Abstract
In order to track the assembly of murine leukemia virus (MLV), we used fluorescence microscopy to visualize particles containing Gag molecules fused to fluorescent proteins (FPs). Gag-FP chimeras budded from cells to produce fluorescent spots, which passed through the same pore-size filters and sedimented at the same velocity as authentic MLV. N-terminal myristylation of Gag-FPs was necessary for particle formation unless wild-type Gag was coexpressed. By labeling nonmyristylated Gag with yellow FP and wild-type Gag with cyan FP, we could quantitate the coincorporation of two proteins into single particles. This experiment showed that nonmyristylated Gag was incorporated into mixed particles at approximately 50% the efficiency of wild-type Gag. Mutations that inhibit Gag-Gag interactions (K. Alin and S. P. Goff, Virology 216:418-424, 1996; K. Alin and S. P. Goff, Virology 222:339-351, 1996) were then introduced into the capsid (CA) region of Gag-FPs. The mutations P150L and R119C/P133L inhibited fluorescent particle formation by these Gag-FPs, but Gag-FPs containing these mutations could be efficiently incorporated into particles when coexpressed with wild-type Gag. When these mutations were introduced into nonmyristylated Gag-FPs, no incorporation into particles in the presence of wild-type Gag was detected. These data suggest that two independent mechanisms, CA interactions and membrane association following myristylation, cooperate in MLV Gag assembly and budding.
The Gag genes of retroviruses encode the principal structural proteins. The matrix (MA), capsid (CA), and nucleocapsid (NC) domains of the Gag precursor are common to all retroviruses. Retroviruses bud from cellular membranes, where they acquire a lipid bilayer containing the viral envelope. Murine leukemia virus (MLV) Gag and Gag-Pol precursors assemble at the plasma membrane, forming particles visible by electron microscopy. MLV Gag-Pol comprises 5 to 10% of the assembling virion and is generated by infrequent suppression of the Gag termination codon (reviewed in reference 47). Gag-Pol brings the viral protease and enzymes required for viral genome replication into the virion. The protease then cleaves mature MA, CA, and NC proteins from the Gag precursor after particle assembly. The expression of just the Gag coding region results in the production of noninfectious particles (41). However, mutation of the Gag termination codon to express only Gag-Pol prevents particle formation (16). Myristylation of MLV Gag at its N-terminal glycine is required for Gag plasma membrane association (36). A basic region within MA is also involved in membrane targeting (44). Human immunodeficiency virus (HIV) also uses Gag myristylation (18) and sequences within MA (31, 32, 45, 46) to direct HIV Gag to the plasma membrane. Other retroviruses have evolved distinct assembly strategies. For example, Rous sarcoma virus (RSV) Gag assembles at the plasma membrane even though Gag is not myristylated (38), whereas Mason-Pfizer monkey virus (M-PMV) Gag is myristylated but initially assembles in the cytoplasm (37).
MLV Gag self-assembly in cells is at least in part due to interactions between CA domains (19, 20, 22, 40). Assembly can be partially mimicked by membrane-targeted recombinant MLV CA, which forms a hexamer lattice (3) similar to that formed by HIV type 1 (HIV-1) Gag (30). Recombinant HIV-1 or RSV CA can assemble into tubular structures in vitro (17, 23); additional Gag sequences are required to make virus-like particles (7, 17). MLV Gag-Gag interactions have been monitored by the yeast two-hybrid system (24). Point mutations in the N-terminal half of CA perturb MLV Gag-Gag interactions detected by the yeast two-hybrid system (1); some of the same mutations inhibit the assembly and release of particles from cells (2). In HIV-1, the C-terminal region of CA is important for assembly in cells (8, 15, 35). Mutations in CA of RSV can also block assembly, although only in avian cells (13); in general, NC of RSV is thought to be a more critical region for Gag-Gag interactions (5). NC is also involved in HIV-1 Gag assembly (14, 51, 52), at least in part through an interaction with RNA (11).
MLV Gag self-assembly might occur before membrane association or might begin only when Gag is concentrated and orientated by membrane binding. Measurement of nonmyristylated Gag precursor incorporation into particles by wild-type Gag has been used to distinguish between these possibilities. Schultz and Rein reported that nonmyristylated MLV Gag is not incorporated into particles with wild-type Gag, suggesting that the interaction occurs only after membrane association (39). This finding differs from the rescue of non-membrane-associated Gag by wild-type Gag observed with RSV (49) or with M-PMV (37) although the assembly of these viruses differs from that of MLV. For HIV-1, the rescue of nonmyristylated Gag-Pol by wild-type Gag has been reported (33). This experiment may be complicated by reports that reverse transcriptase can be incorporated into MLV or HIV-1 particles in the absence of Gag (6, 10). However, it has also been shown that the inhibition of myristylation allows the formation of HIV-1 particles when the majority of Gag is not myristylated (27).
The object of this study was to gain a better understanding of MLV assembly. To do so, we labeled the MLV Gag precursor by fusion of a fluorescent protein (FP) to NC. This construct allowed us to quantitate Gag self-assembly, as we could observe the number and brightness of fluorescent particles released from transfected cells. We quantitated the effects of membrane association and CA sequences on MLV assembly.
MATERIALS AND METHODS
Cells and Gag particles.
COS-Sp6, HT1080 (ATCC CCL-121), 293T, 293T-MLV-A, and 293T-MoAmpho cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum, penicillin G (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified atmosphere containing 10% CO2. 293T-MLV-A cells are infected with amphotropic MLV strain 1504 (21), and 293T-MoAmphoA cells are infected with a chimeric virus carrying Moloney MLV Gag-Pol and an amphotropic envelope (generous gift from Jean-Christophe Pages, Genethon, France). Briefly, this chimeric virus was generated by transfection of 293T cells with a pBluescript plasmid containing an NheI-SfiI fragment encoding Moloney MLV Gag (GenBank accession number J02255) and an SfiI-ClaI fragment encoding the strain 4070A amphotropic envelope (12).
