Abstract
The general amino acid permease (Gap1p) of Saccharomyces cerevisiae is a broad range, low affinity permease that imports amino acids in cells growing on poor nitrogen sources. This permease also signals the presence of amino acids through the fermentable growth medium pathway allowing the cell to respond to new sources of nitrogen in the surrounding medium. Yeast with an activated Ras2/cAMP pathway show many phenotypes indicative of altered nitrogen uptake and metabolism; sensitivity to nitrogen starvation, low amino acid pools. We have shown that Gap1p activity is lowered in cells with an activated RAS2val19 allele or elevated cAMP levels whereas cells with inactive ras2 allele lose ammonia repression of Gap1p-mediated transport. This regulation is through a post-transcriptional mechanism; transcription of GAP1 is not affected by cAMP level. A mechanism by which the Ras2/cAMP/PKA pathway controls the ubiquitin-dependent degradation of Gap1p is most consistent with the data.
Keywords: GAP1, general amino acid permease, Gap1p, Ras2/cAMP pathway, post-transcriptional control
INTRODUCTION
The mechanisms by which yeast cells detect, and respond to, available nutrients have been studied extensively. There are two major signaling pathways, the Ras/cAMP pathway and the fermentable growth medium pathway, which signal sugar and nitrogen source availability respectively and converge through the regulation of protein kinase A (PKA; Thevelein and de Winde, 1999). The subsequent activity of PKA substrates controls the cell’s readiness for continued growth in adequate nutrient conditions or its transition to stationary phase in response to starvation (Broach and Deschenes, 1990).
Yeast contain a series of transport molecules responsible for the uptake of nitrogen sources from the medium (André, 1995) some of which also act as receptors signaling the availability of their substrate to the interior of the cell (Holsbeeks et al., 2004). In a useful oversimplification of the regulation mechanisms, one can classify growing cells as being either nitrogen-repressed or –derepressed (Magasanik and Kaiser, 2002). Nitrogen-repressed cells have a ‘good’ nitrogen source available, which can be imported through specific permeases (Mep1 and Mep2 – ammonia, Gnp1 – glutamine). Alternatively, there are permeases that are induced at the transcriptional level by the presence of external substrates (Nielson et al., 2001) that can take up an amino acid either as a nitrogen source for growth or as an additional nutrient to supplement an auxotrophy. These can be either broad-range (e.g.Agp1) or group-specific (e.g.Bap2 – branched chain amino acids) permeases. In contrast, cells growing on a derepressing nitrogen source like proline induce the expression of the general amino acid permease gene (GAP1) as well as the proline specific permease (PUT4). This allows the cell to take up a broad range of amino acids and related compounds as they become available in the medium to the nitrogen-limited cells (Magasanik and Kaiser, 2002).
Yeast cells employ multiple mechanisms to control Gap1 activity in response to different nitrogen sources. Transcription of GAP1 and other nitrogen-repressible genes, is controlled by the GATA type transcription factors (Gln3p, Nil1p, Nil2p, Dal80p) in response to the presence of glutamine or asparagine (Stanbrough and Magasanik, 1995). In de-repressing media, the levels of Gap1p found in the membrane is also regulated by the control of permease trafficking to the vacuole by amino acids. Cells growing on glutamate synthesize Gap1p but the protein is sorted directly to the vacuole and never reaches the plasma membrane (Chen and Kaiser, 2002).
In nutrient conditions where mature permease does localize to the plasma membrane, Gap1p stability, and thus activity, is regulated in multiple ways. One important activation/inactivation mechanism involves the NPR1 and NPI1/RSP5 gene products (Vandenbol et al., 1990). In non-repressing conditions the action of these enzymes results in a steady state activity of the permease, however, once ammonia is added, Gap1p is internalized and degraded by a pathway involving ubiquitination (Springael and André, 1998; Springael et al., 2002). While the molecular mechanism controlling these processes is not understood, one component of the pathway may be Aua1p. aua1 mutants have lost ammonia repression of Gap1p but show no other phenotypes (Sophianopoulou & Diallinas, 1993). Recently, an additional reversible inhibition of Gap1p activity in response to the transport of excess amino acid has been elucidated (Risinger et al., 2006). In this case, it appears that the permease remains within the plasma membrane in an inactive, but unmodified, form.
