Abstract
Moraxella catarrhalis is an important cause of respiratory infections in adults and otitis media in children. Developing an effective vaccine would reduce the morbidity, mortality, and costs associated with such infections. An unfinished genome sequence of a strain of M. catarrhalis available in the GenBank database was analyzed, and open reading frames predicted to encode potential vaccine candidates were identified. Three genes encoding proteins having molecular masses of approximately 22, 75, and 78 kDa (designated Msp [Moraxella surface proteins]) (msp22, msp75, and msp78, respectively) were determined to be conserved by competitive hybridization using a microarray, PCR, and sequencing of the genes in clinical isolates of M. catarrhalis. The genes were transcribed when M. catarrhalis was grown in vitro. These genes were amplified by PCR and cloned into Escherichia coli expression vectors. Recombinant proteins were generated and then studied using enzyme-linked immunosorbent assays with preacquisition and postclearance serum and sputum samples from 31 adults with chronic obstructive pulmonary disease (COPD) who acquired and cleared M. catarrhalis. New antibody responses to the three proteins were observed for a small proportion of the patients with COPD, indicating that these proteins were expressed during human infection. These studies indicate that the Msp22, Msp75, and Msp78 proteins, whose genes were discovered using genome mining, are highly conserved among strains, are expressed during human infection with M. catarrhalis, and represent potential vaccine antigens.
Whole-genome sequencing of bacteria and genome mining is a powerful new approach for identification of novel potential vaccine antigens. Previous approaches for vaccine development used biochemical, immunological, and microbiological methods that allowed identification of the most abundant antigens. A genome mining approach reduces the time and cost required for identification of candidate vaccines. This study focused on identification of putative surface proteins for use as vaccine antigens by mining the genomic sequences of the pathogenic bacterium Moraxella catarrhalis.
M. catarrhalis is an aerobic, gram-negative diplococcus that is an important respiratory tract pathogen in humans (15, 26, 34). M. catarrhalis annually causes 4 to 5 million of the total 25 million episodes of acute otitis media in the United States (27, 33). Between $3.8 billion and $5.7 billion is spent annually in the United States alone on healthcare for children with otitis media (6, 13). A subset of children is otitis prone, experiencing recurrent acute otitis media and chronic otitis media, which are associated with delayed speech and language development (6, 7, 48). Preventing otitis media with a vaccine would result in substantial reductions in morbidity and health care costs, particularly for otitis-prone children.
M. catarrhalis is also a cause of exacerbations of chronic obstructive pulmonary disease (COPD) (34, 43). M. catarrhalis infection is the second most common cause of exacerbations of COPD after nontypeable Haemophilus influenzae infection. COPD affects 24 million Americans, and M. catarrhalis causes 2 to 4 million exacerbations annually (30, 34). Overall, COPD is the fourth leading cause of death in the United States, and the estimated annual direct and indirect health care costs of COPD are $32.1 billion (31). Adults with COPD represent a second group that would benefit from an effective vaccine for M. catarrhalis. Such a vaccine would reduce the morbidity, mortality, and financial costs associated with COPD.
Surface proteins of M. catarrhalis are attractive vaccine antigens (33). An ideal vaccine candidate has several characteristics, including exposure on the bacterial surface, sequence conservation among strains, expression during human infection, and generation of a protective immune response. To date, a limited number of outer membrane proteins have been examined as potential vaccine antigens (1, 4, 11, 16, 17, 21, 28, 37).
In this study we used a bioinformatics approach to identify open reading frames (ORFs) that encode novel proteins predicted to be on the surface of M. catarrhalis. Three genes were studied further by using a variety of methods to assess their sequence conservation among strains and to test the hypothesis that these genes are transcribed and expressed when M. catarrhalis colonizes or infects the human respiratory tract. Due to the conservation of these three novel genes among strains and their expression in the human respiratory tract, the proteins encoded by them are good candidates for further study as vaccine antigens.
MATERIALS AND METHODS
Sequence analysis.
The genome of strain ATCC 43617 was analyzed using the 41 contigs deposited in the GenBank database (accession numbers AX067426 through AX067466). GeneMarkS was used to identify ORFs in the genome sequence (10). Potential lipoproteins were identified using a PERL script developed in house and based on sequences described at the Prosite database (http://us.expasy.org). SignalP was used to identify ORFs with signal sequences characteristic of membrane proteins and secreted proteins (19). Type IV signal peptides were identified with an in-house PERL script. A search of the April 2003 GenBank protein database revealed 13,536 bacterial proteins that were annotated with the terms outer membrane protein, secreted protein, and virulence. These proteins were combined with 199 annotated bacterial lipoproteins from the DOLOP database (5, 29). The combined annotated proteins were converted to a BLAST searchable database using formatdb (http://www.ncbi.nlm.nih.gov/BLAST/download.shtml). Predicted ORFs were compared to this database, and proteins with e values less than 1 × 10−30 were identified.
Construction of microarray.
Primers were made for each of the 348 ORFs predicted by sequence analyses. PCRs were performed using template DNA from M. catarrhalis strain ATCC 43617 to generate products that were spotted on a microarray. Prior to microarray construction, each PCR product was confirmed to be the correct size by agarose gel electrophoresis. Microarray construction and hybridization were done at the Roswell Park Cancer Institute (RPCI) Microarray and Genomics Facility. The PCR products were resuspended in 20 μl of 25% dimethyl sulfoxide and rearrayed in 384-well plates. Slides were then printed using MicroSpot 10K split pins and a MicroGrid II TAS arrayer (BioRobotics, Inc.). The ORF products had ∼150-μm-diameter spots with 300-μm center-to-center spacing. During the print run the humidity, temperature, and dust in the environment were controlled. Each ORF product was printed 12 times on amino-silanated glass slides (Schott Nexterion type A). Each print run contained amplicons corresponding to three genes (genes encoding outer membrane proteins [OMPs] CD, E, and G1b) that showed >95% sequence identity to the reference strain and were spotted ∼600 times. PCR products of irrelevant genes from H. influenzae were included as negative controls. The printed slides were dried overnight and were UV cross-linked (500 mJ) with a Stratalinker 2400 (Stratagene). The slides were hybridized without additional treatment. No indication of DNA loss from the spots was detected at any stage when hybridization in formamide buffers at 55°C was performed (using 4′,6′-diamidino-2-phenylindole [DAPI] staining).
Bacterial strains and culture conditions.
M. catarrhalis strains ATCC 43617, ATCC 25238, and ATCC 25240 were obtained from the American Type Culture Collection (Manassas, VA). Isolate O35E was provided by Eric Hansen. Strains M2, M9, M10, and M11 were sputum isolates from Houston, TX, provided by Daniel Musher. Strains 14, 21, 23, 27, and 48 were sputum isolates from Johnson City, TN, obtained from Steven Berk. Strains 435, 565, 636, and 1089 were sputum isolates from Birmingham, United Kingdom, provided by Susan Hill. Middle ear fluid isolates 2951, 3584, 4223, 4608, 5191, 7169, and 8184 were provided by Howard Faden. Strains 6P29B1 and 7P94B1 were sputum isolates obtained from adults in our COPD study clinic (see below). Chemically competent Escherichia coli strains TOP10 and BL21(DE3) were obtained from Invitrogen.
