speeds were significantly slower than those observed following drug treatment (10μM bicuculline or 10μM-100μM saclofen; data combined and labeled as drug due to both drugs eliciting the same response). There was no change in the average rate of motion for neurons in the region of the ARC following GABAergic manipulation (F). The scales are different between panels E and F and ARC neurons moved notably faster than those in the region of the VMN.
Figure 2.
Dual label images of Thy-1 driven immunoreactive YFP in neurons and GABA receptor subunits. Immunoreactive GABAB receptor subunit R1 (A), and GABAA receptor subunits α1 (D) and β3 (G) were found in both VMN and ARC (not shown) at E15. YFP containing neurons were surrounded by networks of GABA receptor subunit expression (C, F, I). White arrows indicate example cells that appear to show colocalization. For GABABR1 and GABAAβ3 subunits, immunoreactivity within the VMH was apparent in virtually all cells in non-nuclear compartment(s) whereas immunoreactive GABAAα1 subunits were found in a subset of cells (also in non-nuclear cellular compartments). Images were taken on a confocal microscope with a 1.6μm optical slice (scale bars = 50μm).
The distribution of lateral ERα immunoreactive neurons was changed in the GABABR1 knockout while more centrally located cell types (containing immunoreactive SF-1 or BDNF mRNA) were unchanged
To test the hypothesis that alterations in GABA signaling in development affect final cell positioning, we examined brain sections from wild-type and GABAB receptor knockout mice for changes in immunochemically defined cell populations. We looked at the distribution of cells containing immunoreactive ERα throughout the VMN. They were densely located in the most ventrolateral region of the VMN at three different developmental ages - E17, P0, and P4. ERα immunoreactivity in the GABABR1 knockout at P0 appeared to shift dorsal to the expression pattern seen in the wild-type (Fig. 3C and D; genotype × location, F(10,90)= 3.61, p < 0.01). Although cells were located in more dorsal positions, there was no change in the amount of total immunoreactive area [WT- 30,718 +/- 1568 mean pixels2, KO- 23,954 +/- 1271 mean pixels2; (p > 0.10)]; the mean total area (mean pixels2) of ERα expression remained the same. The change in position that became evident at P0 remained at P4; we saw the same effect, with a more dorsal shift of ERα positive cells in the knockout as compared to the wild-type (Fig. 3E and F) with no difference in the total immunoreactive area [WT- 29,785 +/- 3765 mean pixels2, KO- 25,437 +/- 2974 mean pixels2]. At E17, we saw a pattern of immunoreactivity that looked similar to that seen at P0 and P4, however, the difference between genotypes was not statistically significant at this age (Fig. 3A and B) and the total immunoreactive area was unchanged [WT- 27,091 +/- 3836 mean pixels2, KO- 25,559 +/- 5299 mean pixels2]. Figure 4 shows a graphical representation of the dorsal shift seen in immunoreactive ERα. There is no significant difference in any of the rows at E17 (Fig. 4A). At P0, the largest significant difference in ERα immunoreactivity is between 100 and 200μm from the base of the brain (Fig. 4B). A similar pattern is seen at P4 (Fig. 4C), with the largest significant difference between 200μm and 300μm from the base of the brain. There were no positional changes (medial/lateral or dorsal/ventral) based on ERα immunoreactive cells in the ARC at any age examined. We also measured the mean total area of ERα in the region of the ARC and found no difference between wild type and GABABR1 knockout animals at any of the three ages [E17; WT- 30,101 +/- 894 mean pixels2, KO- 28,444 +/- 1420 mean pixels2, P0; WT- 30,170 +/- 3299 mean pixels2, KO- 32,570, +/- 5612 mean pixels2, P4; WT- 38,020 +/- 3611 mean pixels2, KO- 41,267 +/- 3660 mean pixels2 (p > 0.10)].
Figure 3.
Images show immunoreactive ERα in sections through the center of the VMN examined at 3 different developmental ages (E17, P0, and P4) in wild-type (A, C, E) and GABABR1 knockout (B, D, F) mice revealed a significant difference in the distribution of ERα positive cells in the ventrolateral quadrant of the VMN. At E17 there was no difference in the position of ERα immunoreactive cells (A and B). At P0 (C and D) and P4 (E and F) there was a more dorsal shift of cells containing immunoreactive ERα in the knockout animal. There was no significant change in ERα expression in the ARC.
Figure 4.
