Abstract
The aim of this investigation was to study the effect of polysaccharide capsule on the gene expression in dendritic cells (DC) during their interaction with Cryptococcus neoformans. To this end, we used an encapsulated virulent strain of C. neoformans and a cap59 gene-disrupted acapsular avirulent strain derived from the same genetic background. DC were exposed to encapsulated and acapsular C. neoformans strains for 4 h and 18 h, and their transcriptional profiles were analyzed using the Affymetrix mouse gene chip U74Av2. A large number of DC genes were up-regulated after treatment with the acapsular strain. In particular, we observed the up-regulation of the genes involved in DC maturation, such as cell surface receptors, cytokines, and chemokines (interleukin-12 [IL-12], IL-2, IL-1α, IL-1β, IL-6, IL-10, tumor necrosis factor alpha, CCR7, CCL17, CCL22, CCL3, CCL4, CCL7, and CXCL10), membrane proteins, and the genes involved in antigen processing and presentation as well as cell cycle or apoptosis. The chemokine gene expression data were confirmed by real-time reverse transcription-PCR, while the expression of cytokine genes was correlated with their secretion. A completely different pattern of gene expression was observed for DC treated with an encapsulated strain of C. neoformans. In particular, no significant induction was observed in the expression of the genes mentioned above. Moreover, a number of genes, such as those coding for chemokines, were down-regulated. These results suggest that the polysaccharide capsule shrouding the cell wall of C. neoformans plays a fundamental role in inducing DC response, highlighting the molecular basis of the true nature of immune silencing exerted by capsular material.
Cryptococcus neoformans is an opportunistic encapsulated yeast that causes pulmonary, cerebral, and disseminated infections primarily in patients with defective T-cell immunity, such as those with AIDS, hematological malignancies, and organ transplants (33). The polysaccharide capsule is a major virulence factor of C. neoformans, the concept of which was initially established by studying several acapsular mutants obtained by chemical mutagenesis. These acapsular strains were avirulent in mice, and they were readily ingested by phagocytes, but their ingestion could be inhibited by the addition of purified polysaccharide (3). The effect of the C. neoformans polysaccharide capsule on the host cells can be summarized as follows: it interferes with phagocytosis, blocks the recruitment of inflammatory cells, increases costimulatory molecules, suppresses the delayed-type-hypersensitivity response, and reduces the antibody production in response to fungal infection (11, 43).
Dendritic cells (DC) are professional antigen-presenting cells that can initiate the innate and adaptive immune response against invading pathogens, thus enabling the decoding of microbe-associated information which then results in qualitatively different adaptive T-helper responses in vitro and in vivo (2, 8).
Mouse DC internalize C. neoformans cells via the mannose receptor and the Fc receptor for immunoglobulin G (FcγR). The cryptococcal antigens are then presented to T cells, which undergo proliferation and activation (40). The protective immunity against C. neoformans has been attributed to a T-helper 1 type response (17) with consequent phagocytic-cell activation and direct antifungal activity (33).
The role of capsular material in DC activation and maturation was also investigated using the encapsulated and acapsular strains of C. neoformans. Acapsular strains are easily phagocytosed by immature DC, triggering the up-regulation of several costimulatory molecules, thereby promoting DC maturation. Conversely, encapsulated yeast cells are unable to produce an increase in costimulatory molecules, thus inhibiting maturation (44). These results show how the capsule interferes with DC activation and maturation, thereby hampering an efficient T-cell response. In addition, the presence of the inflammatory process plays a key role in restricting C. neoformans to the local level, preventing dissemination (12). Accordingly, the absence of inflammation observed for immunocompromised hosts represents the negative outcome of cryptococcosis (12).
Microarray technology, which allows the simultaneous analysis of the expression of thousands of genes, has been used to investigate immune system and pathogen interplay. These studies have included analyses of the responses of particular tissues and cells, e.g., macrophages, to a wide range of pathogens, such as bacteria, fungi, helminthes, and protozoan parasites. The majority of these studies have concentrated on the responses of human or mouse cell lines and primary cells to different challenges (30).