M. catarrhalis strains were grown on brain heart infusion plates at 37°C with 5% CO2 or in brain heart infusion broth with shaking at 37°C. E. coli strains were grown at 37°C on Luria-Bertani (LB) plates, in LB broth, or in terrific broth supplemented with the appropriate antibiotics (MoBio Laboratories, Carlsbad, CA).
Competitive hybridization of genomic DNA.
Genomic DNA was labeled using the fluorescent nucleotide analog Cy5 (strain ATCC 43617) or Cy3 (test strains). One microgram of genomic DNA was random primer labeled using a BioPrime DNA labeling kit (Invitrogen, Inc.) for 3 h at 37°C with the appropriate dye (Cy3 or Cy5). After ethanol precipitation, the probes were resuspended in H2O and combined, and unincorporated Cy dye was removed by passage over a Qiagen spin column. The labeled probes were dried and stored at −20°C until hybridization.
Hybridization to the microarrays was conducted under controlled conditions. The probes were combined and resuspended in 110 μl of hybridization solution (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 40 μg salmon sperm DNA, 0.25% sodium dodecyl sulfate [SDS]), heated to 95°C for 5 min, and placed on ice. The entire probe was added to the array, and hybridization was performed for 16 h at 55°C using a GeneTAC hybridization station (Genomics Solutions, Inc.). After hybridization, the slide was washed with decreasing concentrations of SSC and SDS, which was followed by one wash with 0.1× SSC, one rinse with 95% ethanol, and centrifugal drying for 3 min.
The hybridized slides were scanned using a GenePix 4200A scanner to generate high-resolution (10-μm) images for both the Cy3 and Cy5 channels. Image analysis was performed using the raw images and ImaGene, version 4.1, from BioDiversity, Inc.
Each spot was defined by a circular region. The size of the region was programmatically adjusted to match the size of the spot. A 2- to 3-pixel buffer region around the spot was ignored. There was another 2 to 3 pixels outside the buffer region that was considered the local background for the spot. Each spot and its background region were segmented using a proprietary optimized segmentation algorithm that excluded pixels not representative of the rest of the pixels in the region. The background-corrected signal for each cDNA was the mean signal of all the pixels in the region minus the mean local background. The output of the image analyses was two tab-delimited files, one for each channel, containing all of the fluorescence data.
The output of the image analyses was then processed by using a program developed in house. Spots whose levels were not significantly above the background level or had a poor coefficient of variation were excluded. For each spot, a ratio was calculated using the background-subtracted mean signal of the two channels, one representing reference strain ATCC 43617 and one representing the test strain. The ratios were normalized on the log scale across clones known to have high homology. Replicate measurements were averaged on the log scale. The final log2 ratio was converted back to a linear ratio.
RT-PCR.
Bacterial RNA was isolated using a Qiagen RNeasy kit and a Qiashredder column (Qiagen, Valencia, CA) by following the manufacturer's instructions, with an additional incubation with RNase-free DNase I (Promega) for 30 min at 37°C. Reverse transcriptase PCR (RT-PCR) was performed using a Qiagen OneStep RT-PCR kit and RNaseOut inhibitor (Invitrogen, Carlsbad, CA). Primers were designed to amplify ∼500-bp fragments of msp75 and msp78 and full-size msp22 (Table 1). To eliminate the possibility of contaminating DNA, parallel reactions were performed with TaqI DNA polymerase (HotMaster mixture; Eppendorf, Hamburg, Germany). Following amplification, samples were electrophoresed in 1.5% agarose gels and stained with ethidium bromide.
TABLE 1.
Primer sequences
| Gene | Expt | Direction | Primer sequencea |
|---|---|---|---|
| msp22 | Clone gene | Forward | 5′ ATATATATCCATGGAACAGCTAGGGACTGCCACC 3′ |
| msp22 | Clone gene | Reverse | 5′ TCTCTAGGATCCAGAACCACACTGGCTGGCCATTTC 3′ |
| msp22 | RT-PCR | Forward | 5′ AACAGCTAGGGACTGCCACC 3′ |
| msp22 | RT-PCR | Reverse | 5′ CTTCAGGGTCTGTCCATATCTC 3′ |
| msp75 | Clone gene | Forward | 5′ ATATGGATCCGCAAGCCTGTTTGATTG 3′ |
| msp75 | Clone gene | Reverse | 5′ GCGCGAATTCTTATTCGCTGATATCC 3′ |
| msp75 | RT-PCR | Forward | 5′ GATACACACAAGGAAGATTTG 3′ |
| msp75 | RT-PCR | Reverse | 5′ CATAGATACGGTTGGCACACAC 3′ |
| msp78 | Clone gene | Forward | 5′ ATATGGATCCAGCGGACAAAGCCGCC 3′ |
| msp78 | Clone gene | Reverse | 5′ GCGCGAATTCTCAGTTTGGCTTGGT 3′ |
| msp78 | RT-PCR | Forward | 5′ CATTTACCGCACCGGGTCATAC 3′ |
| msp78 | RT-PCR | Reverse | 5′ CTTCTTGGGTATCAATTGCTTG 3′ |
Restriction enzyme sites are underlined.
Expression and purification of recombinant proteins.
Selected putative surface proteins were chosen for further study based on the data mining strategy described above and their sequence conservation in strains. Genes were amplified by PCR from M. catarrhalis ATCC 43617 genomic DNA using gene-specific primers (Sigma-Genosys, The Woodlands, TX) (Table 1). The primers included restriction enzyme sites for NcoI, BamHI, or EcoRI at the ends of the amplified genes to allow directional cloning into either pRSETB (Invitrogen, San Diego, CA) for msp75 and msp78 or pCATCH, kindly provided by Paul Cullen and Ben Adler, for msp22 (18). Cloning into these vectors resulted in fusion proteins expressing a six-His tag under the control of an isopropyl-β-d-thiogalactosidase (IPTG)-inducible promoter. Additionally, the pCATCH plasmid was chosen for msp22, which encodes a putative lipoprotein, because this plasmid contains E. coli lipoprotein signal sequences that code for attachment of an N-terminal lipid moiety during processing, allowing expression of the msp22 protein with an amino-terminal lipid (3, 18).
Amplification of msp22 was performed using Taq HiFi polymerase (Invitrogen) and an Eppendorf Mastercycler personal thermal cycler with the following program: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 90 s. Amplification of msp75 and msp78 was performed using Vent polymerase (New England Biolabs) with the following programs: for msp75, 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 90 s; and for msp78, 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 90 s. Aliquots from the PCR mixture were subjected to agarose gel electrophoresis. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Chatsworth, CA) and were cloned into either pRSETB or pCATCH.