Graphic representation of the area of ERα immunoreactivity in GABABR1 wild-type and knockout mice at 3 developmental ages. Bar graphs illustrate the immunoreactivity seen in each row based on a grid system with the first row along the base of the brain (n=6 for each genotype at each age). At P0 (B), the peak of immunoreactivity in the wild-type animal is between 150 and 200μm from the base of the brain and the peak of immunoreactivity in the KO animal is between 250 and 300μm from the base of the brain (scale bars = 100μm). At P4 (C), the peak of immunoreactivity in the wild-type is around 250μm from the base of the brain while the peak in immunoreactivity for the knockout animal ranged from 300-400μm from the base of the brain. There were significant differences between the wild-type and knockout animals at these two ages (genotype × position interaction, p<0.05). There were no significant differences at E17 (A).
To determine if GABA signaling through the GABAB receptor affects all cell types within the VMN, we also examined changes in the position (using the grid system as described above) or expression (total immunoreactive area) of neurons expressing immunoreactive SF-1 protein or BDNF mRNA (by in situ hybridization). SF-1 and BDNF are localized in cells of the more dorsal and central regions of the VMN (Tran et al., 2006). In the GABABR1 knockout the expression of these two markers was unchanged in both amount and position indicating that not all phenotypically identified cells of the VMN have altered positions in the GABABR1 knockout (Fig. 5).
Figure 5.

BDNF and SF-1 expression in the VMN did not change in the GABABR1 knockout mouse at P0 (scale bars = 100μm). Wild-type (A, D) and GABABR1 knockout (B, E) mice were examined for changes in expression of BDNF mRNA (A-C) and immunoreactive SF-1 protein (D-F) located in the central and dorsomedial regions of the VMN. There were no apparent changes in total expression levels or the distribution of cells containing either marker.
To determine if the change in position seen in ERα positive cells in the ventrolateral quadrant of the VMN (vlVMN) of GABABR1 knockout mice was due to a disruption in radial fibers, sections containing regions of the VMN and ARC were processed for Thy-1 YFP (green) immunoreactivity and RC2 immunoreactive radial fibers (red). Tissue sections were taken from a mouse line generated by crossing the GABABR1 heterozygote with a Thy-1/YFP mouse resulting in mice that were knockouts, heterozygotes, or wild-type for the GABABR1 gene and contained YFP expressing Thy-1 neurons. There were no apparent alterations of radial fibers in the GABABR1 knockout mouse (Fig. 6B) as compared to those in wild-type littermates (Fig. 6A). The presence of intact radial fibers suggests that the altered positions of ERα immunoreactive neurons in GABABR1 knockout mice was not due to the lack of or the severing of fibers that guide neuronal migration.
Figure 6.
Dual label images show immunoreactive Thy1/YFP (green) and RC2 (red) in the GABABR1 wild-type (A) and knockout (B) mouse. Radial fibers were intact in the knockout mouse suggesting that altered neuronal movement seen in Thy1/YFP-positive neurons was not due to severed or misdirected radial fibers (scale bars = 100μm; standard epifluorescence microscopy). GABABR1 (C) and ERα (D) appear to localize in the same cells (example at white arrow). As GABABR1 is a receptor found on the cell surface and ERα is a nuclear receptor it is difficult to conclude definitive colocalization. Images C, D, and E were taken using a confocal microscope with an optical slice of 1.6μm.
A small subset of Thy-1 YFP-positive neurons express islet-1 and ERα in the VMN and ARC
To determine if neurons containing YFP also expressed ERα, we utilized tissue sections from E15 Thy-1/YFP mice to look for colocalization. In the VMN (Fig. 7C) we found 3.3% +/-1.2 (n = 4) of YFP neurons colocalized with immunoreactive nuclear ERα. In the ARC (Fig. 7F), there was more colocalization of ERα and YFP (6.7% +/- 2.4, n = 3), which may be due to the abundance of YFP neurons in the ARC at this age. ERα and islet-1 immunoreactive neurons localize to the ventrolateral quadrant of the VMN where most islet-1 neurons are also ERα immunoreactive (Davis et al., 2004b). There was no significant difference in the amount of YFP-positive neurons that colocalized with islet-1 versus ERα in the VMN (7.9% +/- 2.9) or the ARC (13.9% +/- 7.5) (white arrows in Fig. 7I). Despite the low levels of YFP-ERα and YFP-islet-1 colocalization at E15, it is still possible that a number of the YFP neurons viewed in live video microscopy become estrogen receptor positive at later points in development. However, at the time points examined, they are not estrogen receptor positive and may represent other cell phenotypes within the VMN and ARC.
Figure 7.
