Abstract
The ability to bind extracellular matrix proteins is a critical virulence determinant for skin pathogens. Haemophilus ducreyi, the etiological agent of the genital ulcer disease chancroid, binds extracellular matrix components, including fibronectin (FN). We investigated H. ducreyi FN binding and report several important findings about this interaction. First, FN binding by H. ducreyi was greatly increased in bacteria grown on heme and almost completely inhibited by hemoglobin. Second, wild-type strain 35000HP bound significantly more FN than did a dsrA mutant in two different FN binding assays. Third, the expression of dsrA in the dsrA mutant restored FN binding and conferred the ability to bind FN to a non-FN-binding Haemophilus influenzae strain. Fourth, an anti-DsrA monoclonal antibody partially blocked FN binding by H. ducreyi. The hemoglobin receptor, the collagen-binding protein, the H. ducreyi lectin, the fine-tangle pili, and the outer membrane protein OmpA2 were not involved in H. ducreyi FN binding, since single mutants bound FN as well as the parent strain did. However, the major outer membrane protein may have a minor role in FN binding by H. ducreyi, since a double dsrA momp mutant bound less FN than did the single dsrA mutant. Finally, despite major sequence differences, DsrA proteins from both class I and class II H. ducreyi strains mediated FN and vitronectin binding. We concluded that DsrA is the major factor involved in FN binding by both classes of H. ducreyi strains.
Haemophilus ducreyi is the cause of the sexually transmitted ulcer disease chancroid (3, 32, 33, 39, 46). H. ducreyi is thought to initiate infection by entering the skin through small abrasions acquired during sexual intercourse (3, 33, 41). Thus, attachment of H. ducreyi to extracellular matrix (ECM) proteins of the skin may be an important initial step in the infection process, as shown for other bacterial skin infections (8, 15, 37).
Little is known about the mechanisms employed by H. ducreyi in the first critical steps of attaching to host skin. In vitro, H. ducreyi binds to fibrinogen, fibronectin (FN), type I and III collagen, gelatin, and laminin, but not elastin (2, 10). FN may have a role in the interaction of H. ducreyi with foreskin fibroblasts (4). During the later pustule stages of human experimental chancroid infection, H. ducreyi localizes with inflammatory infiltrates present in pustules and colocalizes with collagen and fibrin (9, 11). In naturally acquired chancroid, where patients are seen mostly at the last ulcerative stage, H. ducreyi colocalizes with neutrophils and fibrin (12). These data suggest that ECM binding by H. ducreyi may contribute to chancroid pathogenesis.
Bacterial Oca (oligomeric coiled adhesion) proteins (25), also referred to as trimeric autotransporters (17), comprise a multifunctional family with demonstrated roles in binding to ECM proteins, binding to various eukaryotic cells, mediating invasion of cells, and resistance to killing by serum complement (21). Oca proteins are expressed on the bacterial surface of the cell as homotrimers exhibiting a lollipop shape, with head, neck, and stalk structures (21, 25). Different Oca family members share similarity in the C-terminal domain, the region that is believed to traverse and anchor the protein in the bacterial outer membrane (25). However, Oca N-terminal domains vary widely, even among family members expressed by closely related species or within a species (25).
H. ducreyi expresses two Oca proteins, namely, DsrA (ducreyi serum resistance A) and NcaA (necessary for collagen adhesion A). Both were identified by their C-terminal similarity to the prototypical Oca family protein YadA. DsrA was first described as critical for high-level resistance to killing by normal human serum (20) and was later shown to be an adhesin for HaCat keratinocytes and to bind to vitronectin (VN) (16). DsrA is also essential for H. ducreyi infection in the human challenge model of chancroid (13). NcaA mediates type I collagen binding by H. ducreyi and is also important for virulence of H. ducreyi in both the porcine and human models of chancroid (22).
Recently, we identified two classes of H. ducreyi strains, classes I and II (47), based on the presence of variant forms of several outer membrane components, including DsrA, NcaA, DltA (ducreyi lectin A), and lipooligosaccharide (LOS). The DsrA proteins from the two classes of H. ducreyi strains have almost identical C-terminal domains, sharing 88.5% identity in the last 86 amino acids. However, these proteins have divergent N-terminal domains, sharing little amino acid sequence similarity (47).
FNs are high-molecular-weight disulfide-linked glycoprotein dimers found in both soluble and insoluble forms in human blood and skin. The modular structure of FN promotes its involvement in cell adhesion by binding to host cell integrins and a plethora of other molecules present in the ECM and in the blood (27, 34). Bacteria express MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), adhesins that bind ECM proteins of the host, such as FN (29). FN binding by MSCRAMMS has been studied widely in the gram-positive genera Staphylococcus and Streptococcus (38), and recent reports document the presence of such adhesins in a broad variety of diverse genera, including gram-negative bacteria and mycobacterial species (29). FN binding by bacteria occurs via different bacterial components (38) and is redundant in many microorganisms, which may indicate the importance of this function in bacterial pathogenesis.
Given the importance of FN binding in other bacterial systems, the aim of this study was to investigate the role of DsrA in FN binding by H. ducreyi.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are shown in Tables 1 and 2, respectively. H. ducreyi strains were routinely maintained by minimal subculture (no more than five passages) on heme agar containing 50 μg/μl of hemin and 1× GGC (0.1% glucose, 0.01% glutamine, 0.026% cysteine) (45) or 1% IsoVitaleX (Becton Dickinson, NJ) and incubated at 34.5°C in 5% CO2. Preliminary FN binding experiments (Fig. 1) and construction of mutants were conducted with bacteria grown on chocolate agar (CA) plates supplemented with 5% FetalPlex (a fetal bovine serum substitute; Gemini, CA) and 1× GGC. The following antibiotics were used when appropriate: 1 μg/ml chloramphenicol (Cm), 100 μg/ml kanamycin (Kan), and 100 μg/ml streptomycin (Sm). Haemophilus influenzae strain KW20 Rd clones expressing H. ducreyi genes from plasmid pLSKS were routinely maintained on heme plates containing 50 μg/μl of hemin, 1% IsoVitaleX, and 100 μg/ml of Sm.
TABLE 1.
Bacterial strains used in this study
| Strain | Description or genotype | Source or reference |
|---|---|---|
| H. ducreyi strains | ||
| 35000HP | wt; human-passaged (HP) variant of strain 35000HP | 7, 23 |
| FX504 | 35000HP hgbA::CAT | 19 |
| FX517 | 35000HP dsrA::CAT | 20 |
| FX521 | 35000HP ncaA::Kan | This study |
| FX533 | 35000HP dltA::Kan | 31 |
| FX534 | 35000HP dsrA::CAT dltA::Kan | 31 |
| FX536 | 35000HP ncaA::Kan dsrA::CAT | This study |
| FX538 | 35000HPdsrA::Kan | This study |
| 35000HP-SMS1 | 35000HP ftpA::mTn3 (Cm) | 5 |
| FX541 | 35000HP ftpA::mTn3 (Cm) dsrA::CAT | This study |
| 35000HP-SMS2 | 35000HP momp::ΩKan2 | 44 |
| FX544 | 35000HP momp::ΩKan2 dsrA::CAT | This study |
| 35000HP-SMS3 | 35000HP ompA2::ΩKan2 | This study |
| FX545 | 35000HP ompA2::ΩKan2 dsrA::CAT | This study |
| HMC21 (V-1168) | wt strain from Seattle, WA | 18 |
| FX528 | HMC21 dsrA::CAT | This study |
| HMC50 (010-2) | wt strain from Jackson, MS | 18 |
| FX530 | HMC50 dsrA::CAT | 1 |
| HMC54 (425) | wt strain from Dominican Republic | 18 |
| FX529 | HMC54 dsrA::CAT | 1 |
| H. influenzae strain | ||
| KW20 Rd | Host for H. ducreyi clones | American Type Culture Collection |
TABLE 2.