Chemically competent E. coli TOP10 cells were transformed with the recombinant plasmids. Colonies were picked from LB agar plates containing 30 μg/ml kanamycin (msp22) or 60 μg/ml carbenicillin (msp75 and msp78). Plasmids were confirmed to have the gene insert by PCR and by sequencing at the RPCI Biopolymer Facility.
The recombinant plasmids were purified using a Qiagen plasmid mini-purification system according to the manufacturer's instructions. The plasmids were transformed into chemically competent E. coli BL21(DE3) for expression. To express recombinant Msp22 (rMsp22), a 10-ml culture in LB medium with 60 μg/ml kanamycin was inoculated and allowed to grow overnight at 37°C with shaking. To express rMsp75 and rMsp78, a 10-ml culture in LB medium with 100 μg/ml carbenicillin was inoculated and allowed to grow overnight at 37°C with shaking. The following day, 200 ml of terrific broth containing either 120 μg/ml kanamycin (rMsp22) or 300 μg/ml carbenicillin (rMsp75 and rMsp78) was seeded with the overnight culture and grown until the optical density at 600 nm (OD600) was 0.6. Recombinant protein was expressed by adding 1 mM IPTG, followed 15 min later by 0.15 mg/ml rifampin. After 4 h of incubation at 37°C, cultures were centrifuged at 13,000 × g at 4°C for 20 min.
Cell lysis was performed by adding 8 M urea with 0.1 M NaH2PO4 (pH 8.0) to the bacterial pellet and mixing the preparation for 30 min at room temperature. Cleared lysate was obtained by centrifugation at 50,000 × g at 4°C for 20 min. The lysate was then added to TALON Co2+ metal affinity resin (BD Biosciences, Palo Alto, CA) (prewashed with 8 M urea) by mixing with a nutator at room temperature for 1 h. The lysate and resin were centrifuged at 4°C for 5 min at 3,000 × g. The unbound lysate was saved, and the resin was washed four times with lysis buffer. After the final wash, the recombinant proteins were eluted from the resin using 10 column volumes of lysis buffer containing 250 mM imidazole. The resulting supernatant was diluted to obtain a final concentration of 4 M urea with 0.5 M Tris (pH 8.0).
The resulting diluted supernatant was diafiltered using an Amicon stirred ultrafiltration cell and a 10,000-molecular-weight-cutoff filter (Millipore, Bedford, MA) against 10 volumes of buffer Z1 (0.01% Zwittergent 3-14, 0.05 M Tris, 0.01 M Na2EDTA; pH 8.0) under a nitrogen atmosphere. Once in buffer Z1, the protein solution was concentrated to 1 ml using an Amicon Ultra-15 centrifugal filter unit (Millipore, Bedford, MA). Protein concentrations were determined using a BCA protein assay kit (Pierce, Rockford, IL).
PCR and sequencing.
The primers and PCR conditions described above were used to amplify the three genes using HotMaster mixture (Eppendorf) with the following 25 isolates of M. catarrhalis: O35E, M2, M9, M10, M11, 14, 21, 23, 27, 48, 435, 565, 636, 1089, 2951, 3584, 4223, 4608, 5191, 7169, 8184, ATCC 25238, ATCC 25240, 6P29B1, and 7P94B1. Sequencing of each of the three genes for 10 clinical isolates was performed at the RPCI Biopolymer Facility (isolates O35E, M10, 14, 27, 435, 565, 2951, 8184, ATCC 25240 and 7P94B1).
COPD study clinic.
The COPD study clinic at the Buffalo Veterans Affairs Medical Center is an ongoing prospective study that was started in 1994 (34, 43). To be included in this study, patients must have chronic bronchitis as defined by the American Thoracic Society (8) and must be willing to attend the study clinic monthly. Patients with asthma, malignancies, or other immunocompromising illnesses were excluded. Patients were seen monthly and at times when an exacerbation was suspected. At each visit clinical criteria were used to determine whether patients were experiencing an exacerbation or whether they were clinically stable as previously described (43). Additionally at each visit, serum and expectorated sputum samples were collected. Bacteria present in the sputum were identified using standard techniques. Sera, sputum supernatants, and bacteria obtained from sputum cultures were stored at −80°C. These samples were used to analyze human antibody responses to the purified recombinant proteins before and after acquisition and clearance of M. catarrhalis. All patient data and material were collected and processed in compliance with Veterans Affairs Western New York Healthcare System Institutional Review Board guidelines.
ELISA.
Samples of human serum and sputum were obtained from the COPD study clinic. Thirty-one preacquisition and postclearance sera from adults with COPD who acquired M. catarrhalis were studied by using enzyme-linked immunosorbent assays (ELISAs) to detect the development of new immunoglobulin G (IgG) antibodies in serum to the protein following clearance of the strain (34). Preacquisition and postclearance sputum supernatants were similarly studied to detect new IgA antibody responses. The change in the antibody level from preacquisition to postclearance samples was calculated using the following formula: percent change = [(postclearance optical density − preacquisition optical density)/preacquisition optical density] × 100.
ELISAs were carried out by coating the wells of a 96-well microtiter Immunolon 4 plate (Thermo Labsystems, Franklin, MA) with recombinant purified protein. The protein concentrations were determined in preliminary assays in order to optimize conditions. Wells were coated with either 1 or 5 μg of protein. Following overnight incubation at room temperature, plates were washed four times with phosphate-buffered saline (PBS)-0.5% Tween 20. Plates were then blocked using 5% nonfat dry milk in PBS (MPBS) for 1 h at room temperature. After washing, primary antibody (i.e., serum or sputum) diluted in MPBS was added to the wells. The starting dilutions were approximately 1:200 for serum samples and 1:100 for sputum supernatants. Twofold dilutions were prepared on the plate for each sample to obtain a total of three dilutions per primary antibody for serum or two dilutions per primary antibody for sputum. Uncoated and coated control wells received MPBS without primary antibody as negative controls. The plates were incubated at 37°C for 2 h. After the 2-h incubation, the plates were again washed four times with phosphate-buffered saline-0.5% Tween 20. Secondary horseradish peroxidase-conjugated antibody (1:3,000 dilution of goat-anti-human IgG or 1:2,000 dilution of goat-anti-human IgA [KPL, Gaithersburg, MD]) diluted in MPBS was then added. After incubation at 37°C for 1 h, the plates were washed, and developing reagent was added to the wells and allowed to react for 15 min in the dark. Color development was stopped using 4 N H2SO4. The absorbance at 450 nm was determined using a Bio-Rad model 3550-UV microplate reader.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers are EU339315 for msp22, EU339314 for msp75, and EU339313 for msp78.
RESULTS
Identification of ORFs that encode putative surface proteins.