In this study, we examined the role of capsular material of C. neoformans in the gene expression profiles of DC. Our previous study pointed out that capsular material of C. neoformans limits DC maturation and activation (44). By analyzing the gene expression profiles, we attempted to characterize the diverse molecular events that are influenced by the presence or absence of capsular material during DC-C. neoformans interaction. To this end, we used mouse myeloid DC line D1 (37) and two strains of C. neoformans, an encapsulated virulent strain (JEC21) and an acapsular avirulent strain (C566) that was constructed by disruption of the CAP59 gene. Mouse myeloid DC line D1 was challenged with both strains. Gene expression profiling revealed that the acapsular strain not only induces maturation of DC but also is an efficient inducer of genes involved in cytokine and chemotaxis activity and immune response as well as T-cell regulation. The presence of capsular material precludes or hampers activation of the genes involved not only in DC maturation but also in the induction of cytokines, chemotaxis, and inflammation that represent crucial events confining C. neoformans to the local level.
MATERIALS AND METHODS
Strains.
The D1 cell line, a long-term growth-factor-dependent immature myeloid (CD11b+ CD8α+) DC line of mouse splenic origin, was provided by P. Ricciardi-Castagnoli (University of Milan—Bicocca, Milan, Italy). The cell line was cultured in Iscove's modified Dulbecco's medium (Sigma, St. Louis, MO) containing 10% heat-inactivated fetal bovine serum (Gibco-BRL, Gaithersburg, MD), 100 IU of penicillin, 100 μg/ml of streptomycin, 2 mM l-glutamine (all from Sigma), and 50 μM β-mercaptoethanol, with 30% supernatant from R1 medium (supernatant from NIH 3T3 fibroblasts transfected with granulocyte-macrophage colony-stimulating factor). JEC21 is an encapsulated virulent strain of C. neoformans, and C566 is an acapsular prototrophic strain obtained from the F1 progeny of a cross between the encapsulated strain JEC33 (MATα lys2), a JEC21 derivative, and an acapsular strain, TYCC33 (MATa cap59 ura5). TYCC33 was constructed by disruption of the CAP59 gene in an ade2 ura5 mutant, B-4530, derived from strain JEC20, which is isogenic to JEC21 (6). C. neoformans was cultured for 2 days on yeast extract-peptone-dextrose (YPD; 1% yeast extract, 2% peptone, 2% glucose) agar at 30°C. C. neoformans (yeast) cells were harvested and heat inactivated at 60°C for 30 min before use.
Phagocytosis assay.
Yeast cells were opsonized with 20% normal mouse serum in phosphate-buffered saline and 10 mM EDTA. Opsonization was performed by incubating the yeast cells at 37°C for 1 h, after which the cells were washed and used for phagocytosis assays. The D1 cell line, at a concentration of 2 × 105, was added to the eight-well Lab-Tek Chamber slide 2 h before stimulation. Adherent DC were then stimulated at a ratio of 1:2 with opsonized encapsulated or acapsular strains for 2 and 4 h. After incubation, the chambers were removed and the slides were washed with RPMI, air dried, fixed, and stained with the Hemacolor kit for cells (Merck). One hundred macrophages were examined per well, and the phagocytic indices (number of cells ingested per macrophage) were determined.
Stimulation of DC by C. neoformans.
For microarray assays, the D1 cell line (3 × 106 cells) was either not stimulated or stimulated at a ratio of 1:2 with yeast cells of either encapsulated or acapsular strains (6 × 106 yeasts cells were first opsonized with normal mouse serum [obtained from naive C57BL/6 mice] at 37°C for 30 min, washed, and incubated with the D1 cell line). The samples were incubated for 4 h and 18 h at 37°C under 5% CO2. For the cytokine assays, 106 D1 cells/ml and CD11c+ DC from naive mice were either not stimulated or stimulated at a ratio of 1:4 with yeast cells of encapsulated and acapsular strains. As a positive control, lipopolysaccharide (LPS) was used at a concentration of 10 μg/ml. Samples were incubated for 18 h at 37°C under 5% CO2. For each experiment, three samples were included at each time point for each strain.
Isolation of CD11c+ cells.
C57BL/6 mice were obtained from Jackson Laboratory. Spleen cell suspensions were prepared by Liberase CI (Roche, Lewes, United Kingdom) and DNase I digestion. DC-enriched fractions were prepared by labeling splenocytes with anti-CD11c MACS beads (Miltenyi Biotec, Bisley, United Kingdom), followed by a positive selection using LS magnetic columns (Miltenyi Biotec). CD11c+-enriched preparations were further checked for purity by flow cytometry.
