Abstract
In Vibrio cholerae, the second messenger cyclic di-GMP (c-di-GMP) positively regulates biofilm formation and negatively regulates virulence and is proposed to play an important role in the transition from persistence in the environment to survival in the host. Herein we describe a characterization of the infection-induced gene cdpA, which encodes both GGDEF and EAL domains, which are known to mediate diguanylate cyclase and c-di-GMP phosphodiesterase (PDE) activities, respectively. CdpA is shown to possess PDE activity, and this activity is regulated by its inactive degenerate GGDEF domain. CdpA inhibits biofilm formation but has no effect on colonization of the infant mouse small intestine. Consistent with these observations, cdpA is expressed during in vitro growth in a biofilm but is not expressed in vivo until the late stage of infection, after colonization has occurred. To test for a role of c-di-GMP in the early stages of infection, we artificially increased c-di-GMP and observed reduced colonization. This was attributed to a significant reduction in toxT transcription during infection. Cumulatively, these results support a model of the V. cholerae life cycle in which c-di-GMP must be down-regulated early after entering the small intestine and maintained at a low level to allow virulence gene expression, colonization, and motility at appropriate stages of infection.
Vibrio cholerae, the causative agent of the diarrheal disease cholera, is a highly motile gram-negative bacterium that is indigenous to aquatic environments and can survive in both marine water and freshwater. There is evidence showing that V. cholerae lives in association with other aquatic organisms, including cyanobacteria, copepods, and insect egg masses (18, 49). Furthermore, the ability of V. cholerae to survive in the environment likely depends on the ability to form biofilms (52, 56). A large-scale shift in gene expression occurs upon entering and exiting the human gastrointestinal tract (24, 32, 46), but the triggering signals and the mechanism of regulation of these switches are unknown. It is unclear how V. cholerae adapts to and grows within these very disparate environments.
Bis-(3′-5′)-cyclic di-GMP (c-di-GMP), first identified as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus (40, 41, 53), has been recognized as an important bacterial second messenger involved in the regulation of a number of processes. Examples include extracellular polysaccharide biosynthesis in Salmonella enterica serovar Typhimurium, Yersinia pestis, Pseudomonas aeruginosa, and V. cholerae (14, 15, 20, 47, 50); motility in S. enterica serovar Typhimurium, P. aeruginosa, Escherichia coli, Caulobacter crescentus, and V. cholerae (1, 17, 21, 29, 47); differentiation in C. crescentus (1, 36); and virulence in S. enterica serovar Typhimurium, P. aeruginosa, Y. pestis, and V. cholerae (16, 20, 23, 51).
Biosynthesis and degradation of c-di-GMP are performed by three protein domains. Diguanylate cyclases (DGCs) containing a GGDEF domain, named for conserved residues, synthesize c-di-GMP from two GTPs (4, 36, 43). In turn, c-di-GMP is hydrolyzed by EAL domain phosphodiesterases (PDEs), including VieA, which is described below (9, 10, 45, 48). Recent evidence shows that the protein domain HD-GYP can also degrade c-di-GMP (42). Many bacteria encode multiple GGDEF, EAL, and HD-GYP domains, which are commonly found in conjunction with regulatory and sensory domains, as well as together on the same protein. For example, V. cholerae encodes 12 EAL, 30 GGDEF, 9 HD-GYP, and 10 GGDEF-EAL domain proteins (12, 13).
We propose that c-di-GMP plays a key role in regulating the changes in V. cholerae gene expression that occur during the shift from aquatic to host environments. Previous work by our laboratory studying the dual function protein VieA led to a model by which c-di-GMP inversely regulates biofilm formation and virulence of classical-biotype V. cholerae. High c-di-GMP resulting from inactivation of VieA PDE results in increased expression of vps (vibrio exopolysaccharide synthesis) genes and increased biofilm formation (48, 50). A similar effect is seen for V. cholerae overproducing the DGC encoded by VCA0956 (50). Conversely, c-di-GMP inhibits motility: mutation of vieA abrogates motility in soft agar medium (37). In addition, VieA PDE activity is necessary for virulence in the infant mouse model of V. cholerae colonization (51). VieA positively regulates in vitro expression of the major virulence gene transcriptional activator toxT and the genes encoding cholera toxin, ctxAB (51). Based on these data, c-di-GMP is poised to coordinate changes in gene expression during the shift from the aquatic environment to the host. This model is bolstered by the fact that the operon encoding VieA, vieSAB, is induced during infection (7) and contributes to ctxAB expression in the infant mouse (27), providing a mechanism by which c-di-GMP levels can be reduced upon entry into the host. Expression of vieSAB occurs at an early stage of infection (26), indicating that c-di-GMP must be reduced rapidly and early to allow maximal virulence gene expression.
Interestingly, VieA does not regulate biofilm formation, motility, or virulence in the El Tor biotype of V. cholerae (2, 27; R. Tamayo and A. Camilli, unpublished data), which is the biotype that causes present-day cholera; the classical biotype upon which the above-described model was based is essentially extinct in nature. However, c-di-GMP does control biofilm formation and motility in the El Tor biotype (3). There is also evidence that one putative PDE, CdgC (VCA0785), regulates virulence gene expression in vitro in an El Tor strain (30); however, VCA0785 is dispensable for virulence in an infant mouse model of colonization (Tamayo and Camilli, unpublished). Therefore, the role of c-di-GMP in virulence of El Tor biotype V. cholerae is unclear.
We hypothesize that c-di-GMP inversely regulates biofilm and virulence genes in El Tor as in the classical V. cholerae biotype. This could be accomplished by 1 or more of the other 21 EAL domain or 9 HD-GYP domain proteins, besides VieA, encoded by V. cholerae El Tor. Here, we characterize the activity, expression, and function of the GGDEF-EAL domain gene VC0130, herein named cdpA. We further define the role of c-di-GMP in regulation of virulence in El Tor by ectopically producing a DGC during infection. This work has allowed us to better understand the role of c-di-GMP in the virulence of V. cholerae at early and late stages of infection.
MATERIALS AND METHODS
Bacterial growth conditions.
Unless otherwise noted, E. coli, V. cholerae El Tor biotype strain C6709, and derivative strains were grown in Luria-Bertani (LB) broth at 37°C with aeration. Antibiotics were used where appropriate as follows: 100 μg/ml streptomycin (Sm), 50 μg/ml ampicillin (Ap), 50 μg/ml kanamycin (Km), and 1 μg/ml tetracycline (Tc). Strains used in this study are described in Table 1.