Particles were produced by transfection of 293T cells with Lipofectamine (Invitrogen) according to the manufacturer's instructions. To produce Gag or Gag-Pol particles, 8 μg of Gag or Gag-Pol plasmids was transfected singly or in 1:1 (wt/wt) admixtures. After 48 h, cell supernatants were passed through a 0.45-μm-pore-size filter and either used for microscopy or concentrated 80-fold by centrifugation at 100,000 × g for 1 h for Western blots or 16-fold by centrifugation at 2,000 × g overnight for velocity gradients. At the same time, cells were monitored by fluorescence-activated cell sorting to check the transfection efficiency and the level of Gag-yellow FP (YFP) expression.
Construction of Gag and Gag-Pol plasmids.
Gag and Gag-Pol plasmids were derived from pHIT60, which encodes Moloney MLV Gag-Pol (43). Gag-FP chimeras were generated by subcloning a SacII-BamHI fragment from pHIT60 encoding the C terminus of Gag into pBluescript SK(+) or pBluescript SK(−) by using the following primers (underlining indicates SacII and Bam HI sites): 5′-TCGGATGGCCGCGGGACGGCACCTTTAACCGAGACC-3′ and 5′-TAGCGGATCCTGCCGCCGCCGTCATCTAGGGTCAGGAGG-3′. Then, BamHI-EcoRV fragments encoding YFP or cyan FP (CFP) variants of the Aequorea victoria green fluorescent protein (GFP) (48) were inserted by using the following primers (underlining indicates Bam HI and Eco RV sites and lowercase letters indicate unique NaeI and NheI sites): 5′-ATTGCGGATCCGGCGGCGGTGGAgctagcAAGGGCGAGGAGCTGTTCACCGG-3′ and 5′-TTCCGATATCgccggcTTACTTGTACAGCTCGTCCATGCCGAGAGTGAT-3′. This fusion suppressed the stop codon of Gag NC and the FP start codon, inserted a G4SG4 linker between the Gag and FP coding sequences, and generated unique NaeI and NheI sites at the ends of the FP coding sequence. The NC protease cleavage site located upstream of the TAG amber codon was maintained. Then, an XhoI-EcoRV fragment was inserted into pHIT60 digested with XhoI-PmlI.
Mutants were generated from Gag-YFP or wild-type Gag-Pol by using a QuickChange site-directed mutagenesis kit (Stratagene) and the following primers (bold type indicates mutations): G2A-Gag, 5′-TTGTCTGAGAATATGGCCCAGACTGTTACCACTCC-3′; Gag-Pol-RT, 5′-TGACCCTAGATGACCAGGGAGGTCAGGGTCAGG-3′; P102S, 5′-CCCCTCGAGCGCTCAGACTGGGATTACACC-3′; P150L, 5′-GGGCCCAATGAGTCTCTCTCGGCCTTCCTAGAG-3′; R119C, 5′-CCTAGTCCACTATTGCCAGTTGCTCCTAGCG-3′; and P133L, 5′-GCGGGCAGAAGCCTCACCAATTTGGCC-3′. The P133L mutation was generated in plasmids with the R119C mutation.
Gag-STOP and G2A-Gag-STOP chimeras were generated by subcloning an XhoI-BamHI fragment from pHIT60 encoding the C terminus of Gag into Gag-YFP and G2A-Gag-YFP by using the following primers: 5′-TTCCCCTCGAGCGCCCAGACTGGGATTACACC-3′ and 5′-GCCGGATCCTCCGCCGCCctaGTCATCTAGGGTCAGG-3′.
pCNCeGFP was derived from pHIT111 (43) by replacement of a KpnI fragment with a KpnI fragment from pLNCX (26). The enhanced GFP from pEGFP-N1 (Clontech) was then excised with XmaI-HpaI and inserted into the HpaI site.
Immunoblotting.
Cell proteins were extracted in radioimmunoprecipitation buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 8.0]) with protease inhibitor cocktail (Boehringer Ingelheim, Bracknell, United Kingdom) for 15 min on ice. Insoluble material was pelleted at 10,000 × g for 30 min at 4°C, and then protein in the supernatant was measured (protein quantification assay; Bio-Rad, Hemel Hempstead, United Kingdom). A total of 15 to 30 μg of cell extract or 10 μl of concentrated cell supernatant (see above) was separated by SDS-10% polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (enhanced chemiluminescence nitrocellulose membranes; Amersham, Little Chalfont, United Kingdom). Blots were blocked overnight in phosphate-buffered saline containing 5% nonfat milk and then probed for 1 h in phosphate-buffered saline- 0.1% Tween 20- 5% milk with goat anti-Rauscher leukemia virus (RLV) P30 (1:2,500 dilution; Quality Biotech Inc.) or mouse anti-GFP (1:1,000 dilution; antibody JL-8; Clontech) and horseradish peroxidase-conjugated secondary antibodies (1:2,500; Harlan). Enhanced chemiluminescence (kit from Amersham) was used to develop the blots.
Sedimentation velocity.