Images show a subset of neurons co-localize immunoreactive Thy-1/YFP with islet-1 or ERα. The ventrolateral VMN contains a small population of ERα immunoreactive neurons at E15 (A) while the ARC contains a larger population of ERα cells (D). For the most part these cells co-localize (white arrows) in only a small number Thy/YFP neurons in the VMN (C) and ARC (F). Islet-1 is also expressed at developmental ages in the ventrolateral VMN and ARC. At E17 there was a subset of cells containing immunoreactive islet-1 (G) that were also Thy-1/YFP-positive (H, I). White arrows indicate examples of cells that colabel with YFP (scale bars = 50μm).
Discussion
The diencephalon can be parceled into major regions based on transcription factor expression (Puelles and Rubenstein, 2003, Lim and Golden, 2007) and some information is emerging relative to expression of transcription factors within the hypothalamus (Caqueret et al., 2006). Much less, however, is known concerning the details by which specific nuclear groups emerge from early transcriptional coding (McClellan et al., 2006, Xu and Fan, 2007). The VMN presents a unique opportunity to study the formation of a group of cells with a defined involvement in various aspects of neuroendocrine physiology and behavior. Previous studies have described the expression of the transcription factor SF-1 restricted within cells of the VMN while GABAergic elements surround the developing nucleus, providing a potential source of spatial patterning (reviewed McClellan et al., 2006). The results of the current study emphasize GABA’s role in VMN development in contrast to the neighboring ARC and the role of GABAB receptors in VMN development. Figure 8 presents a model illustrating VMN development with and without alterations in GABAA, GABAB, or SF-1 function. To simplify this complicated system, the model focuses on the positioning of cells containing immunoreactive ERα and the movement characteristics of migrating neurons. As illustrated (Fig. 8B), loss of SF-1 causes the largest shift in cell locations with cells containing immunoreactive ERα becoming located more medial (Dellovade et al., 2000, Majdic et al., 2002), in concert with a more medial pattern of immunoreactive glutamic acid decarboxylase (GAD) and GABA (Dellovade et al., 2000). By contrast loss of sensitivity to GABA, either by losing GABAA receptor function (Dellovade et al., 2001) or GABAB receptor function (current study) results in changes at the lateral edges of the VMN (Fig. 8C, D). The actions of GABA cannot explain all of VMN formation (e.g., no influence of GABA receptor modifications on medial and centrally located cells containing immunoreactive SF-1; Fig. 5 and Fig. 8). As an effective signaling molecule, however, it likely plays a key role in fine-tuning the positions of some VMN neurons and ultimately their connectivity.
Figure 8.

In “normal” VMN development (A) cells migrate laterally, away from the third ventricle. Based upon the cell phenotype, VMN neurons occupy different subdivisions of the nucleus. As illustrated, SF-1 containing neurons occupy the dorsal and central regions of the nucleus while ERα containing neurons occupy the ventrolateral quadrant. Most cell movement throughout the nucleus is radial but there is some dorsal ventral movement, therefore ERα neurons may move in both directions before ending up in their final positions. Dotted arrows indicate movement speed of neurons migrating away from the third ventricle. Based on video microscopy data we know that VMN neurons moving away from the third ventricle move at an average rate of 15μm/hour. When SF-1 signaling is absent there is a medial shift in the positioning of ERα immunoreactive cells coincident with a medial shift of GABAergic neurons and fibers (Dellovade et al., 2000). Panels C and D illustrate changes when GABA signaling is inhibited. When GABAA signaling is reduced (C) there is an increase in the overall size of the VMN (Dellovade et al., 2001), ERα immunoreactivity in the ventrolateral quadrant is spread out and shifted slightly more dorsal. Based on data in the current study a lack of GABAB receptor signaling (D) does not change SF-1 expression throughout the dorsal and central VMN but does result in changes in the ventrolateral ERα population.
The results of the current study provide evidence that endogenous GABA signaling affects the rates of cell motion in the region of the developing VMN, but does not alter the rate of motion in the ARC. GABA receptor antagonist administration led to an increase in the rate of motion in cells of the VMN suggesting that normal GABA signaling decreases or inhibits cell motion. These results were seen in fluorescently labeled Thy-1/YFP cells, located near the ventricular zone, which represent a subpopulation of VMN neurons. At E14/15, the ages used for live video microscopy (Figure 1) and dual label immunocytochemistry (Figure 2), the VMN and ARC are becoming distinct nuclear regions through the movement, positioning, and differentiation of various cell types. There is little evidence that these cells within the VMN, or those that will project to the VMN have begun to extend processes. Evidence of VMN projections had been described at 1 week postnatal with processes projecting from the amygdala to the VMN (Choi et al., 2005). Early signs of dendritic development in the rat VMN were also seen as primarily postnatal (Crandall et al, 1989). An alteration in migratory responses could be related to alterations in connectivity as suggested in discussions of roles for Reelin in hippocampal development (Del Rio et al., 1997). Determining influences on connectivity is a topic that needs to be addressed based on the current results.