As the Ras2/cAMP and FGM pathways control yeast cell growth in response to nutrients and permease molecules provide those nutrients, it is not surprising that specific links have been found between these pathways. Certain permeases have been shown to signal the presence of their substrate generating an intracellular signal and response Holsbeeks et al., 2004). For example, Gap1p has been shown to be involved in the regulation of PKA targets in response to the addition of amino acids to the medium (Donaton et al., 2003). As activated Ras2/cAMP pathway mutants show phenotypes suggestive of altered amino acid metabolism and transport, it is of interest to determine if there is feedback control of amino acid transport by Ras2/cAMP/PKA activities. Preliminary studies have suggested that cAMP levels affect amino acid transport (Amitrano et al., 1997; Saenz et al., 1997). In this paper we demonstrate that elevated cAMP levels decrease Gap1p activity and this control is exerted at a post-transcriptional step in Gap1 regulation. Also, that ammonia inactivation of Gap1p requires a functional Ras2p. We suggest that the Ras2/cAMP pathway is involved in regulating the ubiquitin-dependent degradation of Gap1p.
MATERIALS AND METHODS
Yeast strains and plasmids
The strains used in this study are listed in Table 1. The construction of strains HR125Δgap1 and TC41-1Δgap1 was as described previously (Schreve & Garrett, 1997). The plasmids pSE (promoter GAP1-lacz) and pPDE2 (phosphodiesterase 2 over-expression plasmid) were the kind gifts of Mike Stanbrough and Mike Wigler respectively (Stanbrough et al., 1995; Sass et al., 1986). All DNA manipulations in yeast and bacteria were done using standard protocols (Sherman et al., 1986; Sambrook et al., 1989).
Table 1.
Yeast Strains
| Strain | Genotype | Reference |
|---|---|---|
| SP1 | MATa ura3–52 leu2–3,112 his3Δ1 trp1-289 ade8 can1. | Toda et al, 1985. |
| T3-24C | MATα ura3–52 leu2–3,112 his3Δ1 trp1–289 ade8 can1, ras2::URA3 | Toda et al, 1985. |
| TK161-R2V | MATa ura3–52 leu2–3,112 his3Δ1 trp1–289, ade8 can1 RAS2val19 | Toda et al, 1985. |
| Y294 | MATa ura3–52 leu2–3,112 his3Δ1 trp1–289 | Fedor-Chaiken et al, 1990. |
| Y420 | MATα ura3–52 leu2–3,112 his3Δ1 trp1–289 Ras2::GAL10-RAS2val19* | Fedor-Chaiken et al, 1990. |
| Y1020 | MATa ura3–52 leu2–3,112 his3Δ1 trp1–289 RAS2val19-URA3 | Fedor-Chaiken et al, 1990. |
| HR125 | MATa ura3–52 leu2–3,112 his3–532 his4 | Russell et al, 1993. |
| TC41-1 | MATa ura3–52 leu2–3,112 his3–532 his4 Cyr1::URA3 cam | Russell et al, 1993. |
Y420 is a ras2 mutant when grown in glucose-containing media as the GAL10-RAS2val19 construct is not induced.
Growth conditions
Yeast strains were routinely grown in standard yeast media (Sherman et al., 1986; Wickerham, 1946). Minimal medium contained the following supplements: L-histidine (20mg/l), L-leucine (30mg/l), L-tryptophan (20mg/l), L-lysine (30mg/l), adenine (20mg/l) and uracil (20mg/l). For the determination of balanced growth rates on different nitrogen sources, the culture was repeatedly diluted into fresh media during mid-exponential phase and the doubling time determined from at least three growth phases. The amino acid acting as principal nitrogen source was added at 0.01% (w/v) and the supplements were added at 0.1x normal concentration.
Amino Acid Transport and β-galactosidase Assays
L-14C-amino acid uptake was assayed by the method of Woodward and Cirillo (1977), except that cycloheximide was omitted from the buffer. Cells grown in minimal ammonia media were harvested during log phase (OD550 = 0.1 – 0.3; 0.058 – 0.174 mg dry weight/ml), washed with water and resuspended in 10mM potassium phthalate (pH = 5.5), 2% glucose to an OD550 of approximately 0.8. After incubation at 30oC for 30 minutes 14C-labelled amino acid was added. Samples were withdrawn at 30 second intervals, filtered, and washed with 20 ml ice cold water. Filters were placed in vials and 5 ml liquid scintillation fluid was added. Radioactivity was determined on a Wallac LSC. β-galactosidase activity was determined as described previously (Miller, 1972; Schreve et al., 1998).