Continuous linear sucrose gradients (5 ml) were poured with a gradient maker (BioComp Instruments, Fredericton, New Brunswick, Canada); 5 and 20% sucrose solutions in 50 mM sodium phosphate buffer (pH 7.4) containing 2 mM dithiothreitol, 20 μg of aprotinin/ml, and 2 μg of leupeptin/ml were used. Gradients were overlaid with 400 μl of concentrated virus and centrifuged at 50,000 × g for 1 h at 4°C in a Sorvall AH-650 rotor. Twelve fractions (0.4 ml each) were collected from the bottom of the tubes. For Western blot analysis, 300 μl of each fraction was diluted in 1.2 ml of ice-cold 50 mM sodium phosphate buffer (pH 7.4) with 2 μg of bovine serum albumin (Promega) and 10% trichloroacetic acid. Fractions were incubated at −20°C for 2 h and then centrifuged at 10,000 × g for 30 min at 4°C. The pellets were washed once in ice-cold 80% acetone and then resuspended in 20 μl of SDS loading buffer (0.5 M Tris-HCl [pH 6.8], 1% SDS, 10% glycerol, 0.1% bromophenol blue, 1 mM EDTA, 10 mM dithiothreitol, 20 μg of aprotinin/ml, 2 μg of leupeptin/ml, and 10 μg of phenylmethylsulfonyl fluoride/ml). The pH was adjusted to 7, and samples were analyzed by immunoblotting.
Immunostaining of particles.
Immunostaining was performed as described previously (34). Briefly, 50 μl of supernatant was incubated with 8 μg of Polybrene (Sigma, Poole, United Kingdom)/ml on slides for 90 min at 37°C. Dried particles were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 for 15 min at room temperature, and then washed three times with Hanks' balanced salt solution (HBSS; Invitrogen). Samples were incubated for 1 h at room temperature with goat anti-RLV (1:2,000 dilution in 2% donkey serum), washed three times with HBSS, and then incubated for 1 h with Texas red-conjugated donkey anti-goat immunoglobulin G (1:250; Jackson ImmunoResearch, West Grove, Pa.). After three washes with HBSS, the slides were mounted (mounting medium; Dako, Carpinteria, Calif.) and observed by confocal microscopy (MRC 1024 microscope; Bio-Rad, Hercules, Calif.) equipped with a krypton-argon laser. All images were captured by using Kalman filtration and analyzed with Lasersharp software (Bio-Rad). For size estimation, supernatants were sequentially passed through 450-nm (Sartorius, VivaScience, Göttingen, Germany), 200-nm (Sartorius), 100-nm (Whatman, Kent, United Kingdom), and 20-nm (Whatman) filters. After each filtration, 50-μl aliquots were analyzed as described above.
Particle fluorescence.
Two milliliters of supernatant from cells transfected with fluorescent chimeras were filtered (0.45-μm-pore-size filters) and then incubated on glass slides for 90 min at 37°C with 8 μg of Polybrene/ml. The slides were washed twice with HBSS, and the particles were fixed with 4% paraformaldehyde. Images were captured either by confocal microscopy (MRC 1024 microscope equipped with a krypton-argon laser) or by epifluorescence microscopy (Zeiss microscope equipped with a monochromator [Polychrome II; Photonics, Planegg, Germany] and a charge-coupled device [CCD] camera [Princeton Instruments CCD 800; Roper Scientific, Trenton, N.J.]). For YFP and CFP measurements, filters for excitation at 510 and 434 nm and filters for emission at 568 ± 50 and 470 ± 30 nm (mean and range), respectively, were used, and the images were analyzed with Metamorph software (Universal Imaging Corporation, West Chester, Pa.).
Fluorescent particle images were acquired by confocal microscopy (magnification, ×63), and light objects in four fields per sample were counted with Metamorph software. To measure the fluorescence intensity of individual particles, an area of 8 by 8 pixels was drawn around 20 particles, and the value of each pixel was measured with Metamorph software. The fluorescence intensity was designated as follows: particle fluorescence = sum of six highest pixel values − (six × mode for pixel values). This approximation can be used because six or fewer pixels correspond to the peak of fluorescence in each case; the mode represents the local background. For measurements of dual YFP and CFP, areas corresponding to particles on the CFP images were chosen and transferred to the YFP images. For each area, the same six peak pixels were used for the quantitation of YFP and CFP incorporation.
Electron microscopy.
Transfection of COS-Sp6 cells was carried out by electroporation of 30 μg of Gag-YFP plasmid in a buffer containing 20 mM HEPES (pH 7.05),137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM d-glucose with a Gene Pulser at 350 V, 1,250 μF, and infinite ohms (Bio-Rad, Richmond, Calif.). After 2 days, the cells were fixed with 2% paraformaldehyde-1.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for 20 min at room temperature. The cells were then postfixed with 1% osmium tetroxide- 1.5% potassium ferricyanide and treated with tannic acid as described previously (42). After stepwise dehydration in a series of increasing concentrations of ethanol, the cells were removed from the culture dishes by the addition of propylene oxide and then pelleted. Fresh propylene oxide was added to the pellets, which were allowed to stand for 10 min at room temperature. After 1 h of incubation in a 50:50 mixture of propylene oxide and Epon, the pellets were transferred to neat Epon (2 × 2 incubations of 2 h), placed into trays, and polymerized overnight at 70°C. Sections of 60 nm were cut by using an Ultracut E microtome (Reichert-Jung, Vienna, Austria). Sections were stained with lead citrate and viewed by transmission electron microscopy (EM420 microscope; Philips, Eindhoven, The Netherlands).
Infection assay.
To produce infectious viruses, a total of 4 μg of Gag or Gag-Pol plasmids was cotransfected with 2 μg of pCNCeGFP (see above) and 2 μg of pMDG, expressing the vesicular stomatitis virus G protein (28). After 2 days, supernatants were harvested, filtered (0.45-μm-pore-size filters), and concentrated 16-fold by centrifugation at 2,000 × g overnight at 4°C. To determine titers, a total of 5 × 104 HT1080 cells were infected with serial dilutions of the virus plus 8 μg of Polybrene/ml. Infected cells were counted after 6 days by detection of enhanced GFP expression with a FACScan and CellQuest software (Becton Dickinson, Franklin Lakes, N.J.).
RESULTS
Tagging of MLV Gag with YFP.