Plasmids used in this study
| Plasmid | Description or relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| pCR2.1-TOPO | PCR cloning vector; Kanr Ampr | Invitrogen |
| pUC4K | Source of Kanr cassette | 42, 43 |
| pNC40 | Source of CAT cassette | 19 |
| pLSKS | H. ducreyi shuttle vector; Smr | 48 |
| pRSM1791 | Mutagenesis plasmid; β-galactosidase positive; Ampr | 14 |
| pUNCH1256 | dsrA class I from 35000HP with CAT cassette in pRSM1791 | 20 |
| pUNCH1260 | Complete dsrA class I gene from 35000HP in pLSKS | 20 |
| pUNCH1272 | ncaA PCR clone in pCR2.1-TOPO | This study |
| pUNCH1274 | ncaA with CAT cassette in PCR2.1-TOPO | This study |
| pUNCH1275 | ncaA with CAT cassette in pRSM1791 | This study |
| pUNCH1286 | Complete dltA class I gene from 35000HP in pLSKS | 31 |
| pUNCH1296 | Complete dsrA class II gene from CIP 542 in pLSKS | 47 |
| pUNCH1297 | ncaA with Kan cassette in pCR2.1-TOPO | This study |
| pUNCH1298 | ncaA with Kan cassette in pRSM1791 | This study |
| pUNCH1299 | dsrA with Kan cassette insert in pLSKS | This study |
| pUNCH1401 | dsrA with Kan cassette insert in pRSM1791 | This study |
| pUNCH1405 | dsrA with Kan cassette insert in pRSM1791 | This study |
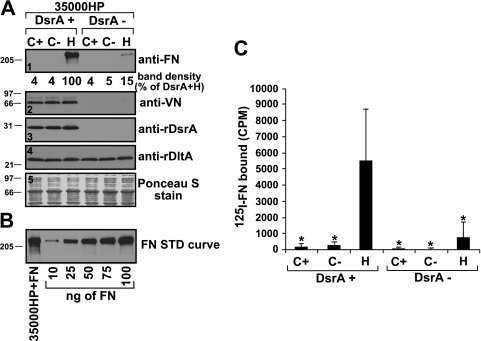
FIG. 1.
H. ducreyi binding to FN, but not to VN, is increased upon growth on heme agar. (A) Unlabeled FN binding assay. Suspensions of H. ducreyi bacterial cells, grown on the indicated media, were mixed with purified FN (panels 1, 3, 4, and 5, first lane) or heat-inactivated normal human serum (source of VN) (panel 2) for 30 min. After being washed to remove unbound ligands, bacterial cells were solubilized in Laemmli sample buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting to detect the bound ligands, as indicated. The experiment presented is representative of at least three other similar experiments. Ponceau S staining (panel 5) of the nitrocellulose membrane indicated approximately equal loading of total proteins for all strains. (B) FN standard curve. Lane 1, strain 35000HP incubated with FN as described for panel A; lanes 2 to 6, FN standard curve using the indicated amounts of purified FN loaded directly into wells. (C) 125I-labeled FN binding assay. Suspensions of H. ducreyi were mixed with purified 125I-labeled FN. After incubation and washing, bacterially associated cpm were determined. Asterisks indicate statistically significant (P < 0.01) differences in FN binding compared to that obtained for the wt strain H. ducreyi 35000HP (DsrA+) grown on heme (H). C+, CA containing 5% FetalPlex; C−, CA without FetalPlex; H, gonococcal medium plates with 50 μg/μl hemin.
Construction of H. ducreyi mutants. (i) Construction of 35000HP ncaA (FX521).
To construct the single ncaA mutant FX521, PCR primers ncaA.01 (GAATTATTTTAAGCAATTTTTTTGC) and ncaA.05 (TTATTGAAAATTATATACAAAGCCTACACC) were used to amplify the ncaA locus, using crude chromosomal DNA from 35000HP as a template and the following amplification parameters: a single denaturation step of 1 min at 95°C and 30 amplification cycles, each consisting of a 1-min denaturation at 95°C, annealing at 52°C for 1 min, and extension at 72°C for 2 min. The approximately 1-kb amplified product was ligated into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) to form pUNCH1272. pUNCH1272 was digested with ClaI, made blunt-ended with T4 DNA polymerase, and ligated to a BglII- and Klenow-treated chloramphenicol acetyltransferase (CAT) cassette from pNC40 (19) to form pUNCH1274. pUNCH1274 was digested with SacI and EcoRV to obtain a 2.0-kb insert containing the mutagenized ncaA locus. The latter insert was treated with Klenow and ligated to NotI- and Klenow-treated mutagenesis plasmid pRSM1791 (14) to form pUNCH1275. Finally, H. ducreyi strain 35000HP was electroporated with 1 μg of pUNCH1275, with selection on CA containing 1 μg/ml Cm (14). Cointegrate colonies were resolved by being streaked heavily onto CA containing 1 μg/ml Cm and 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and white colonies were isolated and confirmed (see below).
(ii) Construction of 35000HP ompA2 (35000HP-SMS3).
A promoterless ompA2 open reading frame (ORF) was amplified by PCR, using DNA from H. ducreyi strain 35000HP and the primers OmpA2-for-5 (5′-TGGATTAGCGCGTAACGATTATAGTG-3′) and OmpA2-rev-5 (5′-GCTCGTCCATCATCATTTGCG-3′). A Ω-Km2 cassette (44) was blunt end ligated into a BssHII site located 775 bp downstream of the start codon of the ompA2 ORF. The insert was ligated into pRSM1791 and electroporated into 35000HP. Cointegrate colonies were resolved as described above, and an ompA2 mutant, designated 35000HP-SMS3, was recovered. Southern blot and PCR analyses confirmed that the Ω-Km2 cassette had been inserted into the ompA2 ORF (data not shown).
(iii) Construction of 35000HP dsrA ncaA (FX536).
pUNCH1272 was digested with ClaI, treated with Klenow, and ligated to an EcoRI- and Klenow-treated Kan cassette from pUC4K (42, 43) to form pUNCH1297. The latter plasmid was thereafter digested with SpeI and NotI, treated with Klenow, and ligated into pRSM1791 to form pUNCH1298. To obtain FX536, strain 35000HP dsrA (FX517) was electroporated with 1 μg of pUNCH1298, and colonies were screened as described above for strain 35000HP ncaA.