The mechanism(s) through which GABA may act as a migratory cue involves direct ligand/ receptor interactions that ultimately influence cells to change rates of motion. The administration of GABA receptor antagonists increased the average speeds of neurons that were already moving in the slice preparation (motion when all frames averaged), but did not alter the likelihood of cell movement (percent of frames with motion). GABA can act through two different receptor subtypes that utilize two different mechanisms, but in the current results, they resulted in the same effect on cell motions. GABAB receptors are metabotropic G-protein coupled receptors (Gi/o) that activate cGMP-signaling pathways, activating potassium channels and altering calcium currents, changing the calcium concentration within cells (Bowery and Brown, 1997). These receptors are different in both structure and mechanism from GABAA receptors, which signal through the movement of chloride along its concentration gradient, changing the membrane potential of cells. However, since chloride moves outward in embryonic neurons causing depolarization and potential calcium entry (Gao and van den Pol, 2000), one possible common mechanism through which GABA may be influencing cell speed is through changes in intracellular calcium levels. The location of GABAA and GABAB receptors on the same cells in hypothalamic neurons has been shown to influence the behavior of the neurons. GABAB receptor mediated inhibition has been shown to affect GABAA receptor mediated calcium spikes in embryonic hypothalamic neurons (Obrietan and van den Pol, 1998). In the current study, there was no additive effect of sequential addition of GABAA and GABAB antagonists further suggesting that the influence on motion was through a common mechanism.
The finding that cell motions are influenced by GABA signaling leads to the hypothesis that GABA signaling plays a role in determining cell positions within the VMN. GABA receptor antagonists influenced movements independent of the location within the developing nucleus. Thy-1/YFP labeled neurons examined in our live video microscopy studies move in two distinct directions; dorsal/ventral parallel to the third ventricle, and laterally along radial glial fibers. The majority of the fluorescently labeled cells examined in this movement analysis were located close to the third ventricle. When the data was analyzed separately for location (along the third ventricle or slightly more lateral) there were no differences in movement speeds between locations or in the orientation of the cells (data not shown). As the majority of VMN neurons express specific subunits of the GABAA receptor and/ or the GABAB receptor (Dellovade et al., 2001, Davis et al., 2002), and the response to drug administration was relatively rapid, it is likely that the drug influence was direct to the moving cells. The specific subpopulation of YFP neurons being tracked for motion appears to be neurons that contain GABA receptors themselves. GABAA and GABAB subunit immunoreactivity appears to be in YFP containing neurons. It is difficult to determine if these are the same cells due to the pattern of immunoreactivity with GABA receptors apparent in non-nuclear cellular compartments and YFP being largely cytoplasmic. Cell movement in the ARC was measured before and after GABA receptor manipulation. Although the ARC also contains neurons expressing GABA receptors (Dellovade et al., 2001), cells in this region did not respond to GABA antagonists. This may be caused by a desensitization to GABA as many neurons in the ARC either synthesize GABA themselves or are surrounded by fibers and neurons that synthesize GABA (Gonzalez-Maeso et al., 2003).