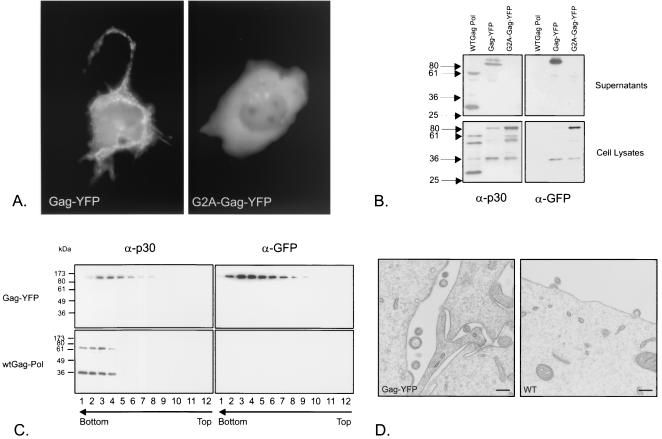
To construct a fluorescent chimera of the Moloney MLV Gag precursor, a yellow variant of A. victoria GFP (YFP) was fused to the C terminus of NC. When a plasmid expressing the chimera was transfected into COS-Sp6 cells, the Gag-YFP fusion was expressed in the cytoplasm and also concentrated at the cell membrane (Fig. 1A). A mutant Gag-YFP where glycine 2 was changed to alanine (G2A-Gag-YFP) lacking the glycine required for the attachment of a myristyl group, which is required for the membrane association of Gag (36), was also constructed. As predicted, G2A-Gag-YFP was expressed in the cytoplasm (Fig. 1A). Immunoblot analysis with an anti-GFP antibody showed that the chimeras were approximately 95 kDa, corresponding to the MLV Gag polyprotein (65 kDa) fused to YFP (30 kDa). They were expressed at levels similar to that of wild-type MLV Gag. Like wild-type MLV Gag, Gag-YFP was detected mostly in cell supernatants, whereas G2A-Gag-YFP was retained in cells (Fig. 1B). The sedimentation velocity of secreted Gag-YFP was slightly lower than that of authentic MLV (Fig. 1C) but faster than that of the soluble protein, demonstrating particle formation. These Gag-YFP particles could be detected by electron microscopy, like wild-type MLV (Fig. 1D). A comparison of particle diameters showed that Gag-YFP particles had a diameter (mean and standard deviation) of 136 ± 27 nm; the diameter for wild-type MLV was 107 ± 16 nm. The diameter of the Gag-YFP particles was compatible with their lower sedimentation velocity. We conclude that the fusion of YFP to the end of NC does not perturb Gag assembly, although the particles formed are slightly larger than those of wild-type MLV.
FIG. 1.
Gag-YFP expression and budding. (A) COS-Sp6 cells were transfected with either Gag-YFP or G2A-Gag-YFP expression plasmids and photographed after 48 h with a fluorescensce microscope, CCD camera, and Metamorph software. The exposure times were 1 s and 150 ms for Gag-YFP- and G2A-Gag-YFP-expressing cells, respectively. (B) Immunoblot of lysates and supernatants of COS-Sp6 cells transfected with MLV Gag-Pol (WTGag Pol), Gag-YFP, or G2A-Gag-YFP expression plasmids and probed with anti-CA (α-p30) or anti-GFP (α-GFP) antibodies. Numbers at left indicate kilodaltons. (C) Sedimentation velocity analysis of supernatants of Gag-YFP-transfected 293T cells by immunoblotting with anti-CA or anti-GFP antibodies. Arrows indicate the direction of the gradient from the lowest (top) to the highest (bottom) density. wt, wild type. (D) Cos-Sp6 cells were transfected with Gag-YFP or MLV Gag-Pol (WT) expression plasmids and processed for electron microscopy. Bars, 200 nm. No virus-like structures were detected in mock-transfected cells.
Direct visualization of fluorescent particles.
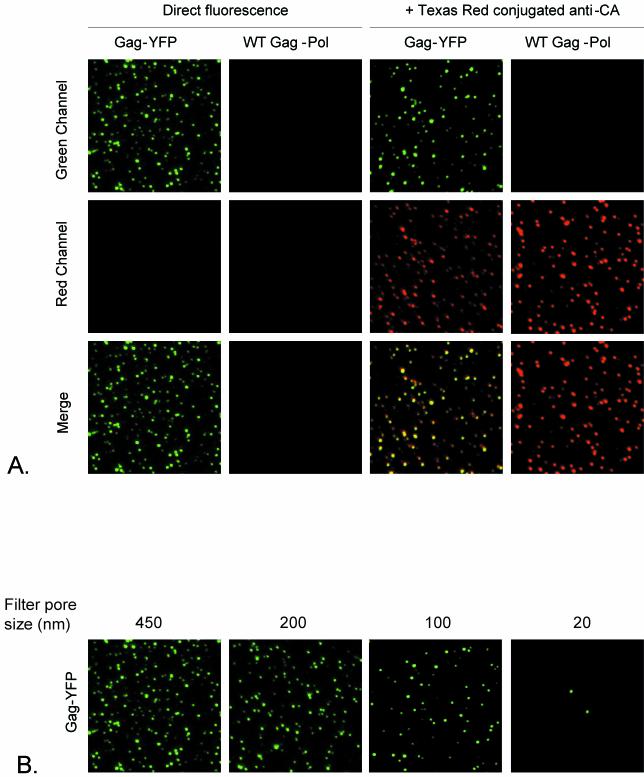
To test whether the Gag-YFP fusion could generate fluorescent particles that could be visualized directly by microscopy, supernatants of cells transfected with the Gag-YFP fusion construct were fixed on glass slides. As shown in Fig. 2A, fluorescent Gag-YFP spots could be visualized in the green channel. This YFP fluorescence was colocalized mostly with MLV Gag, detected with anti-CA antibodies (Fig. 2A), showing that many spots contain both YFP and MLV CA. To examine whether these fluorescent spots represent single virus particles, we estimated their sizes by serial passage through filters of decreasing pore sizes. Removal was 70% following filtration through the 100-nm filter and almost complete after filtration through the 20-nm filter (Fig. 2B). These results are consistent with the sizes of MLV particles estimated by electron microscopy (29) and by filtration of immunostained retrovirus particles (34).