(iv) Construction of a dsrA mutant of H. ducreyi strain 35000HP with a Kan cassette insertion (FX538).
The strain 35000HP dsrA (FX517) contains the Cm cassette inserted into the dsrA gene (20). However, the single mutant 35000HP ftpA (35000HP-SMS1) already had this antibiotic marker inserted into the ftpA (fine-tangle pilus) (5) gene. Thus, in order to construct a double mutant by using strain 35000HP ftpA as a recipient, a single dsrA mutant with a different antibiotic cassette insertion in the dsrA gene was constructed in H. ducreyi strain 35000HP. To do so, pUNCH1260 (20), containing the complete dsrA ORF, was digested with NdeI, Klenow treated, and ligated to an EcoRI- and Klenow-treated Kan cassette from pUC4K to form pUNCH1299. The latter plasmid was thereafter digested with ApaI and XbaI, treated with Klenow, and ligated into NotI- and Klenow-treated pRSM1791 to form pUNCH1401. To produce FX538, 1 μg of pUNCH1401 was electroporated into 35000HP. Mutants were obtained as described above for strain 35000HP ncaA, except that cointegrate colonies were plated on Kan-containing plates.
(v) Construction of 35000HP dsrA ftpA (FX541), 35000HP dsrA momp (FX544), and 35000HP dsrA ompA (FX545).
Mutant strain 35000HP dsrA ftpA was obtained by PCR amplification of the mutated dsrA gene, using plasmid pUNCH1405 (Kan insertion in the dsrA gene in pRSM1791) as the template and the primers 301038-15 (5′-TGGACAGCATTCCACTAACAGTC-3′) and OPA 33 (5′-CATCGTCGAACGCACACTG-3′), using the following PCR conditions: one denaturation cycle at 94°C for 5 min and 30 amplification cycles, each consisting of a 1-min denaturation at 94°C, annealing at 50°C for 1 min, and extension at 72°C for 4 min. The purified PCR product was subsequently electroporated into the 35000HP ftpA mutant without prior cloning into the suicide vector pRSM1791. Colonies were thereafter selected on Cm- and Kan-containing plates.
Mutant strains 35000HP dsrA momp and 35000HP dsrA ompA2 were obtained using the same method as that for strain 35000HP dsrA ftpA, except that plasmid pUNCH1256 (with the Cm antibiotic cassette in dsrA) was used as the template for a PCR with primers 301038-15 and OPA 33. The product obtained from the PCR was also directly electroporated into strains 35000HP momp (35000HP-SMS2) and 35000HP ompA (35000HP-SMS3) to produce the double mutant strains 35000HP dsrA momp and 35000HP dsrA ompA2, respectively.
(vi) Construction of HMC21 dsrA (FX528).
The single dsrA mutant FX528 was constructed in H. ducreyi strain HMC21 exactly as the dsrA mutant strains FX529 and FX530 were, as previously described (1).
Confirmation of the genotypes and phenotypes of the H. ducreyi mutants.
The hgbA (FX504) (6), dsrA (FX517) (13), dltA (FX533) (28), ftpA (35000HP-SMS1) (5), and momp (35000HP-SMS2) (44) mutants have been characterized extensively at the DNA and protein levels. As for the single ncaA (FX521) and dsrA (FX538 [Km insert]) mutants and all of the double dsrA mutants, PCR products obtained using whole-cell lysates as templates and specific primers were appropriately longer due to the presence of an antibiotic cassette inserted in the gene of interest (data not shown). Furthermore, each mutant lacked expression of the appropriate protein in a Western blot assay (Fig. 2, panels 2 to 4). However, outer membrane profiles of 35000HP and the 35000HP ompA2 mutant were similar (data not shown). We were unable to demonstrate a loss of OmpA2 in the mutant via Western blotting due to the lack of a monoclonal antibody (MAb) that specifically recognizes OmpA2 and the fact that the major outer membrane protein (MOMP), which is expressed more abundantly than OmpA2, and OmpA2 comigrate (30). The serum resistance phenotypes for all double dsrA mutants were also evaluated and found to be similar to that of the single dsrA mutant (FX517), except for that of 35000HP dsrA dltA, which has been shown to be more serum susceptible than FX517 (31). All of the mutants used in this study grew well on heme agar, the medium used for the FN and VN binding assays described below.
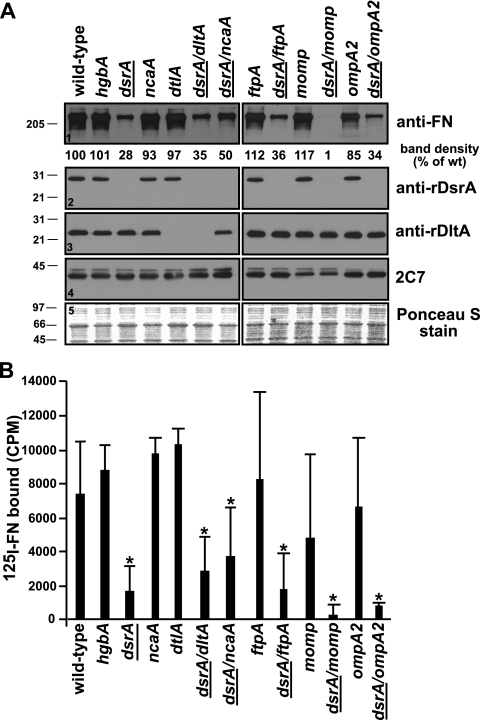
FIG. 2.
Expression of MOMP affects FN binding by H. ducreyi. Both unlabeled (A) and 125I-labeled (B) FN binding assays were repeated, using a panel of mutants constructed in the 35000HP background (Table 1). Asterisks indicate strains with levels of FN binding statistically different (P < 0.05) from the level of FN binding by the wt parent strain 35000HP. Strains not expressing dsrA are indicated with underlining. The results presented in panel A are representative of at least three other similar experiments. Ponceau S staining (panel 5) of the nitrocellulose membrane indicated approximately equal loading of total proteins for all the strains.
Cloning of momp.
The momp gene without its native promoter was PCR amplified using whole cells of strain 35000HP as the template and primers mompF (ATGGCTATCTAGAGGAGTATCAAAA) and mompR (GGGCTTAATCTAGAGACTGAAAAT). XbaI sites (underlined in primers) were included in the design of the primers. PCR was carried out under the following conditions: one denaturation cycle at 95°C for 5 min and 30 amplification cycles, each consisting of a 1-min denaturation at 95°C, annealing at 42°C for 1 min, and extension at 72°C for 2 min, followed by 1 cycle at 72°C for 1 min. The PCR products and the shuttle vector pLSKS (48) were treated with XbaI, and XbaI-treated pLSKS was subsequently treated with shrimp alkaline phosphatase (New England Biolabs, MA) to prevent religation. The restriction enzyme-treated PCR products and plasmid were ligated at room temperature for 30 min, and 1 μl of the ligation reaction mix was transformed into Escherichia coli DH5α and electroporated into H. influenzae strain KW20 Rd and the H. ducreyi dsrA momp mutant. Clones were selected on Sm-containing plates.