RESULTS
Yeast strains with elevated RAS/cAMP pathway activity show phenotypes that indicate altered nitrogen metabolism, including sensitivity to starvation and decreased vacuolar basic amino acid pools (Markwardt et al., 1995). The balanced growth rates of wild-type and activated RAS2 mutants were measured in different amino acids as principal nitrogen source. This data (Table 2) shows that strains containing the RAS2val19 allele are differentially impaired for growth on different amino acids suggesting a defect in either transport or utilization of amino acids in these strains. Alteration of amino acid transport activity in RAS2val19 is also indicated by their hypersensitivity to growth in the presence of the amino acid analogs cycloleucine (125 μg/mL), γ-hydroxyglutamate (500 μg/mL) and norleucine (100 μg/mL) (data not shown). Strains in which RAS/cAMP pathway activity is decreased (ras2) grow at the same rate as their parental strain in these conditions (data not shown).
Table 2.
Growth rates of parental and RAS2 strains on various amino acids as principal nitrogen source
| Balanced growth rate (h−1) of
|
||||
|---|---|---|---|---|
| SP1 strains
|
Y294 strains
|
|||
| Nitrogen source | RAS2 | RAS2Val19 | RAS2 | RAS2Val19 |
| −a | 6.5 | 7.5 | 6.0 | 10.5 |
| Ammonia | 2.5 | 5.5 | 3.25 | 7.25 |
| Alanine | 2.5 | NGb | NG | NG |
| Arginine | 2.25 | 3.5 | NG | NG |
| Aspartic acid | 2.0 | 4.5 | 3.0 | NG |
| Glutamate | 3.75 | NG | 3.25 | 7.5 |
| Glutamine | 3.0 | NG | 3.25 | NG |
| Proline | 3.25 | 6.0 | 3.25 | 7.5 |
| Serine | 3.25 | NG | NG | NG |
| Urea | 3.5 | 5.5 | 3.5 | 7.0 |
No additional amino acid added. The amino acids added to supplement the auxotrophic requirements of the cells sustain this growth rate.
NG = no growth above background.
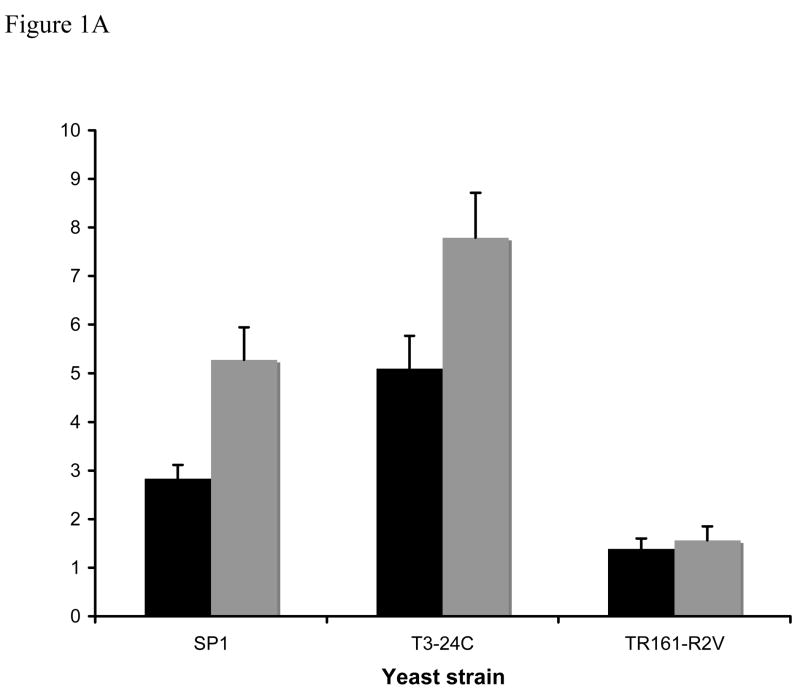
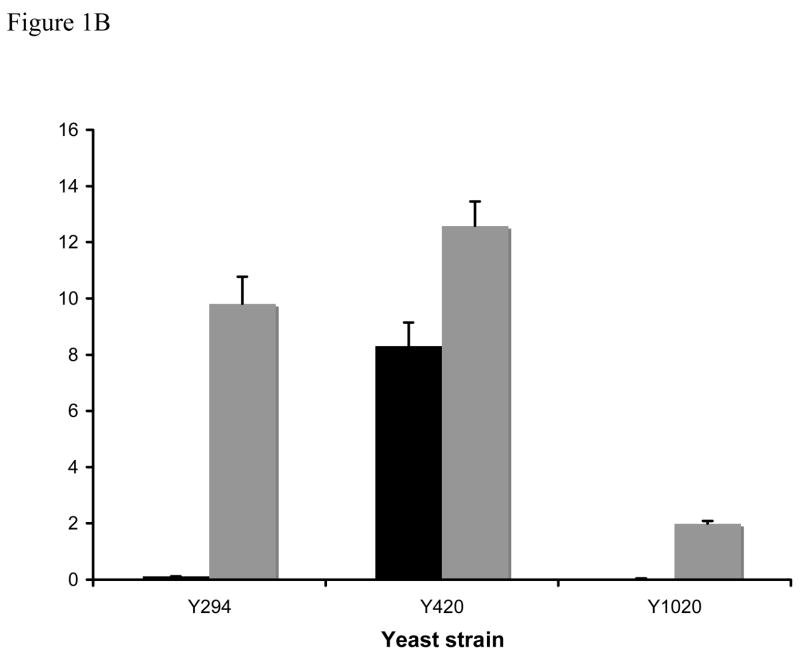
The pleiotropic amino acid-related phenotypes of RAS2val19 strains suggest a general defect in amino acid transport and/or metabolism. We assayed general amino acid permease levels in yeast with normal, elevated and attenuated RAS/cAMP pathway activity in two strain backgrounds. The assays were performed in both repressing (ammonia) and non-repressing (proline) media. In all conditions tested, cells with an overactive RAS/cAMP pathway (TR161-R2V, Y1020) showed lower Gap1 transport as measured by uptake of the Gap1 permease-specific substrate, citrulline than the parental strain (Figure 1). In contrast, ras2 mutants (T3–24C, Y420) showed normal or slightly elevated Gap1-mediated