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| V. cholerae strains | ||
| AC51 | El Tor C6709, Smr | 39 |
| AC66 | C6709 lacZ::res-tet-res | 7 |
| AC1901 | C6709 pMMB67EH::vieA-His6 | This work |
| AC1902 | C6709 pMMB67EH::VCA0956(pVdcA) | This work |
| AC1903 | C6709 lacZ::res-tet-res pMMB67EH | This work |
| AC2131 | C6709 cdpA::pGP704 | This work |
| AC2280 | C6709 ΔvpsR | This work |
| AC2284 | C6709 ΔvpsR pMMB67EH::vieA-His6 | This work |
| AC2286 | C6709 ΔvpsR pVdcA | This work |
| AC2365 | C6709 lacZ::res-tet-res pMMBneo | This work |
| AC2377 | C6709 cdpA::pGP704 pMMBneo::cdpA-His6 | This work |
| AC2380 | C6709 pMMBneo::cdpA-His6 | This work |
| AC2389 | C6709 pMMB67EH::VCA0956(E258A) pVdcA(I) | This work |
| AC2390 | C6709 pMMB67EH::vieA(E170A) | This work |
| AC2395 | C6709 lacZ pVdcA | This work |
| AC2434 | C6709 cdpA::pGP704 pMMBneo | This work |
| AC2435 | C6709 ΔvpsR pMMBneo | This work |
| AC2483 | C6709 toxT::tnpR lacZ::res-tet-res pVdcA | This work |
| AC2484 | C6709 toxT::tnpR lacZ::res-tet-res pVdcA(I) | This work |
| AC2595 | C6709 lacZ::res-neo-sacB-res | This work |
| AC2606 | C6709 cdpA::tnpR lacZ::res-sacB-neo-res | This work |
| AC2655 | C6709 pMMBneo::cdpA(AVGAW)-His6 | This work |
| AC2658 | C6709 cdpA::pGP704 pMMBneo::cdpAΔEAL-His6 | This work |
| AC2659 | C6709 cdpA::pGP704 pMMBneo::cdpA(AVGAW)-His6 | This work |
| E. coli strains | ||
| DH5α | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | Laboratory strain |
| DH5αλPir | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | Laboratory strain |
| SM10λPir | thi recA thr leu tonA lacY supE RP4-2-Tc::Mu λ::pir | Laboratory strain |
| AC575 | DH5α pMMB67EH | 33 |
| AC751 | DH5αλPir pCVD442lac | 11 |
| AC2364 | DH5α pMMBneo | 35 |
| AC1817 | DH5α pMMB67EH::vieA-His6 | 48 |
| AC1835 | DH5α pMMB67EH::vieA(E170A)-His6 | 48 |
| AC1887 | DH5α pVdcA | This work |
| AC2129 | SM10λPir pGP704::′cdpA′a | This work |
| AC2142 | SM10λPir pCVD442lac::ΔvpsR | This work |
| AC2376 | DH5α pMMBneo::cdpA-His6 | This work |
| AC2386 | DH5α pVdcA | This work |
| AC2528 | SM10λPir pRes | 35, 44 |
| AC2599 | SM10λPir pIVET5ncdpA | This work; 44 |
| AC2641 | DH5α pMMBneo::cdpAΔEAL-His6 | This work |
| AC2642 | DH5α pMMBneo::cdpAAVGAW-His6 | This work |
An internal fragment of cdpA was cloned in order to make the plasmid insertion.
DNA manipulations and cloning.
All primers are listed in Table 2, with notable features indicated. VC0130 was disrupted by plasmid insertion. An internal fragment of the gene was amplified using 0130FX and 0130RX and cloned into suicide vector pGP704. This construct was introduced into V. cholerae by conjugation between wild-type (WT) AC51 and AC2129. Integration of the fusion within the EAL domain of the chromosomal copy of VC0130 was confirmed by PCR with primers oriR and 0130PF.
TABLE 2.
Primers used in this study
| Primer name | Sequencea (5′ to 3′) (reference) |
|---|---|
| oriR | CAGCAGTTCAACCTGTTG |
| 67EHF | CGACATCATAACGGTTCTGG |
| 67EHR | TTCACTTCTGAGTTCGGCAT |
| 0130FX | GGTCTAGAAGAGCGGTTTGTATGCGATT |
| 0130RX | GGTCTAGAGCCGGCTCAAACGAGTATAG |
| 0130PF | TAATCGCCCTGAAAGTGACC |
| C0130F | TTCTAGATTTAGGATACATTTTTATGTTTACGGTCTCGC |
| C0130R | TTGCATGCCTAATGGTGATGGTGATGGTGACCCAAGCGTGAAGGT |
| 0130EAL | TTCTAGATTTAGGATACATTTTTATGGCGGTGGTACGTGATGAT |
| vpsRF1 | TGCATGCCTACAACCCAAATCACGC |
| vpsRR1 | AATCAGCAAAACTTACATGAACCTATATTCCTT |
| vpsRF2 | GAATATAGGTTCATGTAAGTTTTGCTGATTTAC |
| vpsRR2 | TTTCTAGAGGTAAACTCAAGCCGATT |
| vpsRF0 | CTCTGTGGCGTTAGAAG |
| vpsRR0 | CCTGTCCTTAGTGATGTG |
| A0956F1 | CCGAGCTCTTTAGGATACATTTTTGTGATGACAACTGAAGAT |
| A0956R1 | GGGCATGCAGTTTAGAGCGGCATGAC |
| A0956EAF | GGCGGTGAAGCGTTTGCACTG |
| A0956EAR | CAGTGCAAACGCTTCACCGCC |
| 0130GEF | TTCTAGATTTAGGATACATTTTTATGACGGACAATTTGGCAC |
| 0130GAF | GCTGTCGGTGCTTGGGCAACGGTTTTT |
| 0130GAR | CCAAGCACCGACAGCAATCGCATACAAACC |
| 0130EAF | TACGCTTGTTTAGTGCGGATTGAA |
| 0130EAR | CACTAAACAAGCGTAGGAAGCCAC |
| 0130CGR | TTGCATGCCTAATGGTGATGGTGATGGTGCTCCTGACGAACACTTTC |
| 0130GAS | GCGATTGCTGTCGGTGCT |
| 0130EAS | ATTGTGGCTTCCTACGCT |
| 3287qF | TTCTCTCCTTTGAGTCGCGAGCAT |
| 3287qR | ATTTGCAGCAACGTCACCTGATGG |
| vpsRqF | TGGATTCAGTACCTGGCTCT |
| vpsRqR | CGCAATCCCGTTAAGGCTAA |
| RPB2FD | CTGTCTCAAGCCGGTTACAA (38) |
| RPB2RV | TTTCTACCAGTGCAGAGATGC (38) |
Restriction sites are underlined; sequence encoding the His6 tag is both underlined and in boldface; boldface text without underlining indicates codons changed to obtain desired amino acid mutations.
For complementation and overexpression studies, the full coding sequence of VC0130 was amplified by PCR using C0130F and C0130R, which introduces a six-histidine (His6) tag to allow detection of the protein by Western blotting. Primers C0130F/0130CGR were used to amplify the COG3287-GGDEF domains of VC0130, omitting the EAL domain. Residues G493 and E496 were mutated to alanines to change GVGEW of the GGDEF domain active site to AVGAW by use of splicing by overlap extension PCR, with the indicated nucleotide changes incorporated into the overlapping primers. Primers C0130F/0130GAR and 0130GAF/C0130R were used to amplify the upstream and downstream regions of homology, respectively. The PCR products were cloned into low-copy-number, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible vector pMMBneo (35). Plasmids were screened by PCR using plasmid-specific primers 67EHF and 67EHR, which flank the multiple cloning site. The incorporation of point mutations in GVGEW was confirmed by sequencing with primer 0130GAS.
VCA0956 (Vibrio DGC A; vdcA) was amplified from C6709 genomic DNA by PCR using primers A0956F1/A0956R1 and cloned into low-copy-number, IPTG-inducible vector pMMB67EH, yielding pVdeA. In addition, VdcA residue E258, which resides in the predicted catalytic site (8), was mutated to alanine by splicing by overlap extension PCR. Using pMMB67EH::vdcA as the template, fragments were amplified using A0956F1/A0956EAR and A0956EAF/A0956R. The fragments were spliced together and amplified with primers A0956F/A0956R. The resulting fragment was cloned into pMMB67EH, yielding pVdcAi, which is expected to produce an inactive VdcA. Ap-resistant colonies were screened by PCR and sequencing with primers 67EHF/67EHR.
For expression in V. cholerae, plasmids were transformed into C6709 (WT AC51 or the lacZ mutant strain AC66, as appropriate) by electroporation. The resulting strains were confirmed to contain the plasmid by PCR using primers 67EHF and 67EHR.
Assays for enzymatic activity.
Stationary-phase cultures of relevant strains were diluted 1:100 in 25 ml LB broth and incubated at 37°C with aeration until mid-exponential phase. IPTG was added to a final concentration of 0.5 mM to induce gene expression and the cultures were incubated 3 h more. Aliquots were collected for Western blot analysis using mouse anti-pentahistidine primary antibodies (Qiagen) and horseradish peroxidase-conjugated sheep anti-mouse secondary antibodies (Amersham). Western blots were developed with ECL detection reagents (Amersham).