The momp gene with its native promoter was obtained the same way as the momp gene without a promoter, except that primer mompF2 (TACCGGTTAATAGGCTCGAGTTT [XhoI site is underlined]) was used instead of mompF, and an annealing temperature of 39°C was used in the PCR. Furthermore, PCR products and plasmid pLSKS were treated with both XbaI and XhoI prior to ligation.
FN and VN binding assays.
Two methods were used to evaluate the role of DsrA in FN binding by H. ducreyi, including an FN binding assay using unlabeled FN and an FN binding assay using 125I-labeled FN. For the unlabeled FN binding assay, suspensions containing bacterial growth from approximately 2 × 108 CFU/ml were prepared in GC broth (GCB) from 16- to 18-h heme agar plates. One milliliter of each suspension was incubated with 500 ng of purified native FN from human plasma, prepared in distilled water as described by the manufacturer (catalog no. F2006; Sigma, MO), at 34.5°C in 5% CO2 for 30 min. The bacterial cells were subsequently washed four times (for 1 h each) in GCB. Between the third and fourth washes, the bacterial cells were transferred to a new microcentrifuge tube. After the fourth wash, the cells were resuspended in Laemmli sample buffer. The bacterial cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions and to Western blotting using an anti-FN antibody (Sigma, MO) as previously described (31). The viability of the bacterial cells after the 4-hour wash in GCB was determined using plate counts. All isolates had >50% viability after incubation for 4 h at room temperature. Only three strains had CFU counts after incubation that were statistically different from the CFU at the beginning of the incubation; they were 35000HP dsrA dlta FX534 (52%; P = 0.0023), 35000HP dsrA momp FX544 (62%; P = 0.014), and 35000HP dsrA ompA FX545 (60%; P = 0.015). We used NIH Image (version 1.63) to perform densitometry on the bands obtained in the unlabeled FN binding assay. The numbers obtained with this program were used to compare the density of the FN binding band for the parent strain to those for the mutant strains. The density is expressed as the percentage of the density of the FN binding band for the dsrA-positive strain in the experiment.
We determined the effect of preincubation of MAbs or purified proteins on the ability of H. ducreyi to bind FN. Purified immunoglobulin G (IgG) MAbs or purified proteins, each at 50 μg/ml, were separately incubated for 30 min at 34.5°C with the bacterial suspensions prior to the addition of FN. The unlabeled FN binding assay was subsequently used to determine FN binding by H. ducreyi.
The VN binding assay was performed the same way as the unlabeled FN binding assay described above, except that 10% heat-inactivated normal human serum was added to the bacterial suspensions as the source of VN. An anti-VN antibody (Complement Technology, TX) was used for detection in the Western blot.
The second assay used 125I-labeled FN to measure FN binding by H. ducreyi. FN was iodinated by preactivation of two mCi of 125I-Na (Perkin-Elmer, MA) in Iodo-Gen tubes (Pierce, IL) and then its addition to 200 μg of purified plasma FN (Sigma). The iodination reaction was allowed to proceed for 9 min on ice and then quenched by the addition of 50 μl of a solution of tyrosine (10 mg/ml). The iodinated product was desalted on a Bio-Gel column (Bio-Rad, CA). The counts per minute (cpm) associated with the iodinated purified FN (125I-FN) was determined using a gamma counter, and the protein concentration was determined using a bicinchoninic acid protein determination kit (Pierce, IL).
After iodination of FN, a dose-response curve was performed to determine the quantity of 125I-FN to use for FN binding experiments. The quantity of 125I-FN at half-saturation was determined to be approximately 15 ng. Based on this information, 15 ng of 125I-FN was added to 200 μl of a bacterial suspension containing 0.5 × 108 CFU in a Multiscreen (Millipore, MA) plate and allowed to incubate at 34.5°C for 45 min. After incubation, the wells were washed six times with 200 μl of phosphate-buffered saline and allowed to dry, and the bacterium-associated cpm was determined using a gamma counter.
Production of MAb 1.82.
MAb 1.82, raised against a recombinant DsrA class I protein, was obtained from the same fusion that generated the anti-recombinant DsrA MAb 4.79, which is described in detail in a previous publication (47). MAb 1.82 binds a conformational epitope of H. ducreyi strain 35000HP but not those of the dsrA mutant FX517. It also immunoprecipitates DsrA from H. ducreyi whole cells (data not shown). The isotype for MAb 1.82 was determined to be IgG2A by use of the SBA Clonotyping system (Southern Biotech, AL).
Statistical analysis.
Statistical analyses were performed using the SigmaStat program (Systat Software Inc., CA). The normal distribution of the data was first determined using SigmaStat. Once this was established, a t test was used to assess the differences between groups of data.
RESULTS
H. ducreyi binding to FN, but not VN, is increased upon growth of H. ducreyi on heme agar.
Two different methods were used to detect FN binding by intact H. ducreyi. In the first assay, hereafter termed the unlabeled FN binding assay, purified plasma FN bound by H. ducreyi was detected in Western blots by use of an anti-FN antibody. In the second assay, hereafter termed the 125I-labeled FN binding assay, purified plasma FN was iodinated prior to incubation with intact H. ducreyi, and gamma counting was used to assess FN bound by H. ducreyi.
When cells were grown on CA, the standard growth medium, H. ducreyi FN binding was marginal and inconsistent. It was possible that the animal FNs in the FetalPlex present in the CA medium inhibited subsequent human FN binding. Therefore, we examined FN binding for H. ducreyi grown on CA with or without the addition of FetalPlex, as well as that for cells grown on heme plates. H. ducreyi grown on heme agar bound FN at levels much greater than that of bacteria grown on CA, regardless of the addition of FetalPlex (Fig. 1A, panel 1, first three lanes). In the 125I-labeled FN binding assay, strain 35000HP grown on heme agar bound 28 and 19 times more FN (Fig. 1C, first three bars) than that grown on CA, with and without FetalPlex, respectively (P < 0.01 for CA with or without FetalPlex compared to heme agar [Fig. 1C). We concluded that the medium used for growth profoundly affected FN binding by H. ducreyi. Therefore, all subsequent FN binding assays used bacteria grown on heme agar.
Since DsrA also mediates H. ducreyi binding to VN (16), the effect of growth medium on VN binding was also investigated. As opposed to FN binding, VN binding by H. ducreyi was not affected by the growth medium. Bacteria grown on CA, with or without the addition of FetalPlex, bound VN as well as those grown on heme plates (Fig. 1A, panel 2).
DsrA is the major OMP mediating FN binding by H. ducreyi.
To determine the role of DsrA in FN binding by H. ducreyi, we compared FN binding by the wild-type (wt) parent strain 35000HP and the dsrA isogenic null mutant FX517 (Fig. 1). The parent strain bound approximately seven times more FN than did the dsrA mutant strain (P < 0.01) (Fig. 1C) in the 125I-labeled FN binding assay. Similar results were observed in the unlabeled FN binding assay, where the dsrA mutant bound very little purified FN compared to strain 35000HP (Fig. 1A, panel 1, lanes 3 and 6).