transport on proline media and lost ammonia repression of the transporter in ammonia-grown cells The two strains studied showed different levels of ammonia repression characteristic of S288C-derived (SP1, partial repression) and Σ1278B-derived (Y294, full repression) strains and in both cases the Gap1 transport levels of ammonia-grown ras2 cells equaled that of proline-grown parental strain. Thus we have demonstrated that mutant RAS2 alleles affect Gap1 activity in two ways; a decrease in amino acid transport in RAS2val19 cells that is not specific to nitrogen source and a loss of ammonia repression in ras2 cells.
Figure 1. General amino acid permease activity in yeast RAS2 mutants.
Strains were grown in either minimal ammonia (black bars) or minimal proline (grey bars) medium and assayed for 14C-citrulline uptake (2.0mM) as described in the methods. A. S288C-derived strains. B. Σ1278B-derived strains. Gap1p activity is measured as nmoles 14C-citrulline/min−1/OD550−1.Results are the average of 4–10 independent assays, error bars denote the standard error.
In order to determine if the decreased Gap1 activity in RAS2val19 cells is the result of elevated cAMP levels we assayed 14C-citrulline uptake in cells engineered to be sensitive to external cAMP levels (Russell et al., 1993). The results in Figure 2 clearly demonstrate that Gap1 activity is decreased with increasing cAMP levels in a cAMP-sensitive strain whereas Gap1 activity is not affected in the isogenic wild-type strain. The observed effect of the RAS/cAMP pathway on amino acid transport appears to be confined to regulation of Gap1 activity, we tested three specific amino acid transporters and found no evidence that they are regulated in a similar manner. The transport of amino acids that are good nitrogen sources like glutamate and glutamine is unaffected by the RAS2val19 allele (Table 3). Similarly, uptake of glutamate and glutamine are not affected by external cAMP concentration in the cAMP-sensitive strain (Table 3). Transport of leucine, a poor nitrogen source, is only slightly reduced in the RAS2val19 mutant. Further evidence that the decrease in Gap1 activity is the result of elevated cAMP levels comes from the finding that overexpression of Pde2 (phosphodiesterase that degrades cAMP) on a multicopy plasmid restores Gap1 activity in TR161-R2V to wildtype levels (SP1 = 2.83 +/− 0.28 nmoles 14C-citrulline/min−1/OD600, TR161-R2V = 1.39 +/− 0.31 nmoles 14C-citrulline/min−1/OD600, TR161-R2VpPDE2 = 3.39 +/− 0.68 nmoles 14C-citrulline/min−1/OD600).