The results of the present study expand on the role GABAB receptors play in the development of the VMN (Davis et al., 2002) by defining changes in VMN cell positions in mice with disruption of the GABAB R1 subunit. The results show significant changes in the positions of cells containing immunoreactive ERα in the ventrolateral VMN in the absence of functional GABAB receptors (Figs. 3, 4, 8D). The R1 subunit of the GABAB receptor contains the ligand binding domain for the GABAB receptor, therefore, the disruption of this subunit globally effects the function of this receptor and its ability to bind GABA (Jones et al., 1998). The pharmacological effects of GABAB antagonism (increased motion; Fig. 1) on the movement of Thy1/YFP cells in vitro is a logical candidate as a mechanism for cell displacement in the GABAB R1 subunit knockout mice. Although immunoreactive ERα and islet-1 was seen in few YFP containing neurons at E15, Thy-1/YFP neurons were found in the ventrolateral quadrant of the VMN. The matching of YFP fluorescence to identified lateral VMN components was incomplete at the ages that motion was examined due to either low expression or antisera ability to detect immunoreactive ERα. The islet-1 immunoreactive cell population at E15 represented a greater population of ventrolateral neurons and therefore more colocalization with YFP may have been due to an increase in the number of islet-1 immunoreactive cells in the ventrolateral VMN. Nonetheless, it is likely that neurons that express ERα are influenced by the surrounding gradient of GABA based on several lines of evidence. First, a previous study showed a more widespread localization of cells containing immunoreactive ERα in mice lacking the β3 subunit of the GABAA receptor (Dellovade et al., 2001; Fig. 8C). Genetic disruption of this subunit has been shown to decrease GABAA signaling by up to 50% (Homanics et al., 1997). Second, in SF-1 knockout mice there is a massive reorganization of the region of the VMN and one of the first things altered is the pattern of GABAergic elements (Dellovade et al., 2000; Fig. 8B). Finally, in the current study, genetic disruption of the R1 subunit of the GABAB receptor resulted in a dorsal shift in cells containing immunoreactive ERα (Fig. 8D). More studies are needed to determine other subgroups of cells that co-label with YFP and that may be influenced by secreted signaling molecules like GABA. Interestingly, cells containing BDNF mRNA or immunoreactive SF-1 that are localized to the dorsomedial and central regions of the VMN were not influenced by the absence of functional GABAB receptors.
The response to antagonist in the current study is further evidence that cells in the region of the VMN respond to endogenous GABAergic signals. Previous work showed that immunoreactive GAD continues to encircle the developing VMN in brain slices up to 3 days in vitro (Tobet et al., 1999). Transcription factors (e.g., SF-1) play key roles in determining pattern formation in the developing nervous system. However, transcription factors do not by themselves signal between cells in development; effector molecules like GABA may play key roles in translating nuclear decisions (e.g., glutamic acid decarboxylase transcription) into cellular behaviors (Edelmann et al., 2007). In the embryonic mouse brain, GABA is found in a number of regions, where it may play role(s) unlike its more widely studied function in adult brain. In development, GABA has been shown to be excitatory (Obrietan and van den Pol, 1995), as well as influencing processes such as neuronal proliferation and migration (Behar et al., 1998). The large amounts of GABA that are synthesized along with the lack of developed synapses lead to the belief that the role GABA plays as a paracrine factor during development of the hypothalamus is unlike its function at the synapse (Taylor et al., 1990, Gao and van den Pol, 2000, Manent and Represa, 2007). GABA receptors are expressed within neurons as early as E13 and are likely capable of responding to GABA that may be secreted at the boundaries of the VMN at these early ages.
In summary, the current study has taken two independent lines of evidence to converge on the potential role of GABAergic stimulation on hypothalamic nuclear development. In vitro evidence showed that the manipulation of GABA receptors altered the motion of a specific subpopulation of neurons within the VMN but not the ARC. This result provides evidence that GABAergic stimulation is relevant to specific regions in the hypothalamus (VMN) while other regions, for reasons yet to be determined, are not responsive. The same result (an increase in motion) was seen upon the addition of either GABAA or GABAB antagonists leading to the conclusion that although the initial mechanisms may differ, GABA action through either receptor may ultimately invoke a common mechanism. In vivo evidence using mice in which the GABAB receptor was disrupted showed that mice lacking this receptor in development exhibited phenotypic changes within the hypothalamus; including, changes in the position of cells in the VMN suggesting significant long term consequences of these developmental actions. These lines of evidence emphasize the importance of GABA and GABA receptors in the development of specific regions or nuclei within the hypothalamus. Further experiments are needed to determine if altered movement eventually leads to altered functional characteristics or synaptic connections within the developing brain.
Supplementary Material
Supplemental Figure - Immunoreactive GABAAα 1 was found in the mouse brain at E15 (A, B, C) and E17 (D and E). The GABAAα 1 affinity purified polyclonal antibody 812-GA1N obtained from Phosphosolutions Inc. (Aurora, CO) labeled cells in the ventrolateral VMN and ARC at E15 (A, high magnification B). Panel C is an adjacent section with antibody buffer instead of primary antibody included in the reaction. At E17, the 812-GA1N antibody (D) again showed immunoreactive GABAAα 1 in the VMN and this pattern was similar to that produced by the GABAAα 1 affinity purified polyclonal antibody AGA-001 made by Alomone Labs (Jerusalem, Israel) (E).