For PDE assays, the pellets were resuspended in 0.5 ml reaction buffer (75 mM Tris, pH 8, 25 mM KCl, 10 mM MgCl2) containing 10% glycerol (48). For DGC reactions, the pellets were resuspended in cyclase buffer (75 mM Tris, pH 7.8, 250 mM NaCl, 25 mM KCl, 10 mM MgCl2) containing 10% glycerol (48). For both assays, bacteria were lysed by sonication.
PDE activity was assessed by incubating lysate with radiolabeled c-di-GMP, enzymatically synthesized as described previously (48). Aliquots were spotted on cellulose-polyethyleneimine thin-layer chromatography (TLC) plates 2 min after the addition of the substrate. Reaction products were separated in 1.5 M KH2PO4, pH 3.65, and visualized by phosphorimaging. DGC reactions were performed as described above, except 19.5 μl of lysates and 0.5 μl of substrate were combined ([α-32P]GTP, 3,000 Ci/mmol; Perkin-Elmer).
2D-TLC of nucleotide extracts.
Two-dimensional TLC (2D-TLC) assays were performed on radiolabeled nucleotide extracts as described previously (5, 50). Where indicated, 1 mM IPTG was added at early exponential phase (optical density at 600 nm [OD600], 0.35) to induce gene expression. To quantify relative levels of c-di-GMP, the amount of c-di-GMP was normalized to the level of GMP in the sample (3, 50). At least two independent experiments were done.
In vitro phenotype assays.
Biofilm and chemotaxis/motility assays were done essentially as described previously (37). Where indicated, 50 μM or 1 mM IPTG was added to induce gene expression.
In vivo competition assays.
Standard in vivo competition assays were done as described previously (7). For competition assays using strains with plasmid-borne enzymes, strains were grown overnight at 37°C on LB agar plates containing Ap. After suspension in LB broth to an OD600 of 0.20, IPTG was added to a final concentration of 50 μM to induce gene expression, and then cultures were incubated for 15 min at 37°C with aeration immediately prior to inoculation. Subsequently the competing strains were combined at 1:1 ratios in LB broth containing 50 μM IPTG. In vitro competition experiments were performed in parallel. Competition indices were determined as the ratio of output mutant to WT CFU to that ratio in the input.
RNA isolation and cDNA synthesis.
For bacteria grown in LB broth, RNA was isolated from mid-exponential-phase cultures (OD600, 0.5 to 0.55) incubated with aeration at 24°C (for biofilm experiments) or 37°C (for in vivo sample comparisons) as described previously (44).
For bacteria grown in biofilms, the biofilms were washed with phosphate-buffered saline to remove unattached bacteria. Biofilm-associated bacteria were recovered by vortexing in 1 ml Trizol and with 0.1-mm zirconia beads (BioSpec). RNA was purified as described previously (44). Aqueous phases from six separate biofilms were pooled into a single sample to obtain sufficient RNA for subsequent experiments.
For quantitative real-time PCR (qPCR) analysis of cdpA expression during infection, infant mice were inoculated intragastrically with 106 WT bacteria or phosphate-buffered saline only (mock). At 22 h postinfection (p.i.), the ilea (most distal 3 cm of the small intestine) were dissected and snap-frozen in a dry ice-ethanol bath. The ileal samples from three mice were pooled into a single preparation and RNA was isolated as described previously (44).
cDNA was made as described previously (44). For broth culture and biofilm qPCR studies, 100 ng RNA was used in each reaction. For RNA obtained from mouse small intestines, as well as broth culture controls, 1 μg RNA was used in each reaction.
qPCR.
Reactions were done with Brilliant SYBR green qPCR master mix (Stratagene) using Stratagene Mv3005P equipment and MxPro qPCR software. Reaction mixtures contained 10 ng or 100 ng template for biofilm or in vivo expression experiments, respectively, and reactions were done as described previously (44). qPCR primer sequences are listed in Table 2. All primer pairs showed efficiencies of 97% or greater (data not shown). At least five samples were tested for each condition and each template sample was tested in triplicate. Controls lacking reverse transcriptase were included. Data were analyzed as described previously (44).
Resolution assays.
Expression of cdpA during infection was monitored using recombination-based in vivo expression technology (RIVET) (6). The res-neo-sacB-res cassette was stably integrated in the lacZ gene by conjugation between AC2528 containing pRes and C6709, yielding C6709 Res (35). The transcriptional fusion cdpA::tnpR was made in C6709 Res by mating with E. coli containing pIVET5ncdpA (35, 44), placing production of the TnpR resolvase under the control of the cdpA promoter.
The cdpA::tnpR strain was inoculated intragastrically into six 5-day-old CD-1 mice as described for competition assays. At 7 and 21 h p.i., three mice were sacrificed, and bacteria were recovered from the small intestines and plated on LB agar containing Sm-Ap and Sm-Km. The percentage of CFU that became Km sensitive was determined at each time point for each mouse. As a control, a strain with a tnpR fusion to ctxA was analyzed in parallel (27). Expression of the fusions in stationary-phase (10-h, 37°C) LB broth culture was analyzed as a control by plating dilutions and quantifying the percent loss of Km resistance as described above.
The effect of ectopically increasing c-di-GMP on virulence gene expression during infection was assessed by expressing vdcA or vdcA(I) during infection in a C6709 derivative previously used to measure in vivo transcription of the major virulence gene regulator toxT (28). This RIVET strain contains a transcriptional fusion of tnpR to toxT and a Tc resistance gene flanked by res sites stably integrated into lacZ. pVdcA and pVdcAi were transformed into this strain by electroporation. The amount of IPTG used, 50 μM, was experimentally determined to induce gene expression and protein detectable by Western blotting but not to affect the stability of the plasmid in the absence of antibiotic selection (data not shown). The pVdcA and pVdcAi RIVET strains were prepared for inoculation as described for the competition assays above, except inocula consisting of 10 μl of the IPTG-induced bacterial suspension were added to 990 μl LB containing 50 μM IPTG. At multiple time points p.i., bacteria were recovered from the small intestines of the mice (three per time point per strain) and plated on LB agar plates containing Sm and Sm-Tc to measure rates of Tc sensitivity and on Sm-Ap to test for retention of the plasmid. Two independent experiments were done; a representative experiment is shown. For all resolution assays, data were analyzed using the Mann-Whitney U test and a P value of <0.05 was considered significant.
RESULTS
Identification of a PDE that decreases the global concentration of c-di-GMP in V. cholerae.
Because the c-di-GMP PDE VieA did not regulate the biofilm formation, motility, or virulence of V. cholerae biotype El Tor (27) (Tamayo and Camilli, unpublished), we sought to identify another EAL domain PDE that could substitute for VieA and modulate the transition from biofilm to colonization of a host by decreasing the c-di-GMP level. We focused on VC0130, because like vieA, it was identified as an in vivo-induced gene in mice and in humans by use of RIVET (31, 35). VC0130 is annotated as encoding a GGDEF family protein but also encodes an EAL domain (Fig. 1A). Interestingly, the GG(D/E)EF motif in the VC0130 protein is degenerate, having the sequence GVGEW. Structural analysis of the DGC PleD from C. crescentus showed that the second glycine is required for catalysis (8). Since this residue is absent from the VC0130 protein, this was expected to preclude DGC activity. The EAL motif is also divergent (ECL); however, other sequences conserved among EAL domains, such as the IDDFGTG and LKLD motifs, are present. We have found that the alanine residue of the VieA EAL motif could be mutated with no effect on PDE activity (Tamayo and Camilli, unpublished). We thus predicted that the VC0130 protein has c-di-GMP PDE activity and so could function similarly to VieA during infection, i.e., up-regulation of VC0130 during infection could reduce intracellular c-di-GMP, resulting in increased virulence factor expression and decreased expression of vps.