RNA extraction, amplification, and labeling.
After 4 h and 18 h of incubation, D1 cells and CD11c+ cells from untreated mice were harvested with RNA extraction buffer and kept in a −80°C freezer until RNA was extracted. Antisense cRNA was prepared according to the recommendations of Affymetrix (Santa Clara, CA). Briefly, total RNA was extracted from frozen cell pellets by using the RNeasy mini kit (Qiagen, Chatsworth, CA). For the microarray assay, 5 μg of RNA was reverse transcribed by using a modified oligo(dT) primer with a 5′ T7 RNA polymerase promoter sequence and the Superscript Choice system for cDNA synthesis (Life Technologies, Gaithersburg, MD). Double-stranded cDNA (1 μg) was transcribed to cRNA with the ENZO kit (Affymetrix). cRNA was purified on an RNeasy affinity column (Qiagen, Chatsworth, CA) and then fragmented to an average size of 50 to 200 bp by incubation for 35 min at 94°C in 40 mM Tris-acetate at pH 8.1, 100 mM potassium acetate, and 30 mM magnesium acetate.
Microarray experiments and analysis.
Hybridization and scanning of arrays were performed according to the manufacturer's (Affymetrix) protocol. Labeled cRNA was hybridized to Affymetrix murine chip U74Av2. The murine genome chip U74Av2 consists of 36,000 mRNA transcripts from mouse and expressed sequence tag cluster sequences including 6,000 known mouse genes. The arrays were read at a resolution of 7.5 μm by a confocal scanner and analyzed with the Microarray Suite 5.0 Gene Expression analysis program (both from Affymetrix). The data for each microarray were initially normalized by scaling all signals to a target intensity of 500. Expression analysis was performed using the default parameter settings. Present calls required a P value of <0.05, and marginal calls required a P value of 0.065 for all probe sets. Probe sets with a P value of >0.065 were marked absent. Each array was analyzed based on the present, marginal, or absent call for each probe set. The data were then uploaded to the National Cancer Institute's mAdb microarray website (http://nciarray.nci.nih.gov) and the absolute expression analysis was performed using the mAdb bioinformatics system generated by the NCI/CCR μArray Center.
Three or four replicates for each array were performed, starting with 21 arrays and 7,582 genes. All data were centered to a signal median, and all absent calls or samples in which the signal was less than 30 were excluded. Signals were also floored at 1. Replicate samples at 4 h and 18 h were grouped and compared by statistical analysis. Transcriptional profiles of D1 cells exposed to encapsulated and acapsular strains were compared to those of nonstimulated cells (NS cells) at both 4-h and 18-h time points. The t test was used to analyze the results. All genes with a P value of <0.05 and with a group mean difference of ≥1 or ≤−1 were considered significantly modulated and used for the next analysis. The logical Boolean comparison analysis was used to compare the data from NS cells against data from the cells exposed to encapsulated strains and the data from NS cells against data from the cells exposed to acapsular strains at both 4 h and 18 h. The final output numbers of significantly modulated genes were 170 at 4 h and 330 at 18 h (see Table S1 in the supplemental material). Each set of data analyzed is statistically highly significant, since it represents the average of three or four replicates. Final data sets are expressed as log2 ratios in signal intensity between the cells exposed to encapsulated strains and NS cells and between those exposed to acapsular strains and NS cells at both 4-h and 18-h time points.
Real-time PCR.
The SuperScript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen, Carlsbad, CA) was used to synthesize cDNA by using the Superscript reverse transcriptase. For PCR amplification, 5 μg of RNA was transcribed and 0.5 μl of cDNA was used per 25 μl of PCR mixture. The TaqMan probes (6-carboxyfluorescein dye labeled) for CCL3, CCL4, CXCL10, CCL7, CCL22, and CCR7 (Applied Biosystems) were used, and the relative changes in gene expression were analyzed by the 2−ΔΔCT method and compared to the expression of β2-microglobulin.
ELISA.
D1 cell line and CD11c+ cells (106 cells/ml) were stimulated at a ratio of 1:4 with an encapsulated or acapsular strain of C. neoformans. After 18 h of stimulation, supernatant fluids were recovered and stored at −20°C. Cytokine presence in culture supernatant fluids was measured by enzyme-linked immunosorbent assay (ELISA) for mouse tumor necrosis factor alpha (TNF-α) and interleukin-12p40 (IL-12p40) (BD Biosciences-Pharmingen, San Diego, CA).