Acknowledgements
This work was supported by NIH grants MH57748 and MH61376 (SAT) and NIH training grant HD07031-29 (KM). We thank J. Gabe Knoll and Dr. Chad Foradori for technical assistance and Brandon Wadas and Cheryl Hartshorn for editing assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Behar TN, Li YX, Tran HT, Ma W, Dunlap V, Scott C, Barker JL. GABA stimulates chemotaxis and chemokinesis of embryonic cortical neurons via calcium-dependent mechanisms. J Neurosci. 1996;16:1808–1818. doi: 10.1523/JNEUROSCI.16-05-01808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar TN, Schaffner AE, Scott CA, O’Connell C, Barker JL. Differential response of cortical plate and ventricular zone cells to GABA as a migration stimulus. J Neurosci. 1998;18:6378–6387. doi: 10.1523/JNEUROSCI.18-16-06378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrendero F, Garcia-Gil L, Hernandez ML, Romero J, Cebeira M, de Miguel R, Ramos JA, Fernandez-Ruiz JJ. Localization of mRNA expression and activation of signal transduction mechanisms for cannabinoid receptor in rat brain during fetal development. Development. 1998;125:3179–3188. doi: 10.1242/dev.125.16.3179. [DOI] [PubMed] [Google Scholar]
- Bless EP, Walker HJ, Yu KW, Knoll JG, Moenter SM, Schwarting GA, Tobet SA. Live view of gonadotropin-releasing hormone containing neuron migration. Endocrinology. 2005;146:463–468. doi: 10.1210/en.2004-0838. [DOI] [PubMed] [Google Scholar]
- Bowery NG, Brown DA. The cloning of GABA(B) receptors. Nature. 1997;386:223–224. doi: 10.1038/386223a0. [DOI] [PubMed] [Google Scholar]
- Canteras NS, Simerly RB, Swanson LW. Organization of projections from the ventromedial nucleus of the hypothalamus: a Phaseolus vulgaris-leucoagglutinin study in the rat. J Comp Neurol. 1994;348:41–79. doi: 10.1002/cne.903480103. [DOI] [PubMed] [Google Scholar]
- Caqueret A, Boucher F, Michaud JL. Laminar organization of the early developing anterior hypothalamus. Dev Biol. 2006;298:95–106. doi: 10.1016/j.ydbio.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Chanas-Sacre G, Thiry M, Pirard S, Rogister B, Moonen G, Mbebi C, Verdiere-Sahuque M, Leprince P. A 295-kDA intermediate filament-associated protein in radial glia and developing muscle cells in vivo and in vitro. Dev Dyn. 2000;219:514–525. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1078>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Davis AM, Henion TR, Tobet SA. Gamma-aminobutyric acidB receptors and the development of the ventromedial nucleus of the hypothalamus. J Comp Neurol. 2002;449:270–280. doi: 10.1002/cne.10293. [DOI] [PubMed] [Google Scholar]
- Davis AM, Seney ML, Stallings N, Zhao L, Parker KL, Tobet SA. Loss of steroidogenic factor 1 alters cellular topograpy in the mouse ventromedial nucleus of the hypothalamus. J Neurobiol. 2004a;60:424–436. doi: 10.1002/neu.20030. [DOI] [PubMed] [Google Scholar]
- Davis AM, Seney ML, Walker HJ, Tobet SA. Differential colocalization of Islet-1 and estrogen receptor alpha in the murine preoptic area and hypothalamus during development. Endocrinology. 2004b;145:360–366. doi: 10.1210/en.2003-0996. [DOI] [PubMed] [Google Scholar]
- De Block M, Debrouwer D. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal Biochem. 1993;215:86–89. doi: 10.1006/abio.1993.1558. [DOI] [PubMed] [Google Scholar]
- Del Rio JA, Heimrich B, Borrell V, Forster E, Drakew A, Alcantara S, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Derer P, Frotscher M, Soriano E. A role for Cajal-Retzius cells and reelin in the development of hippocampal connections. Nature. 1997;385:70–74. doi: 10.1038/385070a0. [DOI] [PubMed] [Google Scholar]
- Dellovade TL, Davis AM, Ferguson C, Sieghart W, Homanics GE, Tobet SA. GABA influences the development of the ventromedial nucleus of the hypothalamus. J Neurobiol. 2001;49:264–276. doi: 10.1002/neu.10011. [DOI] [PubMed] [Google Scholar]