We previously reported that the H. ducreyi lectin DltA strongly binds to a number of N-linked glycoproteins, including FN, in a ligand blot format (31). However, in that study, DltA did not have a major role in FN binding by whole cells of H. ducreyi. This conclusion is supported by the data presented in Fig. 1A, where it is shown that even though the levels of expression of DltA are identical between the parent and the dsrA mutant strains (Fig. 1A, panel 4), the FN binding phenotypes are different. Thus, binding of FN by H. ducreyi did not correlate with expression of DltA.
The data presented above strongly suggest that DsrA is involved in FN binding by H. ducreyi. However, the data also show that there might be a second component involved in FN binding, since there was residual FN binding by H. ducreyi dsrA mutant strain FX517 in the absence of DsrA in both the unlabeled and 125I-labeled FN formats of the FN binding assay (Fig. 1A, panel 1, and C). To determine if another component(s) of the outer membrane of H. ducreyi was involved in FN binding, double mutants that did not express dsrA and other outer membrane protein (OMP) genes, such as dltA, ncaA, ftpA, momp, and ompA2, were constructed. Single and double mutants (Table 1) were then tested for FN binding.
All strains expressing dsrA bound FN comparably to parent strain 35000HP in the unlabeled FN binding assay (Fig. 2). Among strains not expressing dsrA (underlined dsrA in Fig. 2), all dsrA double mutants, save strain 35000HP dsrA momp, bound FN comparably to the single dsrA mutant in both assays. Most importantly, FN binding by strain 35000HP dsrA momp was not detected in the unlabeled FN binding assay (Fig. 2A, panel 1) and was barely detectable in the 125I-labeled FN binding assay (Fig. 2B), which correlated with the absence of expression of momp. The values obtained in the 125I-labeled FN binding assay for strains that had significantly different FN binding from the wt were not statistically different from the values obtained for the single dsrA mutant FX517 (P = 0.402 for the dsrA dltA mutant, P = 0.109 for the dsrA ncaA mutant, P = 0.948 for the dsrA ftpA mutant, P = 0.115 for the dsrA momp mutant, and P = 0.238 for the dsrA ompA2 mutant).
H. ducreyi expresses two OmpA analogues, MOMP and OmpA2 (30). MOMP is estimated to be five times more abundant than OmpA2 in the outer membrane (44), and both proteins migrate as a 37- to 39-kDa doublet and a heat-modifiable 43-kDa species (30, 40). MAb 2C7 binds to both MOMP and OmpA2 (30, 40). The single momp mutant primarily lacked expression of the 37- to 39-kDa doublet (Fig. 2A, panel 4), as described previously (30, 44). However, the ompA2 mutant did not exhibit altered binding of 2C7 (Fig. 2A, panel 4) due to the fact that it expresses MOMP.
To further define the role of MOMP in FN binding by H. ducreyi, we cloned the momp gene under the control of the lac promoter in the shuttle vector pLSKS and expressed it in E. coli DH5α, in H. influenzae strain KW20 Rd, and in the H. ducreyi double dsrA momp mutant. We were unable to observe expression of the momp gene in E. coli and H. influenzae, even in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside), and although the double dsrA momp mutant expressed this construct, it did not do so to wt levels (data not shown). Furthermore, the dsrA momp mutant complemented with the momp construct did not bind more FN than the mutant complemented with an empty vector (data not shown). In hopes of increasing the expression level of momp, and therefore confirming a role for momp in FN binding by H. ducreyi, we attempted to clone the momp gene into pLSKS with its native promoter. However, we were unable to isolate a clone for any of the three strains described above.
To confirm that the mutation in the dsrA gene was responsible for the observed FN binding phenotype of the dsrA mutant FX517, it was complemented in trans with plasmid pUNCH1260 (20), containing intact dsrA from strain 35000HP, and was assayed for FN binding as described above. In the unlabeled FN binding assay, expression of dsrA class I from pUNCH1260 (dsrAI) restored FN binding by the dsrA mutant to levels comparable to that seen for the wt parent strain 35000HP (Fig. 3A, panel 1). Consistent with these results, the 125I-labeled FN binding assay showed that FN binding by the dsrA mutant FX517 expressing dsrAI in trans was significantly increased compared to that of the dsrA mutant complemented with an empty vector (P = 0.003). Taken together, the data presented above suggest that DsrA is the major OMP involved in FN binding by H. ducreyi and that MOMP may have a minor role in this phenotype.
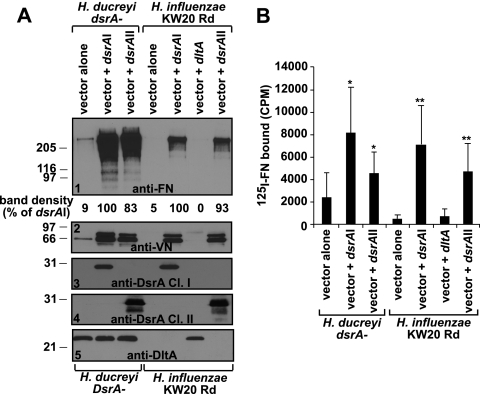
FIG. 3.
Either class of DsrA is sufficient to confer FN binding to the non-FN-binding strain H. influenzae KW20 Rd. The same FN binding assays described in the legend to Fig. 1 (unlabeled [A] and 125I-labeled [B] FN binding assays) were used to test FN binding by the H. ducreyi dsrA isogenic mutant FX517 (DsrA negative) and H. influenzae KW20 Rd expressing either an empty vector (pLSKS), pLSKS containing dsrA class I (dsrAI), pLSKS containing dltA, or pLSKS containing dsrA class II (dsrAII). All strains were grown on heme plates. Single asterisks represent statistically significant results (P ≤ 0.01) compared to the H. ducreyi dsrA mutant expressing the empty vector pLSKS, while double asterisks use H. influenzae KW20 Rd expressing the empty vector pLSKS as the comparison strain (P ≤ 0.01). The results presented in panel A are representative of at least three other similar experiments. Ponceau S staining (not shown) of the nitrocellulose membrane indicated approximately equal loading of total proteins for all the strains.
DsrA is sufficient to confer FN binding to the non-FN-binding H. influenzae strain KW20 Rd.
Escherichia coli is typically used as the host strain for expression of foreign genes and analysis of resulting phenotypes. However, dsrA is unclonable in E. coli due to its toxicity to this species (20). Thus, to study the role of DsrA in the absence of other H. ducreyi factors, FN binding was studied in H. influenzae strain KW20 Rd expressing dsrAI from plasmid pUNCH1260. H. influenzae strain KW20 Rd does not appear to express a homolog of dsrA, since only a homologous pseudogene of YadA with three frameshifts was found in this strain (25, 26). In the unlabeled FN binding assay, expression of dsrAI in H. influenzae KW20 Rd conferred FN binding to this non-FN-binding strain (Fig. 3A, panel 1), although the binding was not at the level seen for the H. ducreyi dsrA mutant complemented with dsrAI. In the 125I-labeled FN binding assay, FN binding was 10 times higher in strain H. influenzae KW20 Rd expressing dsrAI (P < 0.001) than in the H. influenzae strain expressing an empty vector (Fig. 3B, double asterisks). Expression of dsrAI in H. influenzae KW20 Rd also conferred binding to VN to levels seen in the dsrA mutant complemented with dsrAI (Fig. 3A, panel 2), while expression of the dltA gene in H. influenzae did not confer FN or VN binding (Fig. 3A, panels 1 and 2, and B), as expected. These data confirm that DsrA is sufficient to confer FN binding to H. influenzae in the absence of other H. ducreyi components.