Statistical analysis.
For multiple comparison experiments, the analysis of variance (one-way analysis of variance) was performed using Bonferroni's multiple t test. A P value of <0.05 was considered significant.
RESULTS
The mouse DC line D1 is a well-characterized cell line. In this paper, we show that D1 is able to recognize and phagocytize two isogenic strains of C. neoformans with or without a capsule. Following opsonization with normal mouse serum, acapsular and encapsulated strains were added to the D1 cell line and phagocytosis was evaluated after 2 and 4 h. The results from three experiments showed that, following 2 h of incubation, the acapsular strain is easily internalized. In particular, the phagocytic index at 2 h was 1.40 ± 0.20, and the phagocytic index after 4 h of incubation was similar. Moreover, the phagocytic index for the encapsulated strain was also appreciable (0.60 ± 0.04) after 2 and 4 h of incubation.
In order to investigate if the changes in the gene expression profile of DC during interaction with C. neoformans were related to the presence of capsular material, we examined DC gene expression profiles after treatment with two strains of C. neoformans from an isogenic background, one with a capsule and one without capsule. The D1 cell line was stimulated with C. neoformans at a ratio of 1:2 for 4 h and 18 h.
The RNA expression profiles were analyzed using Affymetrix murine chip U74Av2, and the resulting 21-array data set (7,582 genes) were filtered using a P value of <0.05 and a group mean difference in log2 ratio of ≥1 or ≤−1 as described in Materials and Methods. From the data obtained after 4-h incubation, there were 18 and 156 genes which showed >2-fold differences in expression levels in cells stimulated with encapsulated C. neoformans and those stimulated with acapsular C. neoformans, respectively, in comparison to expression in unstimulated cells (see Table S1 in the supplemental material). Among these two groups of genes, four genes were commonly affected in DC exposed to both encapsulated and acapsular C. neoformans strains. In contrast, after 18 h of incubation, the numbers of genes that showed differential expression were 3 and 327 when the unstimulated DC were compared to the cells stimulated with either encapsulated or acapsular strains, respectively, and there was no overlap between these two groups of genes (see Table S1 in the supplemental material).
To evaluate whether the poor stimulatory activity of the encapsulated strain could be due to the suboptimal dose of yeast used in our experimental system, higher doses of both yeasts were tested (effector-to-target cell ratio, 1:5). Results obtained were comparable to those obtained by using lower doses of yeasts (effector-to-target cell ratio, 1:2).
We clustered the differentially expressed genes into different categories based on the proposed gene functions antigen presentation, cytokines, chemokines, transcription factors, and signaling (Table 1). We focused on the genes that are involved in antigen presentation, maturation of DC, cytokine production, and chemotaxis. Most of these genes were significantly up- or down-regulated by the acapsular strain, in contrast to the encapsulated strain, by which only a few genes were modulated.
TABLE 1.
List of differentially expressed genes in DC upon challenging with C. neoformans
| Function/product and gene | Affy idenitification no.a | Log2 ratiob
|
|||
|---|---|---|---|---|---|
| 4 h
|
18 h
|
||||
| Encapsulated/NS | Acapsular/NS | Encapsulated/NS | Acapsular/NS | ||
| Antigen presentation | |||||
| H2-D1 | 97541_f_at | 1.06 | |||
| H2-Q7 | 98438_f_at | 1.02 | |||
| H2-Q8 | 101658_f_at | 1.67 | |||
| H2DMb1 | 101868_i_at | −1.78 | −1.54 | ||
| Relb | 103091_at | 1.53 | 1.78 | ||
| CD86 | 102830_at | 1.02 | |||