- Dellovade TL, Young M, Ross EP, Henderson R, Caron K, Parker K, Tobet SA. Disruption of the gene encoding SF-1 alters the distribution of hypothalamic neuronal phenotypes. J Comp Neurol. 2000;423:579–589. doi: 10.1002/1096-9861(20000807)423:4<579::aid-cne4>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Dielenberg RA, McGregor IS. Defensive behavior in rats towards predatory odors: a review. Neurosci Biobehav Rev. 2001;25:597–609. doi: 10.1016/s0149-7634(01)00044-6. [DOI] [PubMed] [Google Scholar]
- Edelmann M, Wolfe CA, Scordalakes EM, Rissman EF, Tobet S. Neuronal Nitric Oxide Synthase and Calbindin Delineate Sex Differences in the Develoing Hypothalamus and Preoptic Area. Dev Neurobiol. 2007 doi: 10.1002/dneu.20507. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Gao XB, van den Pol AN. GABA release from mouse axonal growth cones. J Physiol. 2000;523(Pt 3):629–637. doi: 10.1111/j.1469-7793.2000.t01-1-00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J, Wise A, Green A, Koenig JA. Agonist-induced desensitization and endocytosis of heterodimeric GABAB receptors in CHO-K1 cells. Eur J Pharmacol. 2003;481:15–23. doi: 10.1016/j.ejphar.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Harrison NL, Lovinger DM, Lambert NA, Teyler TJ, Prager R, Ong J, Kerr DI. The actions of 2-hydroxy-saclofen at presynaptic GABAB receptors in the rat hippocampus. Neurosci Lett. 1990;119:272–276. doi: 10.1016/0304-3940(90)90851-y. [DOI] [PubMed] [Google Scholar]
- Homanics GE, DeLorey TM, Firestone LL, Quinlan JJ, Handforth A, Harrison NL, Krasowski MD, Rick CE, Korpi ER, Makela R, Brilliant MH, Hagiwara N, Ferguson C, Snyder K, Olsen RW. Mice devoid of gamma-aminobutyrate type A receptor beta3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proc Natl Acad Sci U S A. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao W-J, Johnson M, Gunwaldsen C, Huang L-Y, Tang C, Shen Q, Salon JA, Morse K, Laz T, Smith KE, Nagarathnam D, Noble SA, Branchek TA, Gerald C. GABAb receptors function as a heteromeric assembly of the subunits GABAbR1 and GABAbR2. Nature. 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, McMaster G, Angst C, Bittiger H, Froestl W, Bettler B. Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239–246. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- King BM. The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav. 2006;87:221–244. doi: 10.1016/j.physbeh.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Knoll JG, Wolfe CA, Tobet SA. Estrogen modulates neuronal movements within the developing preoptic area-anterior hypothalamus. Eur J Neurosci. 2007;26:1091–1099. doi: 10.1111/j.1460-9568.2007.05751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS. The usual suspects: GABA and glutamate may regulate proliferation in the neocortex. Neuron. 1995;15:1223–1225. doi: 10.1016/0896-6273(95)90002-0. [DOI] [PubMed] [Google Scholar]
- Lim Y, Golden JA. Patterning the developing diencephalon. Brain Res Rev. 2007;53:17–26. doi: 10.1016/j.brainresrev.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- Majdic G, Young M, Gomez-Sanchez E, Anderson P, Szczepaniak LS, Dobbins RL, McGarry JD, Parker KL. Knockout mice lacking steroidogenic factor 1 are a novel genetic model of hypothalamic obesity. Endocrinology. 2002;143:607–614. doi: 10.1210/endo.143.2.8652. [DOI] [PubMed] [Google Scholar]
- Manent JB, Represa A. Neurotransmitters and brain maturation: early paracrine actions of GABA and glutamate modulate neuronal migration. Neuroscientist. 2007;13:268–279. doi: 10.1177/1073858406298918. [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Mitrovic I, Riley RC, Jan LY, Basbaum AI. Immunohistochemical localization of GABA(B) receptors in the rat central nervous system. J Comp Neurol. 1999;405:299–321. doi: 10.1002/(sici)1096-9861(19990315)405:3<299::aid-cne2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- McClellan KM, Parker KL, Tobet S. Development of the ventromedial nucleus of the hypothalamus. Front Neuroendocrinol. 2006 doi: 10.1016/j.yfrne.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Obrietan K, van den Pol AN. GABA neurotransmission in the hypothalamus: developmental reversal from Ca2+ elevating to depressing. J Neurosci. 1995;15:5065–5077. doi: 10.1523/JNEUROSCI.15-07-05065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrietan K, van den Pol AN. GABAB receptor-mediated inhibition of GABAA receptor calcium elevations in developing hypothalamic neurons. J Neurophysiol. 1998;79:1360–1370. doi: 10.1152/jn.1998.79.3.1360. [DOI] [PubMed] [Google Scholar]