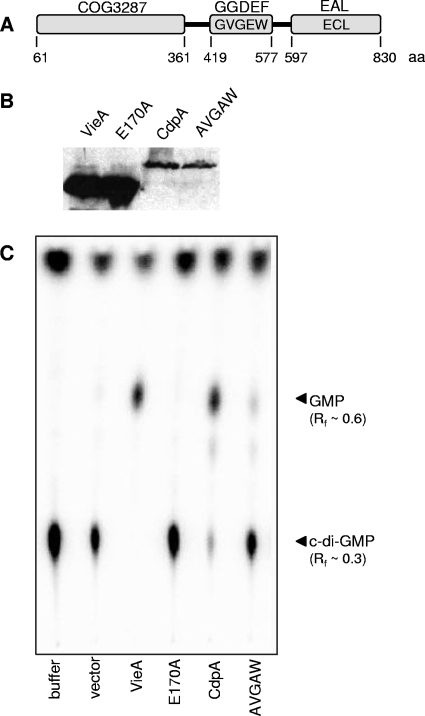
FIG. 1.
cdpA encodes a c-di-GMP PDE. (A) The domain structure of the VC0130 protein, CdpA, is shown. CdpA is a three-domain protein containing an N-terminal domain of unknown function (COG3287), a GGDEF, and an EAL domain. The divergent amino acid sequences present in cdpA at the GG(D/E)EF and EAL motifs are indicated in the relevant domains. The amino acids bounding each domain are noted below. (B) Lysates from AC1817 [VieA], AC1835 [VieA(E170A)], AC2376 [CdpA], and AC2642 [CdpA(AVGAW)] were assayed for protein expression by Western blot analysis. The WT and respective mutant proteins were comparably expressed. No His6-tagged protein was detected in vector-only controls (not shown). (C) Lysates containing the indicated proteins as well as buffer- and vector-only controls were tested for the ability to hydrolyze radiolabeled c-di-GMP. The reactions were analyzed by TLC; products and corresponding Rf values are indicated on the right.
To determine whether the VC0130 gene encodes a DGC, a c-di-GMP PDE, or both, the full-length gene product was marked with a hexahistidine tag at the C terminus and expressed from an IPTG-inducible promoter in E. coli. Production of the proteins by E. coli was confirmed by Western blotting using anti-pentahistidine antibodies (Fig. 1B). Lysates from E. coli producing the VC0130 protein along with VieA, VieA(E170A), and vector-only controls were tested for the ability to hydrolyze c-di-GMP. Whereas the VieA(E170A) and vector negative controls showed no hydrolysis of c-di-GMP, VC0130 protein-containing lysates showed degradation of c-di-GMP (Rf ∼ 0.3) to GMP (Rf ∼ 0.6) (Fig. 1C).
To detect DGC activity, VC0130 protein- or VdcA-containing lysates, as well as a vector-only control, were assessed for the ability to synthesize c-di-GMP using radiolabeled GTP as the substrate. VdcA was chosen as a control for these studies because it was previously shown to have in vitro DGC activity (48, 50). Whereas the reaction products of VdcA showed the presence of c-di-GMP, the VC0130 protein was unable to produce c-di-GMP (data not shown). Thus, as predicted by sequence analysis of the GGDEF and EAL domains of VC0130, the VC0130 protein possesses c-di-GMP PDE activity and so was named CdpA, for cyclic diguanylate PDE A.
Because cdpA has in vitro PDE activity, inactivation of cdpA was predicted to increase the global intracellular concentration of c-di-GMP in V. cholerae. 2D-TLC of radiolabeled nucleotide extracts showed that the cdpA mutant has a level of c-di-GMP almost fourfold higher than that of the respective parent strain (Table 3). The phenotype could be partially complemented by the introduction of cdpA on a low-copy-number, IPTG-inducible expression vector, pMMBneo. Overexpression of cdpA in the mutant by the addition of 1 mM IPTG led to undetectable levels of c-di-GMP. These data further support the assignment of cdpA as a c-di-GMP PDE gene.
TABLE 3.
c-di-GMP levels in cdpA strains
| Strain | Description | Ratio of c-di-GMP to GMP | Fold change in c-di-GMPa |
|---|---|---|---|
| AC61 | WT | 0.054 | N/A |
| AC2131 | cdpA | 0.199 | 3.7 |
| AC2377 | cdpA(pCdpA) | 0.153 | 2.8 |
| AC2377 | cdpA(pCdpA) + IPTG | 0.057 | 1.1 |
Data from one representative experiment are shown. N/A, not applicable.
The GGDEF domain is necessary for PDE activity of CdpA.
CdpA contains a degenerate GGDEF domain in addition to the EAL c-di-GMP hydrolytic domain. Another divergent GGDEF motif (GEDEF) was previously shown to modulate the activity of the tandem EAL domain in CC3396 of C. crescentus (10). We hypothesized that the GGDEF domain could play a similar regulatory role in CdpA. To test this, we assayed the effect of the divergent GGDEF domain (GVGEW) on the PDE activity of CdpA.
A CdpA derivative in which the GVGEW sequence was mutated to AVGAW was made [CdpA(AVGAW)]; the altered residues were shown to be involved in GTP binding in the DGC PleD (8). The ability of CdpA(AVGAW) to hydrolyze c-di-GMP in vitro was determined. E. coli lysate containing CdpA(AVGAW) degraded less c-di-GMP than did that containing WT CdpA (Fig. 1C); both WT and mutant proteins were comparably produced by E. coli (Fig. 1B). Because this method is not quantitative and because it may not reflect the regulation of activity by V. cholerae, other methods were used to determine the role of the GGDEF domain in CdpA activity.
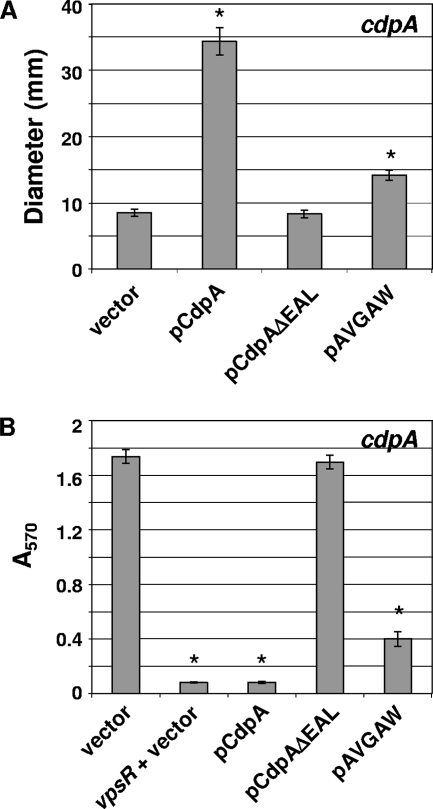
A low concentration of c-di-GMP is necessary for motility in V. cholerae (3, 37). WT cdpA, cdpA(AVGAW), and cdpA lacking the EAL domain [cdpA(ΔEAL)] were expressed from pMMBneo in the cdpA strain and assessed for the ability to increase the motility of V. cholerae on soft agar plates compared to that of the full-length genes. Expression of WT cdpA increased motility fourfold compared to what was seen for the parent strain containing empty vector (Fig. 2A). The vector-only control showed motility comparable to that of the WT parent (compare to WT in Fig. 3B). Overexpression of cdpA(ΔEAL) did not alter motility, supporting the previous findings that the CdpA GGDEF domain does not possess DGC activity and the EAL domain is required for PDE activity. This is in contrast to the reduction of motility seen for the strain containing active DGC (see Fig. 5C below). Expression of cdpA(AVGAW) caused a twofold increase in motility compared to what was seen for the vector-alone strain but was not as effective at enhancing motility as WT cdpA. This suggests that only partial EAL domain activity remains in this mutant derivative.
FIG. 2.