FIG. 2.
Visualization of Gag-YFP fluorescent particles. (A) Supernatants of 293T cells transfected with either Gag-YFP or MLV Gag-Pol (WT Gag-Pol) were fixed with 4% paraformaldehyde on glass slides and either directly visualized by confocal microscopy or further processed for immunostaining as described in Materials and Methods. Images for green and red fluorescence were acquired separately and merged. (B) Estimation of the sizes of Gag-YFP fluorescent particles. Supernatants were sequentially filtered through 450-, 200-, 100-, and 20-nm-pore-size filters. After each filtration, 50-μl aliquots were fixed on glass slides and visualized by confocal microscopy.
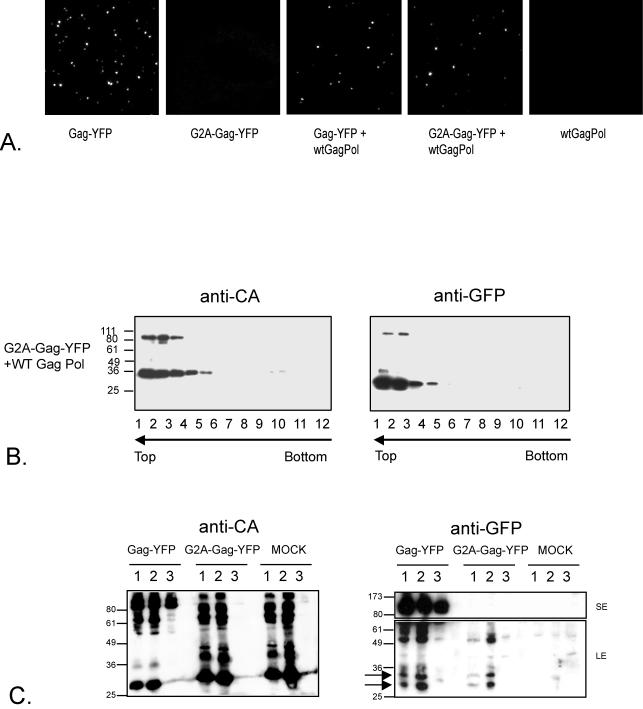
G2A-Gag-YFP is incorporated into particles with wild-type Gag.
G2A-Gag-YFP did not release fluorescent particles into the medium, as would be predicted (Fig. 3A). However, transfection of a mixture of G2A-Gag-YFP and wild-type Gag-Pol led to the release of fluorescent particles into the supernatants of transfected cells (Fig. 3A). The fluorescent spots represented single particles, as judged by filtration (data not shown). The rescued G2A-Gag-YFP polyprotein and smaller proteins corresponding in size to an NC-YFP fusion and YFP cleaved from the precursor by the MLV protease, cosedimented with the wild-type Gag-Pol polyproteins (Fig. 3B). These results demonstrate that nonmyristylated MLV Gag is incorporated into particles in the presence of wild-type Gag, in contrast to a previous report (39). Incorporation of MLV G2A-Gag-YFP into particles was not observed during cotransfection with Gag-Pol expression constructs from the gammaretroviruses PERV-B clone 17 (4; B. Bartosch and Y. Takeuchi, unpublished data) and GALV SEATO strain (M. Bock and J. P. Stoye, unpublished data) or with HIV-1 (28) (data not shown).
FIG. 3.
G2A-Gag-YFP rescue by MLV Gag-Pol. (A) Supernatants of 293T cells transfected with the indicated constructs were fixed on glass slides, and the fluorescent particles were visualized by confocal microscopy. wt, wild type. (B) Supernatants of 293T cells cotransfected with G2A-Gag-YFP and MLV Gag-Pol (WT Gag Pol) were analyzed as described in the legend to Fig. 1C. (C) Supernatants of 293T-MoAmpho (lanes 1), 293T-MLV-A (lanes 2), and 293T (lanes 3) cells, either nontransfected (MOCK) or transfected with Gag-YFP (1:10 diluted) or G2A-Gag-YFP, were harvested, concentrated, and analyzed by immunoblotting with anti-CA or anti-GFP antibodies. A short exposure (SE, 1 min) for the top of the blot and a long exposure (LE, 15 min) for the bottom of the blot are shown. Numbers at left in panels B and C indicate kilodaltons.
The previous study of Schultz and Rein (39) demonstrated that amphotropic MLV failed to incorporate nonmyristylated Moloney MLV Gag. We therefore examined whether our nonmyristylated Moloney MLV Gag-YFP could also be incorporated into amphotropic MLV particles. To do this, 293T cells were chronically infected either with amphotropic MLV (21) or with a chimeric virus carrying ecotropic MLV Gag-Pol and an amphotropic envelope (see Materials and Methods for details). Cells were transfected with G2A-Gag-YFP and, after 2 days, virions were pelleted and analyzed by immunoblotting with anti-CA or anti-GFP antibodies. A diagnostic pair of GFP products cleaved from G2A-Gag-YFP could be observed in both the amphotropic virus and the ecotropic virions (Fig. 3C). Note that 10-fold-more concentrated supernatant was loaded in the G2A-Gag-YFP and mock transfection lanes of Fig. 3C. This relatively insensitive assay is similar to that of Schultz and Rein (39), who used amphotropic MLV to attempt to rescue Moloney MLV G2A-Gag from a transfected cell line.
Measuring the efficiency of G2A-Gag-YFP incorporation.