The genome of strain ATCC 43617 was analyzed using the 41 contigs available in the GenBank database. The unassembled DNA fragments had a total length of 1,913,584 bp, which matched the calculated genome size determined experimentally by pulsed-field gel electrophoresis, 1,750,000 to 1,940,000 bp (22, 38). Using the GeneMarkS program, a total of 1,849 ORFs ranging from 42 to 1946 bp long were obtained. The minimum length of an ORF was chosen to be 225 bp, making the size of the smallest encoded protein 75 amino acids. This method yielded 1,697 ORFs in the M. catarrhalis genome.
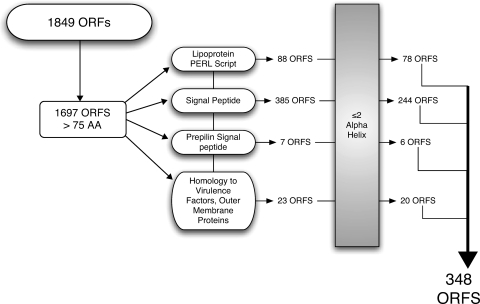
Several approaches were used to determine the ORFs likely to encode proteins that are processed to the bacterial surface, as surface-exposed proteins represent potential vaccine antigens. The outer membranes of gram-negative bacteria are known to have an abundance of lipoproteins, and several lipoproteins in other bacteria are promising vaccine antigens (e.g., OspA of Borrelia burgdorferi and P6 of H. influenzae) (35, 47). Therefore, ORFs were analyzed to identify ORFs that encode lipoproteins, and 88 were identified. Analysis with SignalP, which detects signal sequences for membrane proteins that are cleaved by both signal peptidase I and signal peptidase II, yielded 385 ORFs. Analyzing the ORFs for prepilinlike protein signal sequences that form part of the type II secretion system generated another seven ORFs. In an effort to identify proteins that were previously annotated as potential vaccine candidates but not identified with other search algorithms, all of the ORFs were compared to the annotated GenBank protein database using BLASTP. The proteins with e values less than 1 × 10−30 that had either outer membrane protein, secreted, or virulence as a term in their annotation were also included as potential candidates. This annotation-based homology search revealed another 23 ORFs. ORFs that were predicted to contain two or more α-helix regions were determined to most likely be located in the cytoplasmic membrane and were therefore excluded. Figure 1 shows a flow chart of the strategy that resulted in prediction of 348 ORFs that encode putative surface-exposed proteins.
FIG. 1.
Flow chart of genomic sequence analysis for potential surface proteins. The ORFs were based on analysis of the unannotated genome sequence of M. catarrhalis ATCC 43617. AA, amino acids.
Analysis of putative genes for sequence conservation.
The 348 ORFs found by sequence analyses were amplified by PCR from strain ATCC 43617 template DNA, and the products were spotted on a microarray. Ten genes that encode previously identified OMPs (UspA1, UspA2, TbpA, TbpB, CopB, LbpA, OMP CD, OMP E, OMP G1a, and OMP G1b) were studied to optimize conditions. These 10 genes were predicted to be potentially surface localized by genome analysis. Genomic DNA of the homologous ATCC 43617 strain was labeled with Cy5, and genomic DNA of four different competing strains (ATCC 25240, O35E, 4223, and ATCC 25238) were labeled individually with Cy3. These strains were selected because the sequences of the 10 test genes are known for these strains and thus could be used as controls when the results were analyzed. Microarrays were probed simultaneously with DNA from the reference strain (ATCC 43617) and DNA from each of the four test strains individually, and the red/green ratio was determined to assess the sequence similarity for each ORF compared to the reference strain. As controls, 50 PCRs each were performed to amplify the genes that encode OMPs CD, E, and G1b, which yielded 150 spots with >95% homology between strain ATCC 43617 and test strains for normalization of data.
To identify genes conserved among strains, competitive hybridization experiments using genomic DNA of 12 strains of M. catarrhalis having diverse geographic and clinical origins were performed individually with genomic DNA from the sequenced strain used to construct the microarray (strain ATCC 43617). A cutoff value for the linear ratio between strain ATCC 43617 and each of the 12 clinical strains of >0.8 was used to determine which genes encoded highly conserved proteins. A total of 147 ORFs were identified as ORFs having a signal ratio of >0.8 between strain ATCC 43617 and the competing strain for all 12 strains studied, suggesting that there was sequence conservation of these genes among strains. From these 147 ORFs, three genes were chosen to study in detail. These genes were designated msp22, msp75, and msp78 (Moraxella surface proteins); the numbers in the designations correspond to the molecular masses (in kilodaltons) of the predicted proteins.
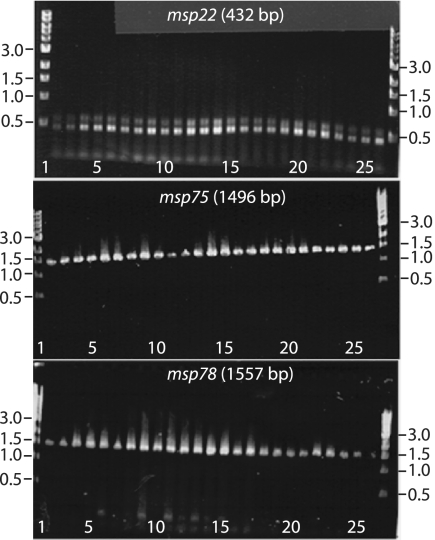
To further assess the sequence conservation of the genes identified by competitive hybridization of the microarray, primers corresponding to the genes of interest were designed. The three genes (msp22, msp75, and msp78) were amplified by PCR from 25 clinical isolates of M. catarrhalis, including the 12 isolates studied using competitive hybridization of the microarray. The primers were based on M. catarrhalis strain ATCC 43617, the strain used for microarray construction. All three genes were present in all 25 clinical isolates tested (Fig. 2). Additionally, each of the PCR products was the expected size for each gene. This result indicated that each of the three genes was present in all strains and that there was no variation in the lengths of these three genes among the diverse strains examined.
FIG. 2.
Ethidium bromide-stained agarose gels showing amplicons from PCRs performed with primers for the genes indicated. The templates were genomic DNA of 25 isolates of M. catarrhalis. Lanes 1 and 27, standard; lane 2, O35E; lane 3, M10; lane 4, M11; lane 5, 14; lane 6, 21; lane 7, 23; lane 8, 27; lane 9, M9; 10, lane 48; lane 11, M2; lane 12, 435; lane 13, 565; lane 14, 636; lane 15, 1089; lane 16, 2951; lane 17, 3584; lane 18, 4223; lane 19, 4608; lane 20, 5191; lane 21, 7169; lane 22, 8184; lane 23, ATCC 25238; lane 24, ATCC 25240; lane 25, 6P29B1; lane 26, 7P94B1. The positions of molecular size standards (in kilobases) are indicated on the left and on the right.