The degenerate GGDEF domain of CdpA is required for optimal PDE activity of the EAL domain. Altered PDE activity of the mutated proteins was determined based on their effects on in vitro c-di-GMP-regulated phenotypes in the cdpA strain, namely, motility and biofilm production. (A) The mean diameters of motility are given in mm for strains containing empty vector, pCdpA, pCdpAΔEAL, or pAVGAW (CdpA containing a mutated GGDEF motif). The medium contained 50 μM IPTG to induce gene expression and Km to maintain the plasmids. (B) The amount of biofilm formed by the above-described strains grown under static conditions in the presence of 50 μM IPTG is shown. In both figures, asterisks indicate significant changes from WT. The pCdpA and pAVGAW strains had significantly different motility (P = 9 × 10−5) and biofilm formation (P = 6 × 10−4) levels.
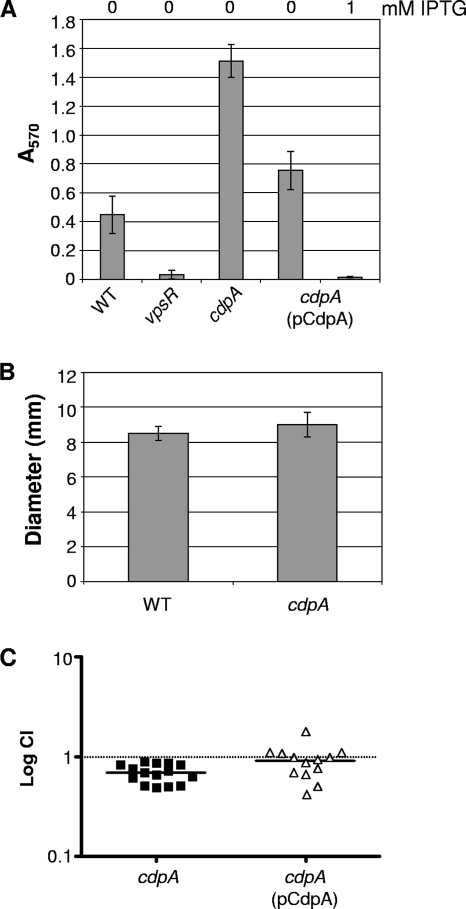
FIG. 3.
CdpA regulates a subset of c-di-GMP-regulated processes. (A) CdpA negatively regulates biofilm production. Shown are the mean A570 values reflecting crystal violet staining of the biofilms formed by WT, the cdpA strain, and the complemented cdpA(pCdpA) strain. The amount of IPTG present in the medium is indicated above each sample. All strains showed significant differences in biofilm formation from WT by Student's t test (P < 0.05). (B) CdpA does not regulate the motility of V. cholerae. Motility was assessed in soft agar medium and is presented as the mean diameter of motility (mm). (C) CdpA is dispensable for V. cholerae colonization of the infant mouse. The cdpA strain was competed against the fully virulent lacZ mutant strain AC66; the cdpA(pCdpA) complementation strain was competed against AC2365 containing empty vector. Each shape represents a CI from a single animal. The horizontal bars indicate the median CI.
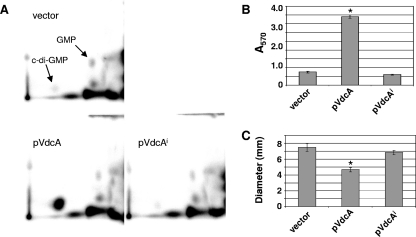
FIG. 5.
Ectopic modulation of intracellular c-di-GMP by expression of vdcA. (A) To directly test the ability of vdcA expression to increase intracellular c-di-GMP, 2D-TLC was done to detect intracellular c-di-GMP in strains containing pVdcA (AC1902), pVdcAi (AC2389), or vector alone (AC1903). The c-di-GMP and GMP spots are indicated. (B) The pVdcA, pVdcAi, and vector-only strains were tested for the ability to activate biofilm formation. The mean intensities of crystal violet staining of the biofilms (A570) are shown. (C) The pVdcA, pVdcAi, and vector strains were tested for decreased motility. The mean diameters of motility in mm from three independent assays are shown. In panels B and C, asterisks indicate significant differences in biofilm and motility, respectively, compared to WT as determined by Student's t test.
The importance of the cdpA GGDEF domain in PDE activity was mirrored when the above strains were tested for V. cholerae biofilm formation, a process that requires a high c-di-GMP concentration (50). Overexpression of cdpA in the cdpA background dramatically inhibited biofilm formation (20-fold) compared to what was seen for the vector control, which produced a WT amount of biofilm (compare to WT in Fig. 3A). Indeed, the expression of cdpA resulted in biofilm production similar to that for the VPS-deficient vpsR mutant, indicating that the c-di-GMP concentration has been dramatically depleted. In contrast, the cdpA(ΔEAL) domain had no effect on biofilm formation (Fig. 2B). Again, these findings are consistent with CdpA being a c-di-GMP hydrolase and with the EAL domain being required for activity. Expression of cdpA-(AVGAW) inhibited biofilm production fourfold, indicating that the GVGEW sequence of CdpA is required for full PDE activity of the EAL domain. The effect of cdpA(AVGAW) expression was intermediate between those of the vector control- and cdpA-expressing strains, suggesting that partial PDE activity remains in the CdpA(AVGAW) mutant.
CdpA affects only a subset of c-di-GMP-regulated phenotypes.
For the section above, we utilized overexpression of cdpA and mutant derivatives to show PDE activity and corresponding effects on motility and biofilm formation. To better assess the physiologic role of CdpA, we tested the cdpA mutant for motility and biofilm formation.
Like mutation of vieA in the classical biotype, mutation of cdpA in El Tor resulted in increased (threefold) biofilm formation (Fig. 3A). Ectopic expression of cdpA complemented the phenotype. CdpA thus functions as a repressor of biofilm formation in the El Tor biotype. This is likely due to CdpA PDE activity, which reduces intracellular c-di-GMP and in this way could impair vps gene expression and biofilm formation.
The effect of CdpA on biofilm formation could occur at the level of gene transcription and/or protein activity. To examine if cdpA expression is increased during biofilm formation, we used qPCR to assess transcript levels. The cdpA transcript level was 4.8-fold higher in biofilm than in broth culture, indicating that cdpA is indeed induced by V. cholerae during biofilm formation (Table 4). For comparison, vpsR, a positive regulator of vps gene expression and biofilm formation (55), was expressed 7.7-fold in biofilm relative to what was seen for broth culture. In addition, RIVET using a cdpA::tnpR transcriptional fusion was adapted for use in measuring cdpA transcription during growth in a biofilm. This method corroborated the result that cdpA transcription is up-regulated in a biofilm compared to growth in broth culture (data not shown).
TABLE 4.
Induction of transcription in biofilm and during infectiona
| Transcript | Comparison | Fold changeb |
|---|---|---|
| vpsR | Biofilm/broth 24°C | 7.0 ± 1.4* |
| cdpA | Biofilm/broth 24°C | 3.9 ± 1.0* |
| cdpA | In vivo/broth 37°C | 8.1 ± 6.0** |
Cycle thresholds for cdpA and vpsR transcripts were normalized to those for rpoB in each sample.
*, P = 0.03 by Mann-Whitney U test; **, P = 0.008 by Mann-Whitney U test.
In the classical biotype, elevated c-di-GMP level reduces motility (37). Consistent with c-di-GMP repressing motility in the El Tor biotype, overexpression of cdpA dramatically increased motility (Fig. 2A). Therefore, we predicted that a null mutation in cdpA would result in reduced motility. However, this was not the case, as the cdpA mutant showed motility identical to that of the parent strain (Fig. 3B).