| CD1d2 | 101897_g_at | 1.18 | |||
| CD14 | 98088_at | 1.12 | |||
| CD83 | 103040_at | 3.42 | |||
| ICAM1 | 96752_at | 1.36 | |||
| Cytokines and receptor | |||||
| Tgfb3 | 102751_at | 1.05 | 1.92 | ||
| Tgtp | 102906_at | 2.90 | |||
| TNF | 102629_at | 3.79 | |||
| Csf1 | 101450_at | 2.50 | |||
| C3 | 93497_at | 1.94 | |||
| Tnfsf9 | 92415_at | 2.04 | |||
| Tnfrsf5 | 92962_at | 1.50 | 1.87 | ||
| Tnfaip2 | 160489_at | 1.58 | |||
| Tnfrsf11a | 101632_at | 1.04 | |||
| Traf6 | 104189_at | 1.86 | |||
| IL-1a | 94755_at | 2.77 | 2.60 | ||
| IL-1b | 103486_at | 1.63 | |||
| IL-1rn | 93871_at | 1.76 | |||
| Ifi203 | 93321_at | 2.03 | |||
| Ifi204 | 98465_f_at | 2.39 | |||
| Ifit1 | 100981_at | 4.63 | |||
| Ifit2 | 103639_at | 4.73 | |||
| Ifit3 | 93956_at | 3.78 | |||
| Ptger4 | 103362_at | 1.84 | |||
| Irg1 | 98773_s_at | 2.63 | |||
| CD47 | 95020_at | 1.43 | |||
| CD68 | 103016_s_at | −1.09 | |||
| Edn1 | 102737_at | 3.37 | |||
| Chemokines | |||||
| CCL3 | 102424_at | 4.23 | |||
| SCYA4 (CCL4) | 94146_at | 4.48 | |||
| SCYA7 (CCL7) | 94761_at | 3.49 | |||
| CXCL10 | 93858_at | 5.02 | |||
| CCL6 | 92849_at | −1.71 | |||
| CXCL5 | 98772_at | −2.29 | |||
| CCL17 | 97783_at | −1.37 | |||
| CCRL2 | 93617_at | 2.00 | |||
| CCL12 | 93717_at | 1.48 | |||
| CCL22 | 102310_at | 1.42 | 2.43 | ||
| CCL5 | 98406_at | 1.96 | |||
| CCR7 | 104443_at | 3.84 | |||
| CXCL2 | 101160_at | 1.85 | |||
| DNA binding and transcription factors | |||||
| Elf1 | 160721_at | −1.19 | |||
| Sqstm1 | 101995_at | −1.28 | |||
| Irf1 | 102401_at | 1.98 | |||
| Idb1 | 100050_at | −1.56 | |||
| Irf4 | 92737_at | 1.36 | |||
| Irf7 | 162202_f_at | 1.14 | |||
| Isg20 | 103432_at | 2.41 | |||
| Nfkb2 | 103614_at | 1.46 | 1.64 | ||
| Nfkbia | 101554_at | 1.16 | 1.73 | ||
| Nfkbiz | 98988_at | 2.51 | |||
| Ilf2 | 93823_at | 1.05 | |||
| Signaling | |||||
| Sema4b | 95387_f_at | −1.20 | |||
| Ifnra2 | 101014_at | 1.13 | |||
| IL-17r | 99992_at | −1.01 | |||
| Igtp | 160933_at | 2.18 | |||
| Ifih1 | 103446_at | 2.70 | |||
| Socs3 | 162206_f_at | 1.31 | 1.09 | ||
| Stat5a | 100422_i_at | 1.31 | 1.28 | ||
| Ikbkb | 93706_at | 1.39 | |||
Affy identification numbers represent codes provided by Affymetrix.
Each number in the columns represents gene expression as log2 of the ratio of cells stimulated with an encapsulated strain to NS cells or of cells stimulated with an acapsular strain to NS cells. Blank entries indicate no significant changes in gene expression. Each data point is the average of three or four different experiments. Data shown have P values of <0.05 and represent a group mean difference of ≥1 or ≤−1.
Genes related to antigen presentation.
Many major histocompatibility complex class II (MHC-II)-related genes, such as H2-D1, H2-Q7, and H2-Q8, were up-regulated by the acapsular strain at 18 h. H2-DMb1 was the only gene in this group whose expression was down-regulated by both yeast strains after 4 h of incubation: the encapsulated strain induced almost a fourfold decrease (ratio of −1.78), and the acapsular strain induced a threefold decrease (ratio of −1.54).
In addition, the acapsular strain up-regulated the expression of the genes that are hallmarks of DC differentiation, such as the c-Relb gene (28, 46), as well as the genes involved in the up-regulation of costimulatory molecules. In particular, the c-Relb gene was up-regulated after 4 and 18 h with log2 ratios of 1.53 and 1.78, respectively. The expression of costimulatory molecule genes, such as the CD86 gene, was also up-regulated with a ratio of 1.02. Other genes involved in antigen presentation, such as the CD14 gene, were up-regulated by the acapsular strain at 4 h. CD83 and intracellular cell adhesion molecule (ICAM) were both up-regulated at 18 h with ratios of 3.42 and 1.36, respectively.