Figure 2. The effect of external cAMP concentration on Gap1p activity in a cyr1 cAMP-sensitive strain.
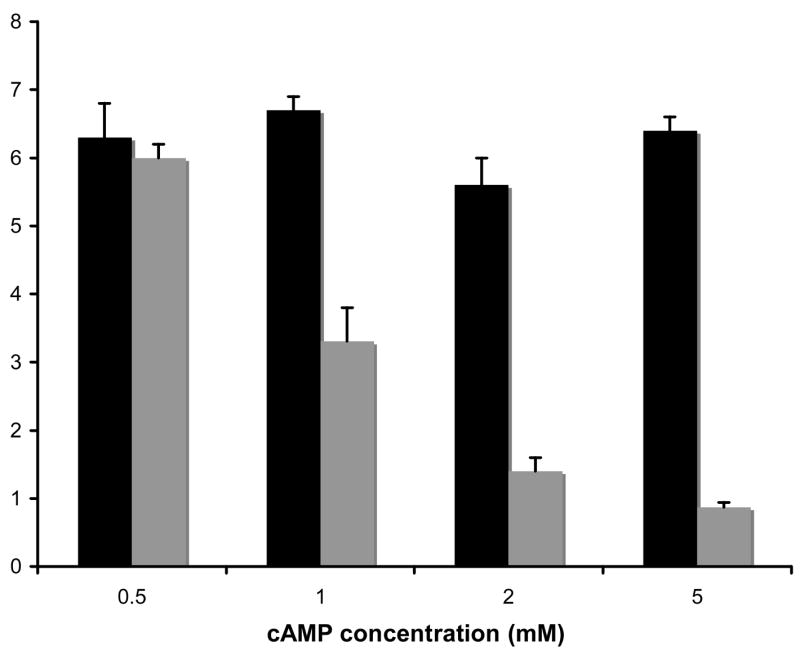
Strains were grown in minimal ammonia medium containing the cAMP concentration given and assayed as described for Fig. 1. Black bars – HR125 (wild-type), grey bars –TC41-1 (cyr1 mutant). Results are the average of three independent assays.
Table 3.
The effect of an activated RAS2 allele or elevated cAMP levels on glutamate, glutamine and leucine transport.
| Strain | Amino acid uptake(nmoles 14C-amino acid/min−1/OD550)
|
||
|---|---|---|---|
| Glutamine | Glutamate | Leucine | |
| SP1 | 8.41 +/− 0.07 | 3.63 +/− 0.18 | 5.30 +/− 0.16 |
| TR161-R2V | 9.30 +/− 0.05 | 3.07 +/− 0.32 | 4.55 +/−0.21 |
| TC41Δgap 0.5mM cAMP | 8.57 +/− 0.43 | 4.53 +/− 0.21 | ND |
| TC41Δgap 2.0mM cAMP | 8.67 +/− 0.34 | 4.23 +/− 0.30 | ND |
Cells were grown on minimal ammonia medium and assayed as described in methods. All assays were performed at least three times.