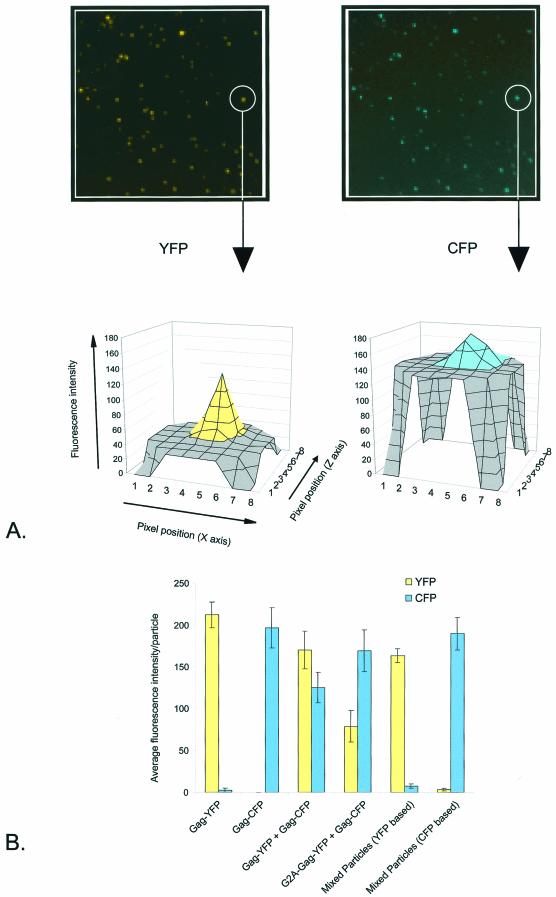
We measured the efficiency of G2A-Gag-YFP incorporation into particles by coexpressing it with wild-type Gag-CFP and then quantitating YFP and CFP in single spots. The method is shown in Fig. 4A. An area of 8 by 8 pixels was drawn around spots on the CFP images and transposed to the YFP images. The sums of the peak pixels for CFP and YFP were then calculated. On average, the ratio of fluorescence in particles released from G2A-Gag-YFP and Gag-CFP cotransfections was 30% YFP to 70% CFP (Fig. 4B). The fluorescence ratio for control particles released from wild-type Gag-YFP and Gag-CFP cotransfections was 60% YFP to 40% CFP. When Gag-YFP and Gag-CFP were transfected separately and the released particles were mixed as a control, coincident fluorescence was not detected (Fig. 4B). We therefore estimated that the loss of the myristylation site reduced G2A-Gag-YFP incorporation to 50% that of wild-type Gag-YFP. In this interpretation, we assumed that the total fluorescence was proportional to the number of FPs. This assumption is not unreasonable, as CCD detection of GFP molecules coupled to beads is linear for the range of 1 to 104 proteins per bead (9).
FIG.4.
Relative efficiencies of Gag-YFP and G2A-Gag-YFP incorporation into particles. (A) Supernatants of 293T cells transfected with Gag-YFP and Gag-CFP were fixed on glass slides and visualized by fluorescence microscopy with a CCD camera as described in Materials and Methods. YFP and CFP signals in the same area were collected at separate settings. An area of 8 by 8 pixels was drawn around an individual particle on the CFP image (unless otherwise stated) and transposed to the YFP image. The value of each pixel in this example was plotted by using Metamorph software. (B) YFP versus CFP incorporation into individual particles, determined as described in Material and Methods. Data are means and standard errors for 20 particle measurements.
Infection by particles incorporating G2A-Gag and G2A-Gag-Pol.
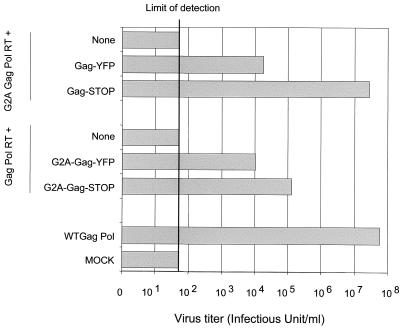
Studies of HIV-1 have shown that a nonmyristylated Gag-Pol precursor can be rescued by wild-type Gag (33). We performed a similar experiment for MLV by mutating the amber termination codon in MLV Gag-Pol to construct G2A-Gag-Pol readthrough (G2A-Gag-Pol-RT) (16). As expected, G2A-Gag-Pol-RT produced no infectious particles above the limit of detection (Fig. 5). However, when Gag precursors were coexpressed with G2A-Gag-Pol-RT, the titer was restored. The titer was restored to the wild-type level when NC of the Gag precursor was not fused to YFP (G2A-Gag-Pol-RT plus wild-type Gag-STOP). The NC-YFP fusion (G2A-Gag-Pol-RT plus Gag-YFP) decreased the titer by over 1,000-fold (Fig. 5). We also examined whether nonmyristylated Gag could be rescued by Gag-Pol-RT, a more stringent assay, as Gag must be incorporated in excess of Gag-Pol. Figure 5 shows that the titer was restored to some extent when NC was not fused to YFP (Gag-Pol-RT plus G2A-Gag-STOP). The NC-YFP fusion (Gag-Pol-RT plus G2A-Gag-YFP) again decreased the titer. This comparison was performed by transfecting 2 μg of the Gag-Pol-RT construct with 2 μg of the Gag construct. Clearly, the alteration of the ratio of Gag-Pol to Gag might improve infectivity; however, the comparison is valid at a single ratio. These data demonstrate that, as in HIV-1 (33), the coexpression of Gag with nonmyristylated Gag-Pol leads to the production of infectious particles. They also show that NC-YFP is poorly functional.
FIG. 5.
Infection assay with Gag mutants. Supernatants of 293T cells nontransfected (MOCK) or transfected with the indicated Gag constructs together with vector pCNCeGFP and a vesicular stomatitis virus G protein-expressing plasmid were assayed for their ability to infect HT1080 cells as described in Materials and Methods. WT, wild type.