A vieA mutation causes a 10-fold reduction in colonization of the infant mouse in the classical biotype (51) but has no effect on colonization in the El Tor biotype. An El Tor CVD110 cdpA mutant was previously shown to have reduced virulence in mice (35); however, this virulence-attenuated vaccine strain has significant genotypic and phenotypic differences from other El Tor strains. To determine whether cdpA plays a role in the virulence of WT El Tor C6709, competition assays were done to compare the colonization of cdpA with WT. The mutant showed a negligible defect (median confidence interval [CI] = 0.7) in colonization compared to the WT (Fig. 3C). The colonization phenotype could be complemented by expressing cdpA in trans. However, this slight decrease in colonization was attributed to a slight growth defect of the cdpA strain (in vitro CI ∼ 0.8).
cdpA expression is not regulated by c-di-GMP concentration.
Since c-di-GMP activates the expression of known biofilm genes, namely, vps genes, we asked whether c-di-GMP, which is expected to be at a high level in biofilm, could act as a positive regulator of cdpA expression. This would result in the hydrolysis of c-di-GMP by CdpA and thus constitute a negative-feedback loop to limit vps expression and biofilm formation.
To test this, qPCR was used to measure the levels of cdpA transcript in V. cholerae containing WT, high, and low levels of c-di-GMP. To achieve this, V. cholerae strains ectopically expressing vdcA (AC1902, high c-di-GMP), vieA PDE (AC1901, low c-di-GMP), or vector alone (AC1903, WT c-di-GMP) were used. All strains were grown with aeration at 37°C in LB broth containing IPTG to induce protein production. No differences were seen in cdpA expression under high and low c-di-GMP levels; thus, c-di-GMP is not a regulator of cdpA transcription under the condition examined (data not shown). In contrast, expression of the vpsR control, which is known to be activated by c-di-GMP, was found to be 2-fold higher in the high c-di-GMP condition and 14-fold lower in the low-c-di-GMP condition.
cdpA expression is induced during a late stage of infection of the infant mouse.
cdpA was originally identified in a RIVET screen for genes induced during infection of the infant mouse (35). The identification of cdpA as a gene induced during growth in a biofilm ostensibly is contradictory to this, as growth in the host and biofilm environments are considered distinct. In addition, we failed to observe a requirement for cdpA in colonization of the mouse.
Because a different strain of V. cholerae was used in the original study, we first sought to confirm that cdpA transcription is induced during infection in the current strain, C6709. qPCR analysis showed that at 24 h p.i., cdpA was induced 7.1-fold in the mouse small intestine compared to what was seen for the mid-log-phase broth culture. Thus, cdpA is induced during infection in this strain.
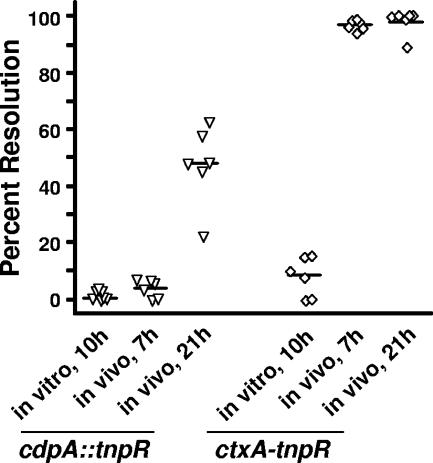
A genetic screen done in our lab suggested that cdpA expression is induced late during the infection process (44). If so, this may explain the lack of a virulence phenotype, since the mouse model of colonization highlights early stages of infection. To address this, the temporal expression of cdpA during infection of the mouse was investigated using the RIVET reporter system. The cdpA::tnpR strain, as well as a ctxA-tnpR control strain, was inoculated into infant mice. At 7 and 21 h p.i., the percentages of Km-sensitive CFU present in the mouse small intestines were enumerated, indicating the percentages of bacteria that had expressed cdpA during infection. At 7 h p.i., only 4% of CFU were Km sensitive, indicating that cdpA was not expressed at early stages of infection (Fig. 4). However by 21 h, 47% of CFU were Km sensitive. In contrast, ctxA, which previously was shown to be expressed early during infection (28), showed high levels of expression (97% Km sensitive) by 7 h. Neither cdpA nor ctxA were expressed during growth in vitro (0.3% or 8% Km sensitive, respectively). These results show that cdpA transcription is activated at a late stage during colonization of the mouse.
FIG. 4.
Expression of cdpA during infection of the mouse. Expression of cdpA during infection of the mouse was measured at 7 h and 21 h p.i. using RIVET. Expression in shaking broth culture (37°C) was included as a control. Expression of ctxA, a virulence gene known to be induced early during infection, was analyzed in parallel. Transcription of cdpA was not induced at 7 h p.i. compared to what was seen in vitro but was activated by 21 h p.i. (P = 0.002). In contrast, transcription of ctxA was induced by an early time point during infection (P = 0.001). In both figures, each symbol represents the percentage of CFU that were Km sensitive (resolved) under each condition at each time point in an individual sample. Horizontal lines indicate the median percent resolved CFU.
c-di-GMP regulates virulence of V. cholerae biotype El Tor.
Mutation of VieA and loss of its c-di-GMP hydrolytic activity results in reduced virulence in the classical biotype (51) but not the El Tor biotype (27) of V. cholerae. Thus, it was unclear if c-di-GMP regulates virulence in the El Tor biotype. Because cdpA is expressed during a late stage of infection, a cdpA mutant is not amenable for use in the analysis of the effect of high c-di-GMP on the virulence of El Tor biotype V. cholerae. Therefore, we further characterized the role of c-di-GMP in the regulation of virulence in the El Tor biotype by use of an alternative strategy. The effect of increased intracellular c-di-GMP on the ability of V. cholerae to colonize the mouse small intestine was assessed by artificially increasing c-di-GMP through the expression of vdcA, previously shown to activate biofilm formation and inhibit motility (3, 50).
To directly test whether expression of vdcA increases intracellular c-di-GMP, 2D-TLC was used to compare the amounts of c-di-GMP in nucleotide extracts from bacteria expressing vdcA and vdcA(I) to that from bacteria with vector alone in the presence of 1 mM IPTG. VdcA(I) is expected to lack DGC activity due to an E258A mutation in the predicted catalytic site. VdcA dramatically increased c-di-GMP relative to the vector control, whereas VdcA(I) did not (Fig. 5A). These experiments confirmed that VdcA increases c-di-GMP in V. cholerae and that the G258A mutation in VdcA(I) abolishes DGC activity.
Assays were performed to ensure that the ectopic overexpression of vdcA, and the consequent increase in c-di-GMP, result in the expected downstream in vitro effects, namely, increased biofilm formation and lowered motility (3, 37, 50). Strains containing pVdcA, pVdcAi, or vector were tested for the ability to form biofilm. Expression of vdcA caused a fourfold increase in biofilm formation compared to what was seen for vector and vdcA(I) controls, indicating the proper activation of biofilm formation by a high concentration of c-di-GMP (Fig. 5B). Motility, on the other hand, is repressed by c-di-GMP (3, 29, 37). VdcA but not VdcA(I) reduced the motility of C6709 compared to the vector control (Fig. 5C). Thus, VdcA modulates c-di-GMP and affects downstream phenotypes appropriately, and VdcA(I) serves as a control for protein levels without modifying c-di-GMP concentration.