DsrA class II confers FN and VN binding in both H. ducreyi dsrA mutant FX517 and H. influenzae KW20 Rd.
We and another group (36, 47) were unable to mutagenize the dsrA and LOS genes from H. ducreyi class II strains. Thus, to circumvent this limitation, we studied the role of the dsrA class II gene (dsrAII) in FN binding by expressing dsrAII from pUNCH1296 in both the null H. ducreyi dsrA mutant FX517 and the non-FN-binding strain H. influenzae KW20 Rd. Expression of dsrAII restored FN binding to wt levels in FX517 (P = 0.008 versus vector alone) and conferred FN binding to the non-FN-binding strain H. influenzae KW20 Rd (Fig. 3A, panel 1, and B) (P = 0.0002 versus H. influenzae KW20 Rd vector alone). dsrAII also restored VN binding in the dsrA class I mutant, as well as conferred VN binding to the non-VN-binding H. influenzae strain KW20 Rd (Fig. 3A, panel 5). These data suggest that DsrA proteins from both classes of H. ducreyi strains mediate FN and VN binding.
Inhibition of H. ducreyi FN binding by antibodies.
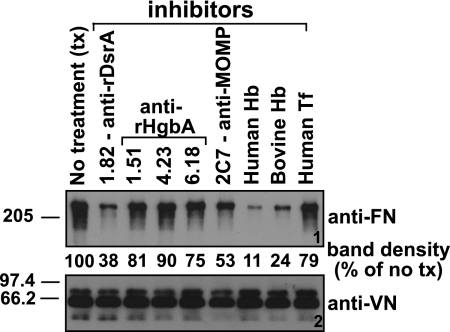
To determine the effect of antibodies on the binding between FN and DsrA, MAbs to H. ducreyi OMPs were preincubated with H. ducreyi cells prior to the addition of FN. FN was then added to the bacterial suspension, and the unlabeled FN binding assay was carried out as previously described (see Materials and Methods). The anti-DsrA MAb 1.82 (this study) and the anti-MOMP MAb 2C7 (11) were selected because they have been shown to bind to the surface of H. ducreyi. Three other MAbs (35), chosen as irrelevant control MAbs for this experiment, were also used. These were MAbs 1.51, 4.23, and 6.18, which are directed against HgbA. As shown in Fig. 4 (panel 1), MAb 1.82 partially blocked FN binding by H. ducreyi, while binding was reduced to intermediate levels by MAb 2C7. None of the other MAbs affected FN binding. VN binding by H. ducreyi strain 35000HP was not affected by any MAb tested (Fig. 4, panel 2).
FIG. 4.
FN binding by H. ducreyi is inhibited by an anti-DsrA MAb and by hemoglobin. The effect of preincubation of MAbs or purified protein on the ability of H. ducreyi to bind FN was measured using the unlabeled FN binding assay (see Materials and Methods). A panel of MAbs (purified IgGs), as well as hemoglobin (Hb) and transferrin (Tf), each at 50 μg/ml, were tested as potential inhibitors of FN and VN binding by H. ducreyi. Ponceau S staining (not shown) of the nitrocellulose membrane indicated approximately equal loading of total proteins for all the strains.
Inhibition of H. ducreyi FN binding by selected proteins.
Earlier, we showed that FN binding by H. ducreyi grown on CA was barely detectable and that FetalPlex could not account for this inhibition (Fig. 1). Based on these data, we hypothesized that hemoglobin (Hgb) present in the CA may inhibit FN binding by H. ducreyi. To test this hypothesis, H. ducreyi strain 35000HP was preincubated with human or bovine Hgb or human transferrin, a protein not present in CA medium, prior to the addition of FN or human serum (a source of VN). While Hgb from both sources greatly reduced FN binding by H. ducreyi (Fig. 4, panel 1), neither affected the binding of VN by H. ducreyi (Fig. 4, panel 2), consistent with the results shown in Fig. 1. As expected, human transferrin did not affect FN or VN binding by H. ducreyi.
Incubation of H. ducreyi with Hgb profoundly affected FN binding by this microorganism (Fig. 4, panel 1). To determine if this could be due to steric hindrance, we evaluated FN binding of the hgbA mutant (FX504) and the parent strain 35000HP upon growth on CA (with Hgb in the medium). We reasoned that the hgbA mutant, which is unable to bind Hgb (19), should be able to bind FN in the absence of Hgb bound to the surface of the cell if Hgb prevents binding of FN by steric hindrance. The hgbA mutant did not bind more FN than the parent strain (data not shown) in the presence of Hgb, suggesting that steric hindrance is not involved in preventing FN binding by H. ducreyi in the presence of Hgb.
DsrA is the major OMP involved in FN binding by other class I H. ducreyi strains.
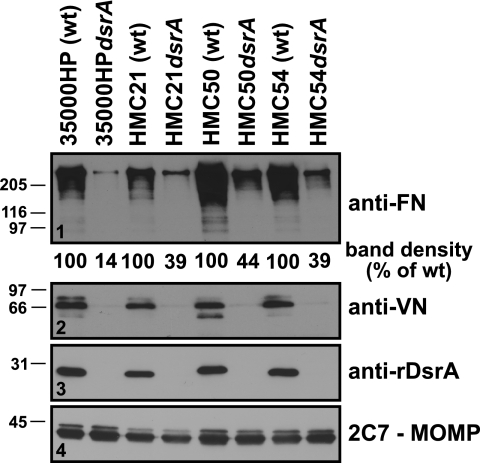
Although class II DsrA proteins are almost identical at the amino acid level (47), class I DsrA proteins are more variable (85% identity) (20). To determine if the differences in amino acid sequence among the DsrA class I proteins could affect FN and VN binding, previously constructed and characterized dsrA mutants FX529 and FX530 (1) and a newly constructed dsrA mutant, FX528, as well as their respective wt parent strains (HMC54, HMC50, and HMC21, respectively), were assayed for FN and VN binding. Each dsrA mutant bound less FN than its respective parent strain, although the level of FN binding was heterogeneous among the wt parent strains (Fig. 5, panel 1). VN binding phenotypes were also as expected, with all wt parent strains tested binding about equally to VN and their respective isogenic mutants unable to bind VN (Fig. 5, panel 2). Thus, in all H. ducreyi strains tested, DsrA was the major FN and VN binding protein.
FIG. 5.
DsrA is the major OMP involved in FN binding by other wt H. ducreyi class I strains. FN binding by three other wt H. ducreyi class I strains and their respective dsrA isogenic mutants was determined using the unlabeled FN binding assay described in the legend to Fig. 1A. Results are representative of similar experiments which were done twice. Ponceau S staining (not shown) of the nitrocellulose membrane indicated approximately equal loading of total proteins for all the strains.
DISCUSSION
DsrA is the major component involved in FN binding by H. ducreyi.