Capsid mutations which prevent G2A-Gag incorporation.
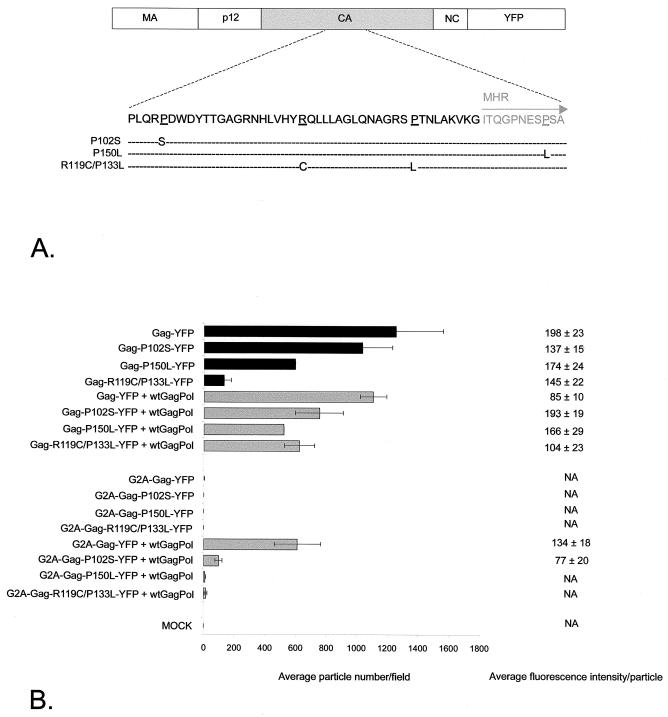
The yeast two-hybrid system was used previously (1, 24) to measure MLV Gag multimerization. These investigators showed that the CA region was critical for Gag-Gag interactions. A P102S mutant could interact with wild-type Gag but not itself, whereas a P150L mutant was capable of a homotypic interaction but could not bind wild-type Gag. Alin and Goff examined the effect of CA mutations on an infectious MLV clone (2). A P102S mutant was able to assemble and bud but was unable to reverse transcribe, whereas a P150L mutant showed a lower level of virion release in some cells and an R119C/P133L mutant was unable to release virions. Our assay allows the effect of mutations on viral particle assembly to be quantitated. We therefore generated these mutations within CA of the Gag-YFP fusion constructs (Fig. 6A). The P102S mutation did not affect the assembly of fluorescent particles, whereas Gag-YFP with the P150L mutation showed reduced assembly and Gag-YFP with the R119C/P133L mutation failed to assemble. However, when the R119C/P133L CA mutant was cotransfected with wild-type Gag-Pol, the number and intensity of fluorescent spots released were similar to those observed with wild-type Gag-YFP (Fig. 6B). We next examined the ability of wild-type Gag-Pol to rescue G2A variants of CA mutants. The G2A-P102S-YFP mutant showed reduced incorporation by wild-type Gag-Pol, and the G2A-P150L-YFP and G2A-R119C/P133L-YFP mutants were not incorporated (Fig. 6B). These data confirm that an intact CA sequence is important for particle assembly. Mutations in CA have a greater impact on assembly when membrane association is prevented.
FIG. 6.
Incorporation of CA mutants. (A) Positions of mutations within CA. (B) Average number of fluorescent particles per field from 293T cells transfected with the constructs shown, as detected by confocal microscopy. Data are means for four fields in one experiment and means and standard errors for at least two separate experiments. wt, wild type; MOCK, mock transfected. The average fluorescence intensity per particle was determined as described in Materials and Methods. Data are means and standard errors for 20 individual particle measurements in a representative experiment. NA, not applicable.
DISCUSSION
Fluorescent protein tagging has allowed us to use microscopy to observe particles produced by MLV Gag and to permit quantitation of the number of particles and measurement of particle fluorescence, which we assume is proportional to the number of fluorescent Gag molecules incorporated. We have used this method to show that nonmyristylated Gag is incorporated into particles in the presence of wild-type Gag at approximately 50% wild-type efficiency. We confirmed our conclusions by infectious rescue assays, showing, for example, that nonmyristylated Gag can form infectious particles with Gag-Pol. This result contrasts that of Schultz and Rein (39), who reported that nonmyristylated Gag was not incorporated into wild-type particles. They used wild-type amphotropic MLV to attempt to rescue nonmyristylated Moloney MLV Gag, which could be detected by a specific antibody. We performed a similar experiment using infectious viruses carrying amphotropic (21) or Moloney MLV Gag to rescue nonmyristylated ecotropic Gag tagged with YFP and detected by an anti-GFP antibody. We observed rescue by both viruses, although the sensitivity of detection of Gag rescue was much lower than that of detection of fluorescent particles, perhaps explaining the negative result of Schultz and Rein (39).
Because the fluorescence microscopy method allowed quantitation of the assembly step, we went on to reexamine two point mutations and one double mutation in MLV CA which had been reported to affect Gag-Gag interactions in the yeast two-hybrid system or in viral assembly (1, 2). As previously described, the P102S mutation did not affect Gag particle formation (2). The P150L mutation reduced particle numbers; in the context of the virus, this mutation was reported to decrease virion release in some assays (2). Finally, the R119C/P133L double mutation, which prevented the release of virions (2), also inhibited Gag particle formation. We next showed that the R119C/P133L mutation could be incorporated into particles in the presence of wild-type Gag-Pol. When the P102S, P105L, or R119C/P133L mutations were introduced into nonmyristylated Gag, rescue by wild-type Gag-Pol was inhibited in each instance. The P102S mutant showed partial rescue, and the P150L and R119C/P133L mutants were not rescued.