ND – not determined
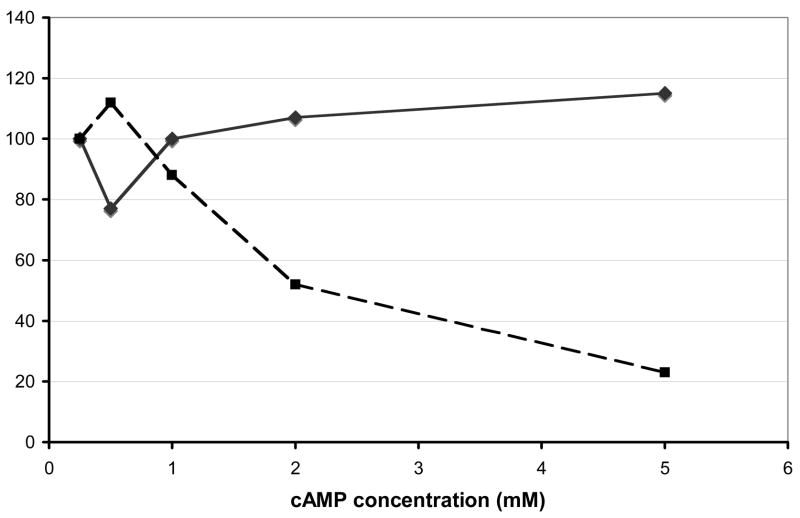
The transport activity of Gap1 has been shown to be regulated in multiple ways both transcriptional and post-transcriptional (Stanbrough and Magasanik, 1995; Springael et al., 2002; Risinger et al., 2006). In order to determine if transcription of GAP1 is affected by cAMP levels we transformed the expression construct pSE (GAP1 promoter fused to lacz) into the cAMP-dependent strain TC41. As shown in Figure 3, while rising cAMP levels caused Gap1 transport activity of cells grown in 5mM cAMP to fall to 20% of that if 0.5mM cAMP grown cells, β-galactosidase levels remained constant. Thus cAMP is not affecting transcription of the GAP1 gene and the loss of activity must be the result of alteration of the activity of a post-transcriptional control mechanism, e.g. permease localization or turnover.
Figure 3. The control of Gap1p activity by cAMP is not through the regulation of transcription.
TC41-1 cells containing the expression plasmid pSE (promoter of GAP1 fused to the E. coli lacz gene) was grown in ammonia medium containing the cAMP levels given. β-galactosidase levels were assayed by the method of Miller (1972). Gap1p activity (filled squares) and β-galactosidase levels (filled triangles, dashed line) are given as a percentage of the level obtained in 0.5mM cAMP where 100% Gap1p activity = 6.2 nmoles 14C-citrulline/min−1/OD550−1 and 100% β-galactosidase = 750 Units.
DISCUSSION
Many of the phenotypes of yeast mutants with an activated Ras2/cAMP pathway are not fully explained by the current model of carbon and nitrogen growth control. It has been shown that amino acid availability is signaled by Gap1p through the fermentable growth pathway to activate protein kinase A resulting in trehalase activation and storage carbohydrate breakdown (Donaton et al., 2003). But many questions remain. Why are activated RAS strains sensitive to nitrogen starvation and unable to accumulate vacuolar amino acids (Markwardt et al., 1995)? Why are they unable to grow on many amino acids as nitrogen source (Table 2) and hypersensitive to some amino acid analogs (data not shown). These phenotypes suggest that amino acid transport and/or metabolism is altered in RAS2val19 strains. They raise the possibility that the Ras2/cAMP/PKA pathway not only assimilates signals of nutrient availability but also provides feedback controls on the acquisition of further nutrients for growth.
There are multiple transporters for most amino acids and they vary in their regulation (André, 1995). The experiments presented here indicate that elevated RAS pathway activity (Fig. 1) or elevated cAMP levels (Fig. 2) decrease the activity of the general amino acid permease and thus impair the ability of the cell to take up a wide range of amino acid substrates. The effect on citrulline uptake is the most marked as Gap1 is the only transporter of this substrate, however a decrease in leucine uptake was also observed consistent with loss of the contribution of Gap1 to uptake of this amino acid. We found no evidence of cAMP control of uptake of the good nitrogen sources glutamate and glutamine.
Previous studies on the effect of cAMP levels on amino acid transport have reached different conclusions. Saenz and colleagues found decreased levels of leucine transport observed in RAS2val19 mutants and concluded these were the result of changes in other permeases (S1 – Bap2p, S2 –Agp1p) and that Gap1p transport was not affected (Saenz et al., 1997). The strains used in that experiment were designated as gap1 mutants because they showed no detectable citrulline transport. However, the initial leucine uptake they observe is three times higher in proline-grown wild-type cells than those grown in ammonia. The difference is leucine uptake is not present in their activated RAS strain indicating that an ammonia-sensitive activity has been lost. This pattern of regulation is typical of an activity subject to ammonia repression like Gap1, not permeases regulated by amino acid induction like Bap2 (S1). While the lack of citrulline transport in their strain does indicate the loss of Gap1 activty, it is also possible that this strain contains a mutant Gap1p permease with an altered active site that does not recognize citrulline but still transports leucine.