Cytokine- and receptor-related genes.
Significant numbers of the genes encoding the innate host defense, which includes complement component C3, proinflammatory, and chemotaxis regulators were up-regulated only by the acapsular strain.
Genes expressing cytokines, such as TNF-α, interleukin 1 (IL-1a and IL-1b), colony stimulating factor 1 (Csf1), and cellular receptors (Tnfsfr5, Tnfsf5, Tnfsf9, and Traf6) were induced early at 4 h by acapsular strain. TNF-α was up-regulated almost 16-fold; IL-1α and IL-1β were up-regulated at 4 h with log2 ratios of 2.77 and 1.63, respectively. Other TNF-α-related genes were promptly induced: induction of the TNF ligand 9 (Tnfsf9) was up-regulated fourfold, the TNF receptor 5 (Tnfsfr5) was up-regulated threefold, and the TNF-α-induced protein 2 (Tnfaip2) was up-regulated more than threefold. At 18 h, the expression of TNF ligands 5 and 9 (Tnfsf5 and Tnfsf9) was comparably up-regulated. Acapsular cells also induced up-regulation of the TNF receptor-associated factor (Traf6) with a ratio of 1.86. In addition, the complement component C3 was up-regulated after 18 h with a ratio of 1.94.
Other genes involved in the immune response, such as the gene encoding prostaglandin E receptor 4 (Ptger4), were up-regulated only by the acapsular strain at 18 h. Moreover, the genes encoding receptors, such as CD68, were down-regulated by the acapsular strain at 4 h, while the endothelin-1 gene (Edn1) and CD47 antigen were up-regulated. Acapsular cells also up-regulated several interferon-related genes, such as Ifi203, Ifi204, Ifit1, Ifit2, and Ifit3, which were induced at 4 h. In contrast, the encapsulated cells did not modulate the expression of these genes. The only gene that was regulated by the encapsulated strain in this category was Tgfb3, which was also up-regulated by the acapsular strain; the levels of up-regulation by the two types of strains were twofold and fourfold, respectively. It is known that transforming growth factor b3 (TGF-β3), a known inhibitor of cell proliferation (27), prevents cell cycle progression and leads to the accumulation of cells in G1 or G0 phase.
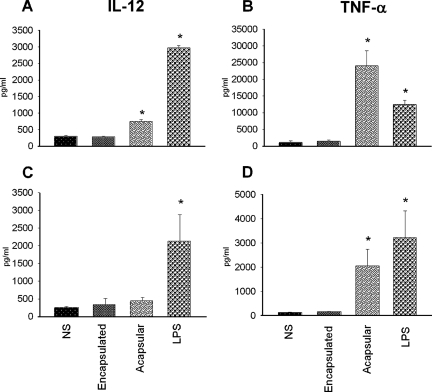
To verify whether the cytokine gene expression induced by the acapsular mutant corresponded to an induction of protein synthesis, we investigated the production of selected cytokines, such as IL-12p40 and TNF-α, after exposing DC to acapsular and encapsulated C. neoformans strains by using an ELISA. Figure 1A shows that acapsular cells induce significantly greater production of IL-12p40 in D1 cells than in NS cells, although the IL-12p40 levels were not as high as those produced following LPS stimulation. On the other hand, IL-12p40 expression in D1 cells was unchanged when stimulated with cells of the encapsulated strain. Moreover, the acapsular strain strongly induced TNF-α production, while the encapsulated strain did not (Fig. 1B). To ascertain whether the results obtained with the D1 cell line were at least partially applicable to ex vivo cells, CD11c+ cells were isolated from the spleens of naive mice and stimulated in vitro with the two strains of C. neoformans and screened for production of the selected cytokines. After 18 h of incubation, the supernatants were recovered and the concentration of IL-12p40 and TNF-α were measured. No significant increase in IL-12p40 production was observed for CD11c+ cells treated with either encapsulated or acapsular strains compared to NS cells (Fig. 1C). Interestingly, acapsular cells stimulated the production of TNF-α significantly, while encapsulated cells did not (Fig. 1D).
FIG. 1.