In this paper, we show four lines of evidence that DsrA is the major factor required for binding by H. ducreyi to the ECM protein FN. First, the single dsrA isogenic mutant strain bound significantly less FN than the wt parent strain 35000HP did. Moreover, the same phenotype was observed for three other class I H. ducreyi strains and their isogenic dsrA mutants. Second, expression of dsrA in trans in the single dsrA mutant restored FN binding to levels comparable to that of the parent strain, confirming that the reduced binding of FN by the dsrA mutant strain FX517 was due to a mutation in dsrA and not another locus. Third, an anti-DsrA MAb partially blocked FN binding by H. ducreyi. Fourth, mutants containing mutations in several outer membrane components bound levels of FN comparable to that of the parent strain, while most double mutants with a mutation in dsrA bound FN at levels similar to that of the single dsrA mutant.
MOMP may have a minor role in FN binding by H. ducreyi.
The data presented in this report demonstrate that DsrA is the major FN binding factor in H. ducreyi. However, residual FN binding by the dsrA mutant strain was also observed. This residual FN binding might be attributed to MOMP, since a double dsrA momp mutant bound less FN than the single dsrA mutant FX517 in both FN binding assays, although the difference in levels of FN binding between the single dsrA and double dsrA momp mutants did not reach statistical significance in the 125I-labeled FN binding assay (P = 0.115).
We attempted to demonstrate a role for MOMP in FN binding by expressing the momp gene in three different hosts, namely, E. coli DH5α, H. influenzae strain KW20 Rd, and the double dsrA momp H. ducreyi mutant (FX544). Although we were able to express momp under the control of the lac promoter in the dsrA momp mutant, it did not reach the levels expressed by the wt strain, and FN binding by the dsrA momp mutant complemented with this construct did not differ from that of the dsrA momp mutant complemented with an empty vector. Furthermore, we were unsuccessful in cloning the momp gene under the control of its native promoter, suggesting that this protein may be toxic when expressed at high levels, even in its natural host.
The DsrA proteins from class I and class II H. ducreyi strains are responsible for FN and VN binding.
Similar to class I DsrA, the DsrA proteins from class II strains were also able to provide comparable FN and VN binding phenotypes to those obtained with class I DsrA. A cloned class II dsrA gene restored FN and VN binding in the isogenic dsrA mutant FX517 to the same levels as those with class I DsrA and conferred binding to the non-FN- and non-VN-binding H. influenzae strain KW20 Rd.
The DsrA proteins from the two classes of H. ducreyi strains share high identity in the C-terminal portion but very low identity in the N-terminal two-thirds of the proteins. The dissimilarity in these proteins raised the possibility that these proteins may have different functions. However, the data presented here and previously (47) suggest that both classes of DsrA have similar functions despite their divergent N termini, since both classes of DsrA mediate serum resistance as well as FN and VN binding. Either FN binding can be attributed to the C-terminal part of the protein, which is 88% identical among the DsrA proteins from the two classes, or features other than the primary sequence in the divergent N-terminal section of the proteins are responsible for this interaction. In order to dissect the multifunctional roles of DsrA in H. ducreyi, experiments are currently under way to construct DsrA proteins that are involved only in adhesive properties and not serum resistance and vice versa. Furthermore, the virulence of these mutants will be examined in the experimental human infection model of chancroid.
Expression of the two classes of DsrA protein in a non-FN-binding H. influenzae strain conferred FN binding, although not to the levels seen for H. ducreyi strain FX517 complemented with dsrAI or dsrAII. The absence of another component(s) may account for the lower levels of FN binding by H. influenzae. This assumption is supported by the results presented in Fig. 3. Expression of dsrA genes from both classes of H. ducreyi strains conferred VN binding by H. influenzae KW20 Rd to levels comparable to those seen in the dsrA mutant complemented with either dsrA gene (Fig. 3, panel 2). However, this was not the case for FN binding, where expression of either dsrA gene in H. influenzae KW20 Rd conferred <50% of the FN binding level of the dsrA mutant complemented with either class of dsrA gene. These data support the conclusion that MOMP may have a role in FN binding by H. ducreyi.
A difference in FN binding was detected between the two FN binding assays upon comparison of the H. influenzae strain KW20 Rd and the H. ducreyi single dsrA mutant expressing the dsrA gene (Fig. 3). FN binding measured by the 125I-labeled FN binding assay was very similar between both H. ducreyi and H. influenzae strains, while there was a substantial difference between these strains in FN binding measured by the unlabeled FN binding assay. We do not understand the reason(s) for this observation. One possibility is that the affinity of FN for dsrA expressed in H. influenzae is less than that for dsrA expressed in H. ducreyi. In this case, the FN bound at the surface of the cell is more likely to be lost during the longer wash in the unlabeled FN assay than it is in the 125I-labeled FN binding assay. This “reduced” affinity for FN binding by H. influenzae expressing dsrA may be due to additional H. ducreyi components which are absent in H. influenzae and that contribute to FN binding.
FN binding by H. ducreyi and virulence.
Historically, limited studies of ECM binding by H. ducreyi have been reported. In the first in vitro ECM binding analysis, all 21 H. ducreyi strains tested bound FN, and a protein was speculated to be involved in this interaction (2). Later, Alfa and DeGagne showed that the addition of purified FN inhibited the binding of H. ducreyi to fibroblasts in a dose-dependent manner (4). A third in vitro study of ECM binding by H. ducreyi confirmed previous studies of FN binding by H. ducreyi, although no role for pili or full-length LOS in FN binding by H. ducreyi could be demonstrated (10). In the human experimental model of chancroid infection, H. ducreyi associates with the ECM proteins fibrin and collagen (9), and in natural chancroid, it is associated with fibrin only (12).
The discrepancy between FN binding by H. ducreyi in vitro and in vivo may be explained in several ways. H. ducreyi has not been visualized in the skin of experimentally infected volunteers in the first 48 h postinoculation (9) and may interact with FN early in the infection process, before organisms can be visualized. Also, the MAb used for the in vivo FN binding studies was developed against cellular FN (MAb 568; Novocastra Laboratories) (9), and it is not known whether this antibody reacts with plasma FN. In the present study, only plasma FN binding by H. ducreyi was examined, so we cannot make any conclusion(s) about the interaction of H. ducreyi with cellular FN.
We have shown that Hgb inhibits FN binding in vitro. Hgb is present at sites of H. ducreyi infection and is an important source of heme/iron for H. ducreyi in human experimental infection (6). Chancroidal ulcers are vascular (33), and blood at the site of infection is most likely a source of Hgb for H. ducreyi. Since H. ducreyi can be visualized only after 48 h of infection, it is possible that Hgb inhibits FN binding by H. ducreyi in vivo, consistent with our in vitro findings. Clearly, further studies are warranted to better understand how hemoglobin and fibronectin interact with the H. ducreyi cell surface.
FN binding by other Oca proteins.