A simple model to explain these data is that two processes, Gag-Gag interactions via CA and the membrane association of Gag, cooperate to facilitate Gag assembly. Thus, mutations that affect CA-CA interactions have a strong effect on assembly when membrane association is inhibited. An experiment supporting this model was described by Bennett and Wills (4). They showed that substitution of the same heterologous myristylation membrane-targeting signal in MLV Gag and RSV Gag allowed the two precursors to coassemble. Thus, Gag molecules that interact only weakly can form particles when efficiently targeted to the same location. Clearly, other domains in Gag are also critical for assembly and release. For example an intact L domain in p12 is required for optimal release (50). MLV NC may also play a role in assembly, as in RSV-MLV hybrid Gag molecules (5).
Quantitation of fluorescent particles provides a rapid assay for the efficiency of incorporation of viral proteins into individual virions. For MLV, envelope incorporation also can be monitored, as FPs can be inserted at the N terminus of gp70 at a point that tolerates an additional domain (12). Fluorescent Gag-FP MLV particles also may be used to track infection, as recently described for HIV-1 carrying a Vpr-FP fusion (25).
Acknowledgments
We thank Charan Koka for generation of the G2A-Gag-YFP chimera, Ian Gerrard for DNA sequencing, and Ariberto Fassati for critical reading of the manuscript.
M.A. is supported by a Marie Curie fellowship. The BBSRC funded part of this work through the Bioimaging Initiative.
REFERENCES
- 1.Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. [DOI] [PubMed] [Google Scholar]
- 2.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 3.Barklis, E., J. McDermott, S. Wilkens, E. Schabtach, M. F. Schmid, S. Fuller, S. Karanjia, Z. Love, R. Jones, Y. Rui, X. Zhao, and D. Thompson. 1997. Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 16:1199-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, R. P., and J. W. Wills. 1999. Conditions for copackaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J. Virol. 73:2045-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchschacher, G. L., L. Yu, F. Murai, T. Friedman, and A. Miyanohara. 1999. Association of murine leukemia virus Pol with virions, independent of Gag-Pol expression. J. Virol. 73:9632-9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carriere, C., B. Gay, N. Chazal, N. Morin, and P. Boulanger. 1995. Sequence requirements for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precursor into retrovirus-like particles. J. Virol. 69:2366-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, C. S., E. Kartalov, M. Unger, S. Quake, and H. A. Lester. 2001. Single-molecule measurements calibrate green fluorescent protein surface densities on transparent beads for use with ′knock-in' animals and other expression systems. J. Neurosci. Methods 105:55-63. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, H. C., F. D. Wang, S. Y. Yao, and C. T. Wang. 2002. Effects of gag mutations on human immunodeficiency virus type 1 particle assembly, processing, and cyclophilin A incorporation. J. Med. Virol. 68:156-163. [DOI] [PubMed] [Google Scholar]
- 11.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosset, F. L., F. J. Morling, Y. Takeuchi, R. A. Weiss, M. K. Collins, and S. J. Russell. 1995. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol. 69:6314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 15.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, K. M., and S. P. Goff. 1988. Expression of the Gag-Pol fusion protein of Moloney murine leukemia virus without the Gag protein does not induce virion formation or proteolytic processing. J. Virol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 18.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5778-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, M., L. Jelinek, S. Whiting, and E. Barklis. 1990. Transport and assembly of Gag proteins into Moloney murine leukemia virus. J. Virol. 64:5306-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, M. S., and E. Barklis. 1995. Structural interactions between retroviral Gag proteins examined by cysteine cross-linking. J. Virol. 69:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartley, J. W., and W. P. Rowe. 1976. Naturally occurring murine leukemia viruses in wild mice: characterization of a new “amphotropic” class. J. Virol. 19:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T. A., G. Blaug, M. Hansen, and E. Barklis. 1990. Assembly of Gag- β-galactosidase proteins into retrovirus particles. J. Virol. 64:2265-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Struct. Fold Des. 8:617-628. [DOI] [PubMed] [Google Scholar]
- 24.Luban, J., K. B. Alin, K. L. Bossolt, T. Humaran, and S. P. Goff. 1992. Genetic assay for multimerization of retroviral Gag polyproteins. J. Virol. 66:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982, 984-986, 989-990. [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 28.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 29.Nermut, M. V., H. Frank, and W. Schafer. 1972. Properties of mouse leukemia viruses. 3. Electron microscopic appearance as revealed after conventional preparation techniques as well as freeze-drying and freeze-etching. Virology 49:345-358. [DOI] [PubMed] [Google Scholar]
- 30.Nermut, M. V., D. J. Hockley, J. B. Jowett, I. M. Jones, M. Garreau, and D. Thomas. 1994. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology 198:288-296. [DOI] [PubMed] [Google Scholar]
- 31.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paillart, J. C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 73:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultz, A. M., and S. Oroszlan. 1983. In vivo modification of retroviral gag gene-encoded polyproteins by myristic acid. J. Virol. 46:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz, A. M., and A. Rein. 1989. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J. Virol. 63:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartzberg, P., J. Colicelli, M. L. Gordon, and S. P. Goff. 1984. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J. Virol. 49:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shields, A., W. N. Witte, E. Rothenberg, and D. Baltimore. 1978. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell 14:601-609. [DOI] [PubMed] [Google Scholar]
- 42.Simionescu, N., and M. Simionescu. 1976. Galloylglucoses of low molecular weight as mordant in electron microscopy. II. The moiety and functional groups possibly involved in the mordanting effect. J. Cell Biol. 70:622-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soneoka, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Mutagenesis analysis of the murine leukemia virus matrix protein: identification of regions important for membrane localization and intracellular transport. J. Virol. 71:5549-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanstrom, R., and J. W. Wills. 1997. Synthesis, processing and assembly of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 48.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 49.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]