Our studies indicate that elevated Ras2p activity or high cAMP levels result in lowered Gap1p permease activity and this could explain the sensitivity to nitrogen starvation observed in these strains. Previous results have have also demonstrated that yeast general amino acid permease is sensitive to cAMP levels (Amitrano et al., 1997). However, in those studies the level of 14C-citrulline uptake increased as cells were grown in 0.25–1.0mM cAMP, but higher levels of exogeneous cAMP were not tested. The low Gap1 activity at low cAMP levels is not consistent with the finding that ras2mutants, which have only partially active Ras pathway have elevated Gap1 levels on all media tested (Fig. 1). Nor with the low Gap1p activity in RAS2val19 mutants we observed (Fig. 1) It is possible that the decreased Gap1p activity at low cAMP observed by Amitano et al (1997) is related to decreased viability and slow growth rate in these conditions and these workers did not assay cAMP concentrations high enough (>1mM) to see the effect on Gap1p activity. Finally, we do not believe that the loss of Gap1 activity we observe at high cAMP is directly correlated with cell viability because, while Gap1 activity decreased steadily from 0.5–5.0mM cAMP, the cells grew at essentially the same rate with no loss of viability from 0.5–2.0mM cAMP.
General amino acid permease is controlled by multiple regulation mechanisms both transcriptional and post-transcriptional. Gene expression studies of a GAP1-lacZ expression construct indicate that there is no repression of transcription in high cAMP conditions (Fig. 3). Compared with 0.5mM cAMP-grown cells, cells grown in 5mM cAMP have lost 80% of their Gap1p activity and yet have slightly higher GAP1-driven β-galactosidase levels. Thus, the Ras2/cAMP pathway must effect one of the post-transcriptional control mechanisms; permease localization, activation or turnover. As the loss of Gap1p activity in RAS2val19 mutants occurs in all media tested and ras2 mutants lose ammonia repression it is most likely that it is the ubiquitination and subsequent turnover of the permease that is being affected. We suggest the following model. In normal cells, growing on adequate nitrogen and carbon sources, the Ras2/cAMP/PKA pathway signals ‘growth’ which includes depression of Gap1p as the permease is not needed when cells have a good nitrogen source. Conversely when cells are starving, Gap1 is up-regulated to allow uptake of any nitrogen sources that become available. Thus, in RAS2val19 mutants the cells are unable to respond to poor nitrogen conditions and lose Gap1p activity. This is achieved by stimulating the ubiquitin-dependent turnover of the permease in a nitrogen-insensitive manner. ras2 mutants maintain high Gap1p levels even in ammonia because the turnover of Gap1p is inhibited in these strains even when ammonia is present in the medium. Thus, the observed loss of ammonia repression in both strains.
In this study we have demonstrated the regulation of Gap1p permease in response to the Ras2/cAMP/PKA levels. Elevated Ras2p or cAMP levels resulted in a decrease in Gap1 transport. Loss of Ras2p or low cAMP resulted in elevated Gap1p activity in ammonia-grown cells. This effect on Gap1 regulation occurs at a post-transcriptional step in the pathway of Gap1p maturation. Studies are underway to further test the model that cAMP levels affect the ubiquitin-dependent turnover of Gap1p in response to the addition of ammonia or amino acids.