To further evaluate sequence conservation of the genes among strains, the sequence of the entire gene for each of the three genes was determined for 10 clinical isolates (O35E, M10, 14, 27, 435, 565, 2951, 8184, ATCC 25240, and 7P94B1). Gene sequences were translated, and the amino acid homology for strains of M. catarrhalis was calculated for each gene using MacVector (Accelerys). The amino acid sequences encoded by each of the three genes were 97 to 99% identical in the 10 strains (Table 2). We concluded that the msp22, msp75, and msp78 genes are highly conserved among strains of M. catarrhalis.
TABLE 2.
Characteristics of three putative surface proteins
| Gene | DNA size (bp) | Predicted protein molecular mass (kDa) | Isoelectric point | % Amino acid homology between strains of M. catarrhalis | Surface location predictor | Amino acid sequence homologue (% identity/% similarity) |
|---|---|---|---|---|---|---|
| msp22 | 432 | 21.6 | 5.11 | 99 | Lipoprotein | Cytochrome c, class II (Psychrobacter sp.) (36/53) |
| msp75 | 1,497 | 74.9 | 4.83 | 97 | Leader, membrane | Succinic semialdehyde dehydrogenase (Psychrobacter sp.) (73/85) |
| msp78 | 1,557 | 77.8 | 5.89 | 99 | Leader | Outer membrane nitrite reductase (Neisseria sp.) (80/90) |
Characterization of genes.
To gain insight into the possible function of Msp22, Msp75, and Msp78, BLASTP homology searches were performed. Table 2 shows characteristics of each of the proteins based on BLASTP searches. Msp22, a putative lipoprotein, has significant homology to cytochrome c′. Msp22 is predicted to contain 152 amino acids and has conserved cytochrome c′ domain architecture with significant homology to COG3903, which includes the pfam PFO1322 or the cytochrome c′ family (also referred to as cytochrome_C_2). Msp22 has the characteristic CXXCH motif at amino acids 142 to 146 that is associated with heme attachment, although in other gram-negative bacteria the cytochromes may be involved in transport of iron and other divalent cations. The genes downstream on the complementary strand have significant homology to genes encoding coproporphyrinogen III oxidase and GTP cyclohydrolase II. This arrangement of cytochrome, coproporphyrinogen oxidase, and GTP cyclohydrolase genes is syngeneic to the genome sequence in Psychrobacter sp. strain PRwf-1, a member of the family Moraxellaceae, as well as genome sequences in members of the genera Acinetobacter, Moraxella, Alkanindiges, and Enhydrobacter.
Msp75 is predicted to contain 499 amino acids and has a high level of homology to succinic semialdehyde dehydrogenase. This protein was identified for study with the algorithm through the BLASTP homology of the msp75 gene with a region of the chromosome of Agrobacterium tumefaciens that is associated with virulence. The localization in the cell was predicted to be in the cytoplasm by psortB.
Msp78, which contains a signal sequence, has high levels of similarity and identity to an anaerobically induced nitrate reductase. Homologues of this protein have been identified as outer membrane proteins. psortB localized the protein to the periplasm but also indicated that the protein has characteristics of a membrane protein. The protein has multiple pfam domains that have been identified, including a multicopper oxidase motif, a cytochrome c motif, and copper binding domains.
Transcription of genes during in vitro growth.
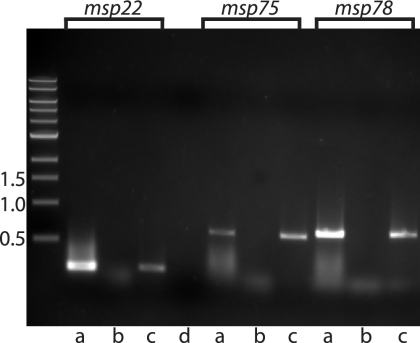
To determine whether the genes that encode Msp 22, Msp75, and Msp78 are transcribed, RT-PCR was performed using RNA isolated from M. catarrhalis strain O35E grown in broth. Figure 3 (lanes a) shows that all three genes are transcribed during growth in vitro. Control assays confirmed that the purified RNA was free of contaminating DNA (lanes b).
FIG. 3.
Results of RT-PCRs with RNA from M. catarrhalis. The primers used in the reactions were primers for the msp22, msp75, and msp78 genes, as indicated at the top. Lanes a, purified RNA amplified with RT; lanes b, purified RNA amplified with TaqI polymerase to exclude DNA contamination; lanes c, purified DNA amplified with TaqI polymerase; lane d, distilled water with RT as a negative control. The positions of molecular size markers (in kilobases) are indicated on the left.
Characterization of purified recombinant proteins.
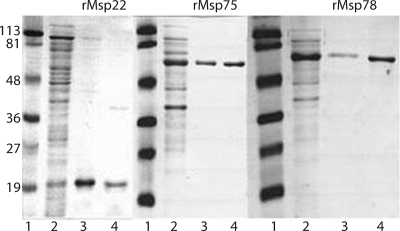
Recombinant proteins were expressed in E. coli BL21(DE3). All three recombinant proteins were purified initially under denaturing conditions. The six-His tag was used to bind the proteins to cobalt affinity resin, which allowed purification. Denatured proteins precipitated at low concentrations when 8 M urea was removed. When the proteins were refolded by diafiltering against buffer Z1, concentrations of approximately 1 mg/ml were obtained. The solubility at a higher protein concentration suggests that the proteins were in a refolded state. The results of a typical purification are shown in Fig. 4. Single bands for each of the proteins were seen when the purified proteins were subjected to SDS-polyacrylamide gel electrophoresis.
FIG. 4.
Coomassie blue-stained SDS-polyacrylamide gel electrophoresis gel showing purification of the rMsp22 (22 kDa), rMsp75 (75 kDa), and rMsp78 (78 kDa) proteins. Lanes 1, standard; lanes 2, unbound supernatant after protein was bound to cobalt resin; lanes 3, cobalt resin after elution of protein with imidazole, showing some protein that remained bound to the resin and did not elute; lanes 4, eluted protein after diafiltration into buffer Z1. The positions of molecular mass standards (in kilodaltons) are indicated on the left.
Human antibody responses.
To determine whether the Msp22, Msp75, and Msp78 proteins were expressed by M. catarrhalis in the human respiratory tract, the three purified recombinant proteins were assayed with 31 serum and sputum pairs from patients who acquired and cleared M. catarrhalis and were found to have developed new serum IgG and sputum IgA antibodies following infection by ELISA and flow cytometry with whole bacteria (34). These 31 pairs were examined for production of antibody to each of the recombinant proteins. Paired preacquisition and postclearance samples were always tested in the same assay.
To determine the cutoff value for a significant change between preacquisition and postclearance serum IgG and sputum IgA levels, 10 control pairs were examined using a previously described method (2, 3, 34, 36). Control samples obtained 2 months apart (the same time interval used for the experimental samples) from patients whose sputum cultures were negative for M. catarrhalis were identified. These samples were subjected to ELISA with purified proteins Msp22, Msp75, and Msp78. Samples that generated OD450 values less than 0.1 were considered not to represent a significant level of antibody, so a value of zero was assigned. The percent changes in OD450 values for the paired control samples were calculated. Table 3 shows the means, standard deviations, and upper limits of the 99% confidence intervals (cutoff values) calculated using control samples for the three proteins. The cutoff values for a significant change were higher for sputum IgA than for serum IgG, as has been observed previously (34).