IL-12p40 and TNF-α production by mouse DC. D1 cell line and CD11c+ cells (106 cells/ml) were stimulated at a ratio of 1:4 with an encapsulated or acapsular strain of C. neoformans. Supernatants of NS and stimulated cells were collected after 24 h and tested for the presence of IL-12p40 and TNF-α. (A and B) IL-12p40 and TNF-α production in the D1 cell line. (C and D) Cytokine production of CD11c+ cells. *, P < 0.05 (cells treated with encapsulated and acapsular yeasts and LPS versus untreated cells).
Chemokine-related genes.
Chemokines are chemotactic factors that are critical for the function of the immune system and can affect a range of additional biological processes, such as angiogenesis and hematopoiesis. We found that the encapsulated strain down-regulated chemokines, such as CCL17, and also did not modulate other chemokine-related genes (Table 1). Conversely, the acapsular strain induced the expression of genes encoding several chemokines. Genes such as CCL3, SCYA4 (CCL4), SCYA7 (CCL7), CCL12, and CXCL10 were highly induced (4 h). In particular, CCL7, CCL3, and CCL4 genes and the CXCL10 gene were up-regulated with ratios of 3.49, 4.23, 4.48, and 5.02, respectively. On the other hand, the chemokine genes, such as CCL6 and CXCL5, were down-regulated with ratios of −1.7 and −2.29, respectively. After 18 h, other chemokines were highly up-regulated by the acapsular strain, in particular, CCL22, CCL5, and CXCL2, with ratios of 2.43, 1.96, and 1.85, respectively. The chemokine receptor CCR7, known to be up-regulated during DC activation/maturation in order for DC to traffic to lymph nodes (13, 15), was up-regulated with a ratio of 3.84.
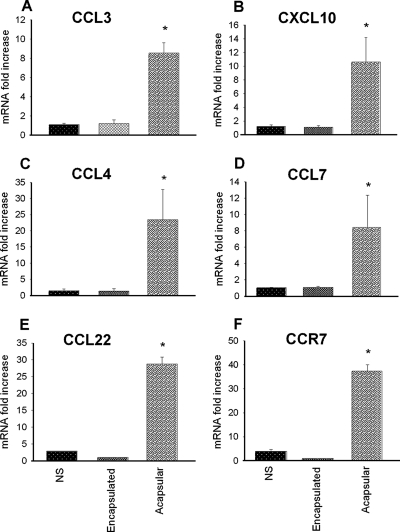
To confirm the microarray data, we performed real-time PCR experiments on genes that showed a >4-fold increase at 4 h and 18 h after stimulation. The same batch of RNA that was used for the array study was transcribed into cDNA and amplified by using TaqMan real-time PCR. Figure 2A shows that the DC treated with acapsular yeast for 4 h showed expression of CCL3 that was 8.56-fold higher than that of the NS cells. In contrast, the encapsulated strain did not influence the transcriptional level of this chemokine in DC. The up-regulation of CXCL10, CCL4, and CCL7 at 4 h after challenging with C. neoformans was also confirmed by RT-PCR. We found that these chemokines were up-regulated by treating DC with the acapsular strain, and the increases were 10.63-, 23.44-, and 7.75-fold, respectively (Fig. 2B, C, and D). Higher expression of the genes for the chemokine CCL22 and the chemokine receptor CCR7 in DC challenged with the acapsular strain for 18 h but not in DC challenged with the encapsulated strain was also confirmed (Fig. 2E and F).
FIG. 2.
Real-time PCR analysis of the chemokine gene expression from D1 cells. Expression levels of CCL3 (A), CXCL10 (B), CCL4 (C), and CCL7 (D) were compared to that of the β2-microglobulin gene from D1 cells 4 h after challenging with C. neoformans. Each data point represents an average of three independent experiments. The relative mRNA levels were calculated using the 2−ΔΔCT method. Expression levels of CCL22 (E) and CCR7 (F) were also measured at 18 h in comparison to the expression level of the β2-microglobulin gene. *, P < 0.05 (cells treated with encapsulated and acapsular yeasts versus untreated cells).