The amino acid sequence between amino acids 53 and 83 in YadA from Yersinia pseudotuberculosis is responsible for FN binding, and this region is absent in Yersinia enterocolitica YadA and H. ducreyi DsrA (24). Furthermore, FN binding by Y. pseudotuberculosis through the 53-83 region of YadA allows invasion through the α5β1 integrin receptor. However, there was residual binding in Δ53-83 YadA of Y. pseudotuberculosis, suggesting that another section(s) of the protein may also bind FN. Experiments are under way to determine the residues of DsrA which are required for plasma FN binding by H. ducreyi.
In conclusion, we have shown that the OMP DsrA is the major mediator of human plasma FN binding by the gram-negative bacterium H. ducreyi. Our results also suggest that MOMP may play a minor role in the interaction of H. ducreyi with FN.
Acknowledgments
We thank Annice Roundtree and Bonnie Olsen for expert technical support and Marcia Hobbs for careful review of the manuscript.
This research was supported by NIH grants AI031496 to C.E. and AI27863-16 to S.M.S.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Abdullah, M., I. Nepluev, G. Afonina, S. Ram, P. Rice, W. Cade, and C. Elkins. 2005. Killing of dsrA mutants of Haemophilus ducreyi by normal human serum occurs via the classical complement pathway and is initiated by immunoglobulin M binding. Infect. Immun. 733431-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeck, D., A. P. Johnson, and H. Mensing. 1992. Binding of Haemophilus ducreyi to extracellular matrix proteins. Microb. Pathog. 1381-84. [DOI] [PubMed] [Google Scholar]
- 3.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa, M. J., and P. DeGagne. 1997. Attachment of Haemophilus ducreyi to human foreskin fibroblasts involves LOS and fibronectin. Microb. Pathog. 2239-46. [DOI] [PubMed] [Google Scholar]
- 5.Al-Tawfiq, J. A., M. E. Bauer, K. R. Fortney, B. P. Katz, A. F. Hood, M. Ketterer, M. A. Apicella, and S. M. Spinola. 2000. A pilus-deficient mutant of Haemophilus ducreyi is virulent in the human model of experimental infection. J. Infect. Dis. 1811176-1179. [DOI] [PubMed] [Google Scholar]
- 6.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 1811049-1054. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 1781684-1687. [DOI] [PubMed] [Google Scholar]
- 8.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer, M. E., M. P. Goheen, C. A. Townsend, and S. M. Spinola. 2001. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect. Immun. 692549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer, M. E., and S. M. Spinola. 1999. Binding of Haemophilus ducreyi to extracellular matrix proteins. Infect. Immun. 672649-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer, M. E., and S. M. Spinola. 2000. Localization of Haemophilus ducreyi at the pustular stage of disease in the human model of infection. Infect. Immun. 682309-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer, M. E., C. A. Townsend, A. R. Ronald, and S. M. Spinola. 2006. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microbes Infect. 82465-2468. [DOI] [PubMed] [Google Scholar]
- 13.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 691488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozue, J. A., L. Tarantino, and R. S. Munson, Jr. 1998. Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol. Lett. 164269-273. [DOI] [PubMed] [Google Scholar]
- 15.Clarke, S. R., and S. J. Foster. 2006. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 51187-224. [DOI] [PubMed] [Google Scholar]
- 16.Cole, L. E., T. H. Kawula, K. L. Toffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect. Immun. 706158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 18.Dutro, S. M., G. Wood, and P. Totten. 1999. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect. Immun. 673317-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins, C., C. J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus of Haemophilus ducreyi. Infect. Immun. 632194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 681608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291209-218. [DOI] [PubMed] [Google Scholar]
- 22.Fulcher, R. A., L. E. Cole, D. M. Janowicz, K. L. Toffer, K. R. Fortney, B. P. Katz, P. E. Orndorff, S. M. Spinola, and T. H. Kawula. 2006. Expression of Haemophilus ducreyi collagen binding outer membrane protein NcaA is required for virulence in swine and human challenge models of chancroid. Infect. Immun. 742651-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond, G. W., C. J. Lian, J. C. Wilt, W. L. Albritton, and A. R. Ronald. 1978. Determination of the hemin requirement of Haemophilus ducreyi: evaluation of the porphyrin test and media used in the satellite growth test. J. Clin. Microbiol. 7243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heise, T., and P. Dersch. 2006. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. USA 1033375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood, D. W., M. E. Deadman, T. Allen, H. Masoud, A. Martin, J. R. Brisson, R. Fleischmann, J. C. Venter, J. C. Richards, and E. R. Moxon. 1996. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22951-965. [DOI] [PubMed] [Google Scholar]
- 27.Hynes, R. O. 1990. Fibronectins. Springer-Verlag, New York, NY.
- 28.Janowicz, D., I. Leduc, K. R. Fortney, B. P. Katz, C. Elkins, and S. M. Spinola. 2006. A DltA mutant of Haemophilus ducreyi is partially attenuated in its ability to cause pustules in human volunteers. Infect. Immun. 741394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18211-223. [DOI] [PubMed] [Google Scholar]
- 30.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 1791764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 723418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, D. A. 2000. Chancroid: from clinical practice to basic science. AIDS Patient Care STDS 1419-36. [DOI] [PubMed] [Google Scholar]
- 33.Morse, S. A. 1989. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 2137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosher, D. R. 1989. Fibronectin. Academic Press, San Diego, CA.
- 35.Patterson, K., B. Olsen, C. Thomas, D. Norn, M. Tam, and C. Elkins. 2002. Development of a rapid immunodiagnostic test for Haemophilus ducreyi. J. Clin. Microbiol. 403694-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post, D. M., R. S. Munson, Jr., B. Baker, H. Zhong, J. A. Bozue, and B. W. Gibson. 2007. Identification of genes involved in the expression of atypical lipooligosaccharide structures from a second class of Haemophilus ducreyi. Infect. Immun. 75113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riesbeck, K., T. T. Tan, and A. Forsgren. 2006. MID and UspA1/A2 of the human respiratory pathogen Moraxella catarrhalis, and interactions with the human host as basis for vaccine development. Acta Biochim. Pol. 53445-456. [PubMed] [Google Scholar]
- 38.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52631-641. [DOI] [PubMed] [Google Scholar]
- 39.Spinola, S. M., M. E. Bauer, and R. S. Munson. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 701667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spinola, S. M., G. E. Griffiths, K. L. Shanks, and M. S. Blake. 1993. The major outer membrane protein of Haemophilus ducreyi is a member of the OmpA family of proteins. Infect. Immun. 611346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan, M. 1940. Chancroid. Am. J. Syph. Gonorrhea Vener. Dis. 24482-521. [Google Scholar]
- 42.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, W. D., Jr., S. P. Wagner, and R. A. Welch. 1992. A heterologous membrane protein domain fused to the C-terminal ATP-binding domain of HlyB can export Escherichia coli hemolysin. J. Bacteriol. 1746771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Throm, R. E., J. A. Al-Tawfiq, K. R. Fortney, B. P. Katz, A. F. Hood, C. A. Slaughter, E. J. Hansen, and S. M. Spinola. 2000. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect. Immun. 682602-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Totten, P. A., and W. E. Stamm. 1994. Clear broth and plate media for the culture of Haemophilus ducreyi. J. Clin. Microbiol. 322019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, C. D., I. Leduc, B. Olsen, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 732387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of the Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 673740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]