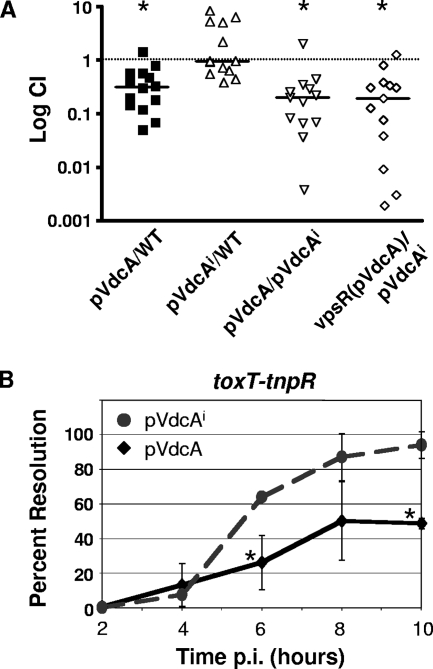
V. cholerae with elevated c-di-GMP was analyzed for virulence using competition experiments in an infant mouse model of colonization. Bacteria containing pVdcA showed a threefold decrease in colonization relative to the vector control (Fig. 6A); the pVdcAi strain showed no such virulence defect. In fact, when the pVdcA strain was competed directly against the pVdcAi strain to control for any defects due to protein level, fivefold-reduced colonization was seen for the bacteria with increased c-di-GMP. Importantly, these defects were not seen when the strains were competed in vitro (data not shown), so attenuation of virulence was not due to a general reduction in growth. Furthermore, the plasmids were maintained at greater than 98% during the course of infection.
FIG. 6.
The effects of increased c-di-GMP on virulence of V. cholerae. (A) The pVdcA and pVdcAi strains were competed against WT containing vector alone in mice. In addition, the VdcA and VdcA(I) strains were competed directly to control for any effects of protein levels. Each symbol represents the CI obtained from an individual mouse. The horizontal bars indicate the geometric mean CI. Asterisks denote statistically significant differences in colonization (P < 1 × 10−4). (B) Transcription of toxT during infection of the mouse in the presence of WT (pVdcAi) and ectopically increased c-di-GMP (pVdcA) was measured using RIVET. The percentage of CFU that were Tc sensitive, and therefore the percentage of bacteria that have expressed toxT, at the indicated time points p.i. are shown. Each data point represents the mean from at least three mice. Asterisks indicate significant differences in resolution rate at that time point as determined by Student's t test.
One possible cause of the virulence defect of the pVdcA strain is the high amount of VPS produced by these bacteria. Such increased VPS synthesis could physically impede colonization or metabolically slow replication in the small intestine. To address this possibility, pVdcA was transformed into V. cholerae lacking vpsR and tested in competition experiments. Deletion of vpsR by itself did not affect colonization (25; also data not shown). The vpsR(pVdcA) strain showed the same attenuation as the pVdcA strain, suggesting that the colonization defect of the pVdcA strain was independent of VPS production.
c-di-GMP inhibits virulence gene transcription during infection of the infant mouse.
Because increased VPS was excluded as a cause for reduced colonization of the mouse small intestine by bacteria with high c-di-GMP, we hypothesized that the attenuation of virulence in the pVdcA strain was a result of reduced virulence gene transcription. In vitro studies using the classical biotype have shown that high c-di-GMP due to loss of VieA PDE activity inhibits the transcription of V. cholerae virulence genes, including ctxAB and the major virulence gene transcriptional regulator toxT (51). However, the effect of c-di-GMP on virulence gene expression during infection has not been directly tested.
VdcA and VdcA(I) were used to investigate the effect of c-di-GMP on virulence gene transcription in vivo. For these experiments, pVdcA and pVdcAi were transformed into an El Tor strain that was previously constructed to monitor toxT transcription using RIVET (28). In this strain, the resolvase gene tnpR is fused to toxT and a res-tet-res cassette is stably integrated in the lacZ locus. Previous studies using this strain have shown that toxT is not expressed during in vitro growth but is induced during infection, leading to production of TnpR, resolution of the res-tet-res cassette, and loss of Tc resistance.
The toxT-tnpR strains containing pVdcA and pVdcAi were determined to produce VdcA and VdcA(I) at comparable levels detectable by Western blotting (data not shown). In addition, these strains were tested for proper regulation of c-di-GMP by use of biofilm assays. Whereas VdcA dramatically increased biofilm formation compared to the vector control, VdcA(I) did not, indicating that c-di-GMP was increased only in the pVdcA toxT-tnpR strain (data not shown).
Expression of toxT-tnpR during infection was determined by measuring the rate of resolution at different time points p.i. At 2 h p.i., toxT was not yet expressed in the pVdcA and pVdcAi toxT-tnpR strains, since almost all bacteria isolated from the small intestines were still Tc resistant (Fig. 6B). By 4 h, toxT was beginning to be expressed, and by 6 h p.i., the pVdcA strain showed a twofold reduction in toxT transcription compared to the pVdcAi strain, which allows the normal regulation of c-di-GMP. Whereas 63% of the pVdcAi bacteria had expressed toxT and lost Tc resistance, only 26% of the pVdcA strain, which has artificially high c-di-GMP during infection, had expressed toxT. Inhibition of toxT transcription by c-di-GMP was seen at subsequent time points as well. The plasmids were maintained in more than 98% of the CFU during infection. In addition, toxT was not expressed by either strain under in vitro growth conditions (data not shown). Thus, elevated c-di-GMP caused a reduction in transcription of toxT during infection.
DISCUSSION
We have proposed a model for V. cholerae in which intracellular c-di-GMP is elevated during persistence in aquatic reservoirs, particularly when V. cholerae is associated in a biofilm, and is reduced upon entry into the host small intestine to allow proper expression of virulence genes. This model was developed based on studies of the PDE VieA in classical-biotype V. cholerae (37, 50, 51). However, a vieA mutation causes neither biofilm, motility, nor virulence phenotype in the El Tor biotype, which causes cholera today. This may be due to differences in levels of vieA expression in the classical and El Tor biotypes. Whole-genome expression studies showed that vieA is expressed at fivefold-higher levels in the classical biotype than the El Tor when grown in vieA-inducing conditions (2). Moreover, VieA regulates 401 genes in the classical biotype but does not significantly affect gene expression in the El Tor biotype (2). VieA thus appears to have lost its role as a major c-di-GMP PDE in the El Tor biotype, at least under the conditions that have been examined.
While VieA, specifically its c-di-GMP PDE activity, in the classical biotype has been shown to be essential for colonization of the small intestine and to activate virulence gene transcription in vitro, the effect of c-di-GMP on virulence and virulence factor expression during infection has not been addressed directly for V. cholerae, specifically El Tor. Herein we investigated the role of c-di-GMP early and late during infection by the El Tor biotype.
The first approach undertaken involved the analysis of cdpA. Like the vieSAB operon containing vieA, cdpA was identified as a gene induced during infection of the mouse (35). In addition, cdpA transcription was found to be induced during infection of humans (31). In contrast to vieA in the classical biotype, cdpA in the El Tor biotype was necessary only for the inhibition of biofilm formation and had no effect on motility or colonization. This was observed for El Tor strain N16961 as well. A cdpA mutation in classical-biotype V. cholerae O395, however, had no significant effect on motility or biofilm (data not shown). This underscores key differences in the ways the two biotypes regulate c-di-GMP and downstream processes.
We show that CdpA possesses PDE but not DGC activity by direct enzymatic assays, as well as in vitro phenotype assays, despite the presence of a GGDEF domain. However, the GGDEF domain was required for the full PDE activity of CdpA. This has been observed previously for other c-di-GMP PDEs that contain tandem GGDEF and EAL domains, including CC3396 of C. crescentus and P. aeruginosa FimX and, more recently, BifA (10, 19, 22). For CC3396, it was demonstrated that rather than catalyzing c-di-GMP biosynthesis, the degenerate GEDEF sequence bound GTP and decreased the Km for c-di-GMP hydrolysis by the EAL domain (10). We propose that this occurs for CdpA as well.
Because cdpA was identified as being induced during infection (35), it was surprising that it regulated biofilm but not colonization. We demonstrated that cdpA is expressed at high levels during growth in a biofilm compared to in shaking culture. That cdpA is expressed during biofilm formation and that there is elevated biofilm formation in the cdpA mutant suggest that c-di-GMP PDE induction plays a role in limiting c-di-GMP and the production of biofilm. Such a control mechanism limiting biofilm formation has yet to be described for any biofilm-forming bacterial species.