TABLE 3.
ELISA cutoff value determination for the paired serum and sputum samples
| Ig | % Change
|
|||||
|---|---|---|---|---|---|---|
| rMsp22
|
rMsp75
|
rMsp78
|
||||
| Mean ± SD | 99% CIa | Mean ± SD | 99% CI | Mean ± SD | 99% CI | |
| Serum IgG | −2.5 ± 8.1 | 18.4 | −40.0 ± 25.5 | 25.8 | −21.1 ± 15.1 | 17.7 |
| Sputum IgA | 61.9 ± 137.4 | 416.4 | 0 ± 0b | 0 | −10.3 ± 31.0 | 69.6 |
99% CI, upper limit of the 99% confidence interval for the control samples. Any percent change in the OD450 greater than the upper limit of the 99% confidence interval between the preacquisition and postclearance values for a protein was considered a significant change.
An OD450 less than 0.1 was considered not to represent a significant level of antibody, and a value of zero was assigned.
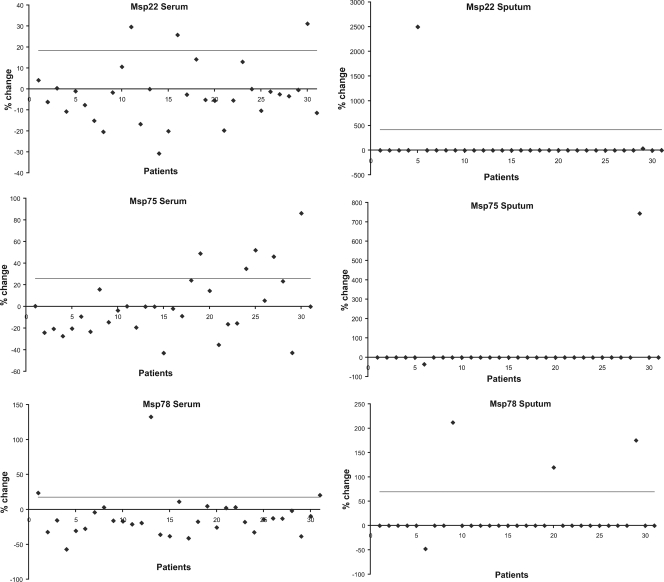
Results from the ELISA analyses of 31 pairs of serum and sputum samples for all three recombinant proteins are shown in Fig. 5. A significant increase in the level of IgG antibodies to individual proteins was seen in 3 to 16% of patients for each of the proteins. Similarly, 3 to 10% of the patients generated a new IgA response to the individual proteins. Overall, 47% of patients generated an antibody response to at least one of the three proteins. These results indicate that Msp22, Msp75, and Msp78 are expressed by M. catarrhalis in the human respiratory tract and are targets of the human systemic and mucosal immune systems in a proportion of adults with COPD.
FIG. 5.
Results of ELISAs measuring serum IgG and sputum IgA to rMsp22, rMsp75, and rMsp78 in serum and sputum supernatants of samples from adults with COPD who acquired and cleared M. catarrhalis. The patients tested are shown on the x axis. The percent changes from preacquisition to postclearance are shown on the y axis. Cutoff values were determined by averaging the difference between 10 control pairs of sera or sputum from patients who had never been colonized with M. catarrhalis, and these values are indicated by the horizontal lines. Samples that generated OD450 values less than 0.1 were considered not to represent a significant level of antibody, and values of zero were assigned.
DISCUSSION
Prymula et al. (40) recently demonstrated that vaccination with capsular polysaccharides from Streptococcus pneumoniae conjugated to a conserved outer membrane protein of H. influenzae (protein D) resulted in protection against otitis media caused by both organisms in clinical trials with young children. This study provides a strong rationale for pursuing the approach of using highly conserved OMPs of H. influenzae and M. catarrhalis as vaccine antigens to prevent otitis media.
In this study, we used a genome mining approach to identify novel vaccine antigens in the pathogenic bacterium M. catarrhalis. Using sequence analyses, a DNA microarray was generated, which allowed determination of genes that were conserved among diverse M. catarrhalis isolates. The main approach used in this study to predict the outer membrane location consisted of determination of potential lipoproteins, identification of various signal sequences, and homology searching. This approach successfully identified ORFs corresponding to 10 previously identified OMPs of M. catarrhalis. The proportion of ORFs predicted to be on the surface (348 of 1,849) matched well with the results of similar analyses of other bacteria (23, 50, 51).
The profiles of the proteins selected for study yielded interesting results. Msp22, a putative lipoprotein, was of particular interest as lipoproteins such as OspA of B. burgdorferi were used in the Lyme disease vaccine and P6 of H. influenzae is a promising vaccine antigen (35, 47). The homology of Msp22 to cytochrome c′ was of interest as the gram-negative bacteria Shewanella oneidensis and Geobacter sulfurreducens have developed electron transfer strategies that require c-type cytochromes, including OmcA, MtrC, OmcE, and OmcS, that are located in the outer membrane (46). Pseudomonas, Paracoccus, and Legionella have cytochromelike proteins that function in pyoverdin production, siderophore production, and iron acquisition/assimilation, respectively (20). Thus, in spite of its homology to cytochrome c′, it is possible that Msp22 does not function as a cytochrome but rather functions as an OMP involved in transport mechanisms. Functional testing has to be done to assess the actual function of Msp22 in M. catarrhalis.
Msp75 has significant homology with the enzyme succinic semialdehyde dehydrogenase, suggesting that this protein is not in the outer membrane. However, there are numerous examples of enzymes that are located in the outer membrane of gram-negative bacteria, including a phosphomonoesterase (P4) and a glycerol-3-phosphodiester phosphodiesterase (protein D) in H. influenzae, an enolase in Pseudomonas aeruginosa, the lipid kinase YegS in Salmonella, and the lipid A acylase PagP and deacylase PagL found in Pseudomonas, Bordetella, and Salmonella (24, 32, 39, 41, 42, 49). Additionally, the M. catarrhalis BRO-1 β-lactamase is in the outer membrane (12). A hydrophobic domain (amino acid residues 150 to 231) may serve as an internal signal sequence for direct translocation of this protein to the outer membrane.
Msp78, which contained a signal sequence, had a high level of homology to the major anaerobically induced nitrate reductase OMP AniA (formerly Pan1) in Neisseria. This is of particular interest as AniA has been shown to be a target of human antibodies in patients with gonococcal disease (14, 25).
Once sequence conservation was confirmed and protein characteristics were studied, the question of whether the genes were transcribed and expressed was addressed. Two approaches were taken. First, transcription of the genes was assessed during in vitro growth using RT-PCR. All three genes were transcribed when M. catarrhalis was grown in vitro (Fig. 3).