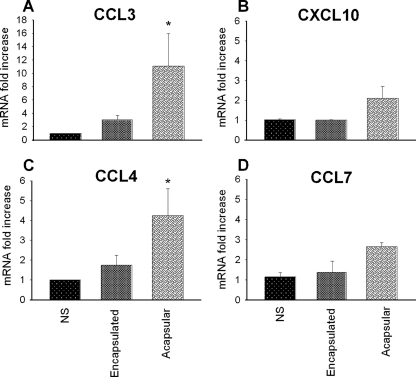
Similar experiments using RT-PCR to quantitate mRNA levels were performed with the CD11c+ cells. These cells were isolated from the mouse spleen and incubated in the presence or absence of C. neoformans cells for 4 h. RNA was extracted from these cells, and the expression levels of CCL3, CCL4, CCL7, and CXCL10 were determined. Only the acapsular strain induced a significant increase in the mRNA expression of CCL3 and CCL4 (Fig. 3). Albeit not significantly modulated, expression levels of CXCL10 and CCL7 were found to have been induced in all the experiments. The encapsulated cells also induced a slight increase in the expression of CCL3, CCL4, and CCL7 genes compared to the expression in NS cells. The mRNA expression of CXCL10 did not appear to be influenced by the encapsulated form compared with the NS sample.
FIG. 3.
Real-time PCR analysis of gene expression of chemokines from CD11c+ cells. RT-PCR of CD 11c+ DC isolated from the spleens of naive mice that were stimulated in vitro for 4 h at a ratio of 1:2 with C. neoformans encapsulated and acapsular strains. Expression levels of CCL3 (A), CXCL10 (B), CCL4 (C), and CCL7 (D) were compared to the expression levels of the β2-microglobulin gene. Each data point represents an average of three independent experiments. The induction levels were calculated by using the 2−ΔΔCT method. *, P < 0.05 (cells treated with encapsulated and acapsular yeasts versus untreated cells).
DNA binding proteins and transcription factors.
At the early time point of 4 h, the encapsulated strain induced the down-regulation of two genes: the E74-like factor 1 gene (Elf1), which is involved in chemokine production, and the sequestosome 1 gene (Sqstm1), which regulates protein degradation. The acapsular strain, however, down-regulated the inhibitor of DNA binding-1 gene (Idb1), which is involved in TGF-β signaling. Other transcription factors, such as interferon-regulatory factors 1 and 7 (Irf1 and Irf7) and interferon-stimulated protein (Isg20), were up-regulated by the acapsular strain. The acapsular strain also up-regulated the expression of genes such as the nuclear factor genes Nfkb2 and Nfkbia. Nfkb2 inhibits the transcriptional activity of NF-κB complexes by sequestering them in the cytoplasm and blocking their binding to DNA sequences (16). The expression levels of nuclear factors Nfkb2, Nfkbiz, and Nfkbia were still threefold higher at 18 h after interaction with the acapsular strain. In contrast, the only DC gene that was up-regulated after 18 h of incubation with an encapsulated strain was the IL enhancer binding factor 2 (IIf2).
Genes associated with signaling.
After the DC were exposed for 4 h to the encapsulated strain, the gene Sema4b, encoding semaphorine domain 4b, was down-regulated with a ratio of −1.2, while the interferon receptor alpha and beta gene (Ifnar2) was up-regulated with a ratio of 1.13. Sema4 provides a costimulatory signal for T cells and is also involved in the regulation of Th1/Th2 responses (4).
The acapsular strain down-regulated the IL-17 receptor gene (IL-17r) in DC, a member of the novel inflammatory cytokine family, with a value of −1.01 at 4 h. This gene, however, was up-regulated at 18 h with a ratio of 1.09. In addition, the acapsular strain up-regulated the expression of interferon-related genes, such as Igtp, with a ratio of 2.18, and interferon induced with helicase C domain (Ifih1), with a ratio of 2.70. The suppressor of cytokine signaling 3 gene (Socs3) was also up-regulated with a ratio of 1.31 by the acapsular strain after 4 h, and the up-regulation decreased to a ratio of 1.09 at 18 h. Socs3 is an inhibitor of cytokine receptor signaling mediated by the Janus kinase family kinases and may protect the host by limiting the excessive production of cytokines (1).
DISCUSSION
In this study, we analyzed changes in murine DC gene expression in relation to the presence of the polysaccharide capsule in C. neoformans during interaction with the fungus. We used two C. neoformans strains with isogenic backgrounds, one of which was acapsular by virtue of the disruption of the CAP59 gene (6). These acapsular cells are completely devoid of capsular material as confirmed not only by India ink preparations but also by