In addition, analysis of the temporal expression of cdpA showed that cdpA transcription occurs only at a late stage of infection, in contrast to ctxA, which was highly expressed soon after entry into the small intestine. Recent work from our laboratory has found that V. cholerae expresses a set of genes specifically at a late stage of infection and that many of these genes confer an advantage for survival in conditions representative of aquatic environments (44). Several putative GGDEF domain genes predicted to encode DGCs, as well as cdpA, were among the late in vivo-induced genes (44). It is possible that cdpA acts to counteract the increased c-di-GMP that would occur as a result of increased DGC activity. We suspected that induction of cdpA expression might occur as a direct response to increasing c-di-GMP in both biofilm and late infection conditions. However, this was not the case, as transcription levels of cdpA were identical in high and low c-di-GMP concentrations.
The identification of cdpA as a gene induced in parallel with putative DGCs late in infection suggests an interesting role for CdpA in the small intestine. Increased production of VPS while in the host may be detrimental, and expression of cdpA may keep VPS production in check until the bacteria have exited the host. Alternatively, or in addition, the purpose of limiting c-di-GMP at this time of infection may be to allow induction of motility and chemotaxis genes. Planktonic V. cholerae is motile in human rice water stool, and recently V. cholerae has been shown to become motile as part of a “mucosal escape response” during which the bacteria detach from the epithelial surface (34). In fact, the expression of chemotaxis and motility genes is up-regulated during this process in rabbit ileal loops (54). This model is supported by the finding that cdpA transcription is up-regulated in human rice water stool as well (31).
These experiments underscore an important mechanism of regulation of c-di-GMP in the cell, namely, differential regulation of transcription of c-di-GMP metabolic genes. Indeed, regulation of enzyme production explains the differences in phenotypes observed as a result of mutating cdpA from what is seen for previously characterized PDE genes such as vieA and bifA. BifA represses biofilm formation and enhances motility of P. aeruginosa, as VieA does in the classical biotype of V. cholerae (22, 50, 51). These results suggest that VieA and BifA are expressed in their respective species under the conditions in which biofilm and motility are assayed. Notably, vieA has been shown to be transcribed in LB broth, as well as in vivo (26). The transcription of cdpA, on the other hand, is regulated differently. Resolution assays monitoring cdpA transcription showed that it is not appreciably expressed in LB broth (data not shown). As the medium used in the motility experiments is a similarly rich medium, it is possible that cdpA is not expressed in that medium, thus resulting in the lack of a motility phenotype for the cdpA mutant. Moreover, the biofilm assays used in this study were done in LB broth, so the induction of cdpA transcription was dependent on static growth as a biofilm, not on the medium.
Because cdpA is not expressed early during infection and has no discernible role in V. cholerae colonization of the small intestine, mutation of this gene was not a suitable strategy for investigating the effect of increased c-di-GMP on early stages of infection by the El Tor biotype. As an alternative approach, c-di-GMP was artificially increased by ectopic expression of vdcA. The advantage of this strategy is that it mimics the deletion of PDE(s) critical to reducing c-di-GMP upon entry into the host. V. cholerae with high c-di-GMP showed reduced colonization of the mouse small intestine. Decreased colonization was attributed to reduced expression of the major virulence gene regulator toxT during infection. These data provide the first direct evidence that high c-di-GMP in El Tor biotype V. cholerae is deleterious to colonization and virulence gene expression during infection of the small intestine, as predicted by the vieA studies with the classical biotype. The limitation of these studies is that, as the level of c-di-GMP that occurs naturally in V. cholerae during infection is unknown, the concentration of c-di-GMP achieved by vdcA expression during infection may not be representative of physiological fluctuations in c-di-GMP.
This work shows that c-di-GMP is inhibitory to the virulence of El Tor biotype V. cholerae at early stages of infection and suggests it may be deleterious to pathogenesis at later stages as well. Which of the 31 EAL or HD-GYP domain putative c-di-GMP PDE(s) mediates the reduction of c-di-GMP in the El Tor biotype upon entry into the small intestine remains unknown, but VieA likely serves this function in the classical biotype. It is also possible that PDEs are already present in V. cholerae and that signals from the host gastrointestinal tract activate enzymatic function. Later during infection, CdpA, and perhaps other PDEs, are produced. PDE expression at this stage may serve to counteract the activities of DGCs made at this time, with the purpose of minimizing VPS production during infection and/or allowing expression of motility and chemotaxis genes.
Acknowledgments
We are grateful to E. Munoz for critically reading the manuscript as well as to the Camilli lab for helpful suggestions and discussion.
This research was supported by NIH grant AI45746 to A.C. and by the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928). R.T. was supported by NIH training grant T32 AI07329-14. A.C. is an investigator of the Howard Hughes Medical Institute.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 471695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 743633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 1883600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247123-130. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 2579759-9769. [PubMed] [Google Scholar]
- 6.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 912634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 10117084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. A. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 403420-3426. [DOI] [PubMed] [Google Scholar]
- 10.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 594310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54264-277. [DOI] [PubMed] [Google Scholar]
- 15.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hisert, K. B., M. MacCoss, M. U. Shiloh, K. H. Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 561234-1245. [DOI] [PubMed] [Google Scholar]
- 17.Huang, B., C. B. Whitchurch, and J. S. Mattick. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 1857068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huq, A., E. B. Small, P. A. West, M. I. Huq, R. Rahman, and R. R. Colwell. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 601026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 5475-88. [DOI] [PubMed] [Google Scholar]
- 21.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303371-382. [DOI] [PubMed] [Google Scholar]
- 22.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 1032839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larocque, R. C., J. B. Harris, M. Dziejman, X. Li, A. I. Khan, A. S. Faruque, S. M. Faruque, G. B. Nair, E. T. Ryan, F. Qadri, J. J. Mekalanos, and S. B. Calderwood. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 734488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 1864864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 1802298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 986889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99625-634. [DOI] [PubMed] [Google Scholar]
- 29.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 30.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombardo, M., J. Michalski, H. Martinez-Wilson, C. Morin, T. Hilton, C. G. Osorio, J. P. Nataro, C. O. Tacket, A. Camilli, and J. B. Kaper. 2007. An in vivo expression technology (IVET) screen for V. cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. USA 10418229-18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 9739-47. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osorio, C. G., J. A. Crawford, J. Michalski, H. Martinez-Wilson, J. B. Kaper, and A. Camilli. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 28212860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinones, M., H. H. Kimsey, and M. K. Waldor. 2005. LexA cleavage is required for CTX prophage induction. Mol. Cell 17291-300. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, A., G. D. Pearson, and J. J. Mekalanos. 1992. Cholera vaccines strains derived from a 1991 Peruvian isolate of Vibrio cholerae and other El Tor strains, p. 43-47. Proc. 28th Joint Conf. U.S.-Jpn. Coop. Med. Sci. Program Cholera Relat. Diarrh. Dis.
- 40.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose biosynthesis in Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149101-117. [Google Scholar]
- 41.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325279-281. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 1871792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schild, S., R. Tamayo, E. J. Nelson, F. Qadri, S. B. Calderwood, and A. Camilli. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2264-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 1874774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoolnik, G. K., M. I. Voskuil, D. Schnappinger, F. H. Yildiz, K. Meibom, N. A. Dolganov, M. A. Wilson, and K. H. Chong. 2001. Whole genome DNA microarray expression analysis of biofilm development by Vibrio cholerae O1 E1 Tor. Methods Enzymol. 3363-18. [DOI] [PubMed] [Google Scholar]
- 47.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 531123-1134. [DOI] [PubMed] [Google Scholar]
- 48.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 28033324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamplin, M. L., A. L. Gauzens, A. Huq, D. A. Sack, and R. R. Colwell. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 561977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 735873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416207-211. [DOI] [PubMed] [Google Scholar]
- 54.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 1001286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]