To assess whether the surface proteins were expressed by M. catarrhalis in the human respiratory tract, immune responses to Msp22, Msp75, and Msp78 in patients with COPD following clearance of M. catarrhalis were assayed. The availability of both serum and sputum samples allowed both systemic and mucosal immune responses to be studied. The prospective design of the COPD study clinic permitted examination of preacquisition samples obtained 1 month before M. catarrhalis was acquired by the patient. This design and the associated samples allowed us to distinguish production of cross-reactive background antibody from production of new antibody to the proteins following clearance. Paired samples that showed a significant increase in antibody level in the postclearance samples compared to the preacquisition samples indicated that patients developed an antibody response to the protein during carriage. The development of an antibody response indicates that the proteins are expressed by M. catarrhalis in the human respiratory tract.
A small proportion of patients with COPD exhibited new antibody responses to each individual protein, suggesting that these proteins are not immunodominant antigens. This result is not surprising. Variability among individuals in the surface bacterial antigens to which antibodies are directed is a hallmark of antibody responses to bacteria in the COPD setting (9, 44, 45). The proportion of patients who developed new antibody responses to Msp22, Msp75, and Msp78 observed in this study parallels the proportion observed for M. catarrhalis OMPs E, G1a, and G1b when similar methods were used (2, 3, 11). The observation that some patients with COPD developed new systemic and mucosal antibodies to Msp22, Msp75, and Msp78 in samples collected 1 month after clearance of M. catarrhalis compared to paired samples collected immediately before acquisition of the organism supports the conclusion that the proteins are expressed by M. catarrhalis in the human respiratory tract.
The absence of an antibody response as determined by ELISA with recombinant proteins does not exclude the possibility that other immune responses to the proteins occurred. New immune responses may have been generated but not detected by ELISA, as we coated recombinant protein onto plastic wells. Other responses to these proteins, including T-cell-dependent responses, antibodies generated toward epitopes that may be denatured, and IgM antibodies, would be missed.
The observation that the proteins examined are not immunodominant antigens does not preclude the possibility that these proteins could become vaccine candidates. One expects differences between the antibody responses to an antigen generated during natural infection and the responses to immunization with the purified antigen combined with an adjuvant. Antigen-adjuvant formulations could be adjusted to induce an Ig isotype and subclass that would be associated with a protective response.
To summarize the current results, the genome of M. catarrhalis was analyzed, and ORFs predicted to encode putative surface proteins were identified. Conservation of three newly detected genes, msp22, msp75, and msp78, was established by competitive hybridization using a microarray, PCR, and sequencing of the genes in 10 isolates. The proteins encoded by these three genes are expressed by M. catarrhalis in the human respiratory tract. New serum IgG or sputum IgA developed in a proportion of patients following acquisition and clearance of M. catarrhalis. Systemic and mucosal antibody responses occurred independent of one another. The conservation among strains, the expression of the proteins in the human respiratory tract, and the human antibody responses to the proteins indicate that Msp22, Msp75, and Msp78 may make good vaccine antigens. Future work will focus on determining if these proteins are capable of generating protective immune responses.
Acknowledgments
We thank Jeffrey M. Conroy and Devin McQuaid, who assisted with the DNA microarray and the cDNA complementation at the RPCI Microarray and Genomics Facility. The DNA sequence was determined by Michelle Detwiler at the RPCI Biopolymer Facility. We thank Sanjay Sethi for his continued work with the COPD study clinic at the Buffalo Veterans Affairs Medical Center.
This work was supported by NIH grant AI28304 (to T.F.M.) from the National Institutes of Allergy and Infectious Diseases and by the Department of Veterans Affairs.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Adlowitz, D. G., T. Hiltke, A. J. Lesse, and T. F. Murphy. 2004. Identification and characterization of outer membrane proteins G1a and G1b of Moraxella catarrhalis. Vaccine 222533-2540. [DOI] [PubMed] [Google Scholar]
- 2.Adlowitz, D. G., C. Kirkham, S. Sethi, and T. F. Murphy. 2006. Human serum and mucosal antibody responses to outer membrane protein G1b of Moraxella catarrhalis in chronic obstructive pulmonary disease. FEMS Immunol. Med. Microbiol. 46139-146. [DOI] [PubMed] [Google Scholar]
- 3.Adlowitz, D. G., S. Sethi, P. Cullen, B. Adler, and T. F. Murphy. 2005. Human antibody response to outer membrane protein G1a, a lipoprotein of Moraxella catarrhalis. Infect. Immun. 736601-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, J. McCracken, G. H., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 654367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American Academy of Pediatrics. 2004. Diagnosis and management of acute otitis media. Pediatrics 1131451-1465. [DOI] [PubMed] [Google Scholar]
- 7.American Speech-Language-Hearing Association. 2007. Causes of hearing loss in children. American Speech-Language-Hearing Association, Rockville, MD. http:/www.asha.org/public/hearing/disorders/causes.htm.