- Prosser HM, Gill CH, Hirst WD, Grau E, Robbins M, Calver A, Soffin EM, Farmer CE, Lanneau C, Gray J, Schenck E, Warmerdam BS, Clapham C, Reavill C, Rogers DC, Stean T, Upton N, Humphreys K, Randall A, Geppert M, Davies CH, Pangalos MN. Epileptogenesis and enhanced prepulse inhibition in GABA(B1)-deficient mice. Mol Cell Neurosci. 2001;17:1059–1070. doi: 10.1006/mcne.2001.0995. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Saper CB, Swanson LW, Cowan WM. The efferent connections of the ventromedial nucleus of the hypothalamus of the rat. J Comp Neurol. 1976;169:409–442. doi: 10.1002/cne.901690403. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- Sperk G, Schwarzer C, Tsunashima K, Fuchs K, Sieghart W. GABA(A) receptor subunits in the rat hippocampus: I. immunocytochemical distribution of 13 subunits. Neuroscience. 1997;80:987–1000. doi: 10.1016/s0306-4522(97)00146-2. [DOI] [PubMed] [Google Scholar]
- Taylor J, Docherty M, Gordon-Weeks PR. GABAergic growth cones: release of endogenous gamma-aminobutyric acid precedes the expression of synaptic vesicle antigens. J Neurochem. 1990;54:1689–1699. doi: 10.1111/j.1471-4159.1990.tb01223.x. [DOI] [PubMed] [Google Scholar]
- Tobet SA, Chickering TW, King JC, Stopa EG, Kim K, Kuo-Leblank V, Schwarting GA. Expression of gamma-aminobutyric acid and gonadotropin-releasing hormone during neuronal migration through the olfactory system. Endocrinology. 1996;137:5415–5420. doi: 10.1210/endo.137.12.8940365. [DOI] [PubMed] [Google Scholar]
- Tobet SA, Henderson RG, Whiting PJ, Sieghart W. Special relationship of gamma-aminobutyric acid to the ventromedial nucleus of the hypothalamus during embryonic development. J Comp Neurol. 1999;405:88–98. doi: 10.1002/(sici)1096-9861(19990301)405:1<88::aid-cne7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Tobet SA, Walker HJ, Seney ML, Yu KW. Viewing cell movements in the developing neuroendocrine brain. Integr Comp Biol. 2003;43:794–801. doi: 10.1093/icb/43.6.794. [DOI] [PubMed] [Google Scholar]
- Tran PV, Akana SF, Malkovska I, Dallman MF, Parada LF, Ingraham HA. Diminished hypothalamic bdnf expression and impaired VMH function are associated with reduced SF-1 gene dosage. J Comp Neurol. 2006;498:637–648. doi: 10.1002/cne.21070. [DOI] [PubMed] [Google Scholar]
- Tran PV, Lee MB, Marin O, Xu B, Jones KR, Reichardt LF, Rubenstein JR, Ingraham HA. Requirement of the orphan nuclear receptor SF-1 in terminal differentiation of ventromedial hypothalamic neurons. Mol Cell Neurosci. 2003;22:441–453. doi: 10.1016/S1044-7431(03)00027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten M, Brawer JR. Cytology of neurons of the hypothalamic ventromedial nucleus in the adult male rat. J Comp Neurol. 1978;178:89–116. doi: 10.1002/cne.901780106. [DOI] [PubMed] [Google Scholar]
- Xu C, Fan CM. Allocation of paraventricular and supraoptic neurons requires Sim1 function: a role for a Sim1 downstream gene PlexinC1. Mol Endocrinol. 2007;21:1234–1245. doi: 10.1210/me.2007-0034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure - Immunoreactive GABAAα 1 was found in the mouse brain at E15 (A, B, C) and E17 (D and E). The GABAAα 1 affinity purified polyclonal antibody 812-GA1N obtained from Phosphosolutions Inc. (Aurora, CO) labeled cells in the ventrolateral VMN and ARC at E15 (A, high magnification B). Panel C is an adjacent section with antibody buffer instead of primary antibody included in the reaction. At E17, the 812-GA1N antibody (D) again showed immunoreactive GABAAα 1 in the VMN and this pattern was similar to that produced by the GABAAα 1 affinity purified polyclonal antibody AGA-001 made by Alomone Labs (Jerusalem, Israel) (E).