Acknowledgments
Thanks to Mike Wigler, Mike Stanbrough, Jeffrey Field for the gift of strains and/or plasmids. Jim Schreve for excellent technical assistance. Wei-Na Lim, Catherine Michels, Michaela Pfieffer are undergraduate students who contributed to the research as part of their senior thesis research. This work was supported by grant 1R15GM54280-02 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amitano AA, Saenz DA, Ramos EH. GAP1 activity is dependent on cAMP activity in Saccharomyces cerevisiae. FEMS Micro Letts. 1997;151:131–133. doi: 10.1111/j.1574-6968.1997.tb12560.x. [DOI] [PubMed] [Google Scholar]
- André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Broach JR, Deschenes RJ. The function of RAS genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Chen EJ, Kaiser CA. Amino acids regulate the intracellular trafficking of he general amino acid permease of Saccharomyces cerrevisiae. Proc Natl Acad Sci. 2002;99:14837–14842. doi: 10.1073/pnas.232591899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaton MCV, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelien JT. The Gap1 general amino acid permease acts as an amino acid sensor from activation of protein kinase A targets in the yeast Sacharomyces cerevisiae. Mol Micro. 2003;50:911–929. doi: 10.1046/j.1365-2958.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M, Deschenes RJ, Broach JR. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61:329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Field J, Vojtek A, Ballester R, Bolger G, Colivelli J, Ferguson K, Gerst J, Kataoka T, Michaeli T, Powers S, Riggs M, Rodgers I, Wieland B, Wigler M. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70kd adenylyl cyclase-associated protein. Cell. 1990;61:319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Grenson M. Study of the positive control of the amino acid permeae and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. Eur J Biochem. 1983;133:141–144. doi: 10.1111/j.1432-1033.1983.tb07439.x. [DOI] [PubMed] [Google Scholar]
- Holsbeeks I, Lagatie O, Van Nuland A, van de Velde S, Thevelein J. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci. 2004;29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Markwardt DD, Garrett JM, Eberhardy S, Heideman W. Activation of the Ras/cyclic AMP pathway in the yeast Saccharomyces cerevisiae does not prevent G1 arrest in response to nitrogen starvation. J Bacteriol. 1995;177:6761–6765. doi: 10.1128/jb.177.23.6761-6765.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; NY: 1972. [Google Scholar]
- Nielson PS, van den Hazel B, Didion T, de Boer M, Jorgenson M, Planta RJ, Kielland-Brandt MC, Anderson HA. Transcriptional regulation of the Saccharomyces cerevisiae amino aicd permease gene BAP2. Mol Gen Genet. 2001;264:613–622. doi: 10.1007/s004380000347. [DOI] [PubMed] [Google Scholar]
- Risinger AL, Cain NE, Chen EJ, Kaiser CA. Activity-dependent reversible inactivation of the general amino acid permease. Mol Biol Cell. 2006;17:4411–4419. doi: 10.1091/mbc.E06-06-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DA, Chianelli MA, Stella CA, Mattoon JR, Ramos EH. RAS2/PKA pathway is involved in nitrogen regulation of L-leucine uptake in Saccharomyces cerevisiae. Int J Biochem Cell Biol. 1997;29:505–512. doi: 10.1016/s1357-2725(96)00102-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TE. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; NY: 1989. [Google Scholar]
- Sass P, Field J, Nikawa J, Toda T, Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreve JL, Garrett JM. The branched-chain amino acid permease gene of Saccharomyces cerevisiae, BAP2, encodes the high-affinity leucine permease (S1) Yeast. 1997;13:435–439. doi: 10.1002/(SICI)1097-0061(199704)13:5<435::AID-YEA95>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Schreve JL, Sin JK, Garrett JM. The Saccharomyces cerevisiae YCC5(YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagines and glutamine. J Bact. 1998;180:2556–2559. doi: 10.1128/jb.180.9.2556-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; NY: 1986. [Google Scholar]
- Sophianopoulou V, Diallinas G. AUA1, a gene involved in ammonia regulation of amino acid transport in Saccharomyces cerevisiae. Mol Micro. 1993;8:167–178. doi: 10.1111/j.1365-2958.1993.tb01213.x. [DOI] [PubMed] [Google Scholar]
- Springael JY, André B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, Nikko E, André B, Marini AM. Yeast Npi3/Bro1 is involved in ubiquitin-dependent control of permease trafficking. FEBS Letts. 2002;517:103–109. doi: 10.1016/s0014-5793(02)02586-3. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Magasanik B. Transcriptional and post translational regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbrough M, Rowen DW, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated gene. Proc Natl Acad Sci. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, deWinde JH. Novel sensing mechanisms and targets for the camp-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Vanderbol M, Jauniaux JC, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permease encodes a protein kinase homolog. Mol Gen Genet. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- Wicherham LJ. A critical evaluation of the nitrogen and assimilation tests commonly used in the classification of yeasts. J Bacteriol. 1946;177:293–301. doi: 10.1128/JB.52.3.293-301.1946. [DOI] [PubMed] [Google Scholar]
- Woodward JR, Cirello VP. Amino acid transport and metabolism in nitrogen-starved cells of Sacharomyces cerevisiae. J Bacteriol. 1977;130:714–723. doi: 10.1128/jb.130.2.714-723.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Garrett JM, Schreve J, Michaeli T. GNP1, a glutamine permease whose overproduction induces growth defects in the yeast Saccharomyces cerevisiae. Curr Gen. 1996;30:107–114. doi: 10.1007/s002940050108. [DOI] [PubMed] [Google Scholar]