- 8.American Thoracic Society. 1995. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 152S77-S121. [PubMed] [Google Scholar]
- 9.Bakri, F., A. L. Brauer, S. Sethi, and T. F. Murphy. 2002. Systemic and mucosal antibody response to Moraxella catarrhalis following exacerbations of chronic obstructive pulmonary disease. J. Infect. Dis. 185632-640. [DOI] [PubMed] [Google Scholar]
- 10.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 292607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhushan, R., C. Kirkham, S. Sethi, and T. F. Murphy. 1997. Antigenic characterization and analysis of the human immune response to outer membrane protein E of Branhamella catarrhalis. Infect. Immun. 652668-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bootsma, H. J., P. C. Aerts, G. Posthuma, T. Harmsen, J. Verhoef, H. van Dijk, and F. R. Mooi. 1999. Moraxella (Branhamella) catarrhalis BRO β-lactamase: a lipoprotein of gram-positive origin? J. Bacteriol. 1815090-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brixner, D. I. 2005. Improving acute otitis media outcomes through proper antibiotic use and adherence. Am. J. Manag. Care 11S202-S210. [PubMed] [Google Scholar]
- 14.Cardinale, J. A., and V. L. Clark. 2000. Expression of AniA, the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae, provides protection against killing by normal human sera. Infect. Immun. 684368-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, D., J. C. McMichael, K. R. van der Meid, D. Hahn, T. Mininni, J. Cowell, and J. Eldridge. 1996. Evaluation of purified UspA from Moraxella catarrhalis as a vaccine in a murine model after active immunization. Infect. Immun. 641900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, D., J. C. McMichael, K. R. VanDerMeid, A. W. Masi, E. Bortell, J. D. Caplan, D. N. Chakravarti, and V. L. Barniak. 1999. Evaluation of a 74-kDa transferrin-binding protein from Moraxella (Branhamella) catarrhalis as a vaccine candidate. Vaccine 18109-118. [DOI] [PubMed] [Google Scholar]
- 18.Cullen, P. A., M. Lo, D. M. Bulach, S. J. Cordwell, and B. Adler. 2003. Construction and evaluation of a plasmid vector for the expression of recombinant lipoproteins in Escherichia coli. Plasmid 4918-29. [DOI] [PubMed] [Google Scholar]
- 19.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2953-971. [DOI] [PubMed] [Google Scholar]
- 20.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, and R. G. Kranz. 2006. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61219-231. [DOI] [PubMed] [Google Scholar]
- 21.Forsgren, A., M. Brant, and K. Riesbeck. 2004. Immunization with the truncated adhesin Moraxella catarrhalis immunoglobulin D-binding protein (MID764-913) is protective against M. catarrhalis in a mouse model of pulmonary clearance. J. Infect. Dis. 190352-355. [DOI] [PubMed] [Google Scholar]
- 22.Furihata, K., K. Sato, and H. Matsumoto. 1995. Construction of a combined NotI/SmaI physical and genetic map of Moraxella (Branhamella) catarrhalis strain ATCC25238. Microbiol. Immunol. 39745-751. [DOI] [PubMed] [Google Scholar]
- 23.Gat, O., H. Grosfeld, N. Ariel, I. Inbar, G. Zaide, Y. Broder, A. Zvi, T. Chitlaru, Z. Altboum, D. Stein, S. Cohen, and A. Shafferman. 2006. Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect. Immun. 743987-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geurtsen, J., L. Steeghs, H. J. Hamstra, J. Ten Hove, A. de Haan, B. Kuipers, J. Tommassen, and P. van der Ley. 2006. Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect. Immun. 745574-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Householder, T. C., W. A. Belli, S. Lissenden, J. A. Cole, and V. L. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2547-559. [DOI] [PubMed] [Google Scholar]
- 27.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19823-833. [DOI] [PubMed] [Google Scholar]
- 28.Liu, D. F., J. C. McMichael, and S. M. Baker. 2007. Moraxella catarrhalis outer membrane protein CD elicits antibodies that inhibit CD binding to human mucin and enhance pulmonary clearance of M. catarrhalis in a mouse model. Infect. Immun. 752818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madan Babu, M., and K. Sankaran. 2002. DOLOP—database of bacterial lipoproteins. Bioinformatics 18641-643. [DOI] [PubMed] [Google Scholar]
- 30.Mannino, D. M. 2002. COPD: epidemiology, prevalence, morbidity and mortality, and disease heterogeneity. Chest 121121S-126S. [DOI] [PubMed] [Google Scholar]
- 31.Mannino, D. M., and A. S. Buist. 2007. Global burden of COPD: risk factors, prevalence, and future trends. Lancet 370765-773. [DOI] [PubMed] [Google Scholar]
- 32.Munson, R. S., Jr., and K. Sasaki. 1993. Protein D, a putative immunoglobulin D-binding protein produced by Haemophilus influenzae, is glycerophosphodiester phosphodiesterase. J. Bacteriol. 1754569-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, T. F. 2005. Vaccine development for non-typeable Haemophilus influenzae and Moraxella catarrhalis: progress and challenges. Expert Rev. Vaccines 4843-853. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease. Burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F., C. Kirkham, and A. J. Lesse. 2006. Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect. Immun. 745169-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, T. F., C. Kirkham, D. F. Liu, and S. Sethi. 2003. Human immune response to outer membrane protein CD of Moraxella catarrhalis in adults with chronic obstructive pulmonary disease. Infect. Immun. 711288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy, T. F., J. M. Kyd, A. John, C. Kirkham, and A. W. Cripps. 1998. Enhancement of pulmonary clearance of Moraxella (Branhamella) catarrhalis following immunization with outer membrane protein CD in a mouse model. J. Infect. Dis. 1781667-1675. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, K. T., E. J. Hansen, and M. A. Farinha. 1999. Construction of a genomic map of Moraxella (Branhamella) catarrhalis ATCC 25238 and physical mapping of virulence-associated genes. Can. J. Microbiol. 45299-303. [PubMed] [Google Scholar]
- 39.Nichols, C. E., H. K. Lamb, M. Lockyer, I. G. Charles, S. Pyne, A. R. Hawkins, and D. K. Stammers. 2007. Characterization of Salmonella typhimurium YegS, a putative lipid kinase homologous to eukaryotic sphingosine and diacylglycerol kinases. Proteins 6813-25. [DOI] [PubMed] [Google Scholar]
- 40.Prymula, R., P. Peeters, V. Chrobok, P. Kriz, E. Novakova, E. Kaliskova, I. Kohl, P. Lommel, J. Poolman, J. P. Prieels, and L. Schuerman. 2006. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study. Lancet 367740-748. [DOI] [PubMed] [Google Scholar]
- 41.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reilly, T. J., D. L. Chance, and A. L. Smith. 1999. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J. Bacteriol. 1816797-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethi, S., N. Evans, B. J. B. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347465-471. [DOI] [PubMed] [Google Scholar]
- 44.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000. A state of the art review. Clin. Microbiol. Rev. 14336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sethi, S., C. Wrona, B. J. Grant, and T. F. Murphy. 2004. Strain-specific immune response to Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 169448-453. [DOI] [PubMed] [Google Scholar]
- 46.Shi, L., T. C. Squier, J. M. Zachara, and J. K. Fredrickson. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 6512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigal, L. H., J. M. Zahradnik, P. Lavin, S. J. Patella, G. Bryant, R. Haselby, E. Hilton, M. Kunkel, D. Adler-Klein, T. Doherty, J. Evans, S. E. Malawista, P. J. Molloy, A. L. Seidner, J. R. Sabetta, H. J. Simon, M. S. Klempner, J. Mays, and D. Marks. 1998. A vaccine consisting of recombinant Borrelia burgdorferi outer-surface protein A to prevent Lyme disease. N. Engl. J. Med. 339216-222. [DOI] [PubMed] [Google Scholar]
- 48.Teele, D. W., J. O. Klein, C. Chase, P. Menyuk, B. A. Rosner, and Greater Boston Otitis Media Study Group. 1990. Otitis media in infancy and intellectual ability, school achievement, speech, and language at age 7 years. J. Infect. Dis. 162685-694. [DOI] [PubMed] [Google Scholar]
- 49.Witkowska, D., J. Pietkiewicz, B. Szostko, R. Danielewicz, L. Maslowski, and A. Gamian. 2005. Antibodies against human muscle enolase recognize a 45-kDa bacterial cell wall outer membrane enolase-like protein. FEMS Immunol. Med. Microbiol. 4553-62. [DOI] [PubMed] [Google Scholar]
- 50.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 691593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, H. L., Y. Z. Zhu, J. H. Qin, P. He, X. C. Jiang, G. P. Zhao, and X. K. Guo. 2006. In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans. BMC Genomics 7293. [DOI] [PMC free article] [PubMed] [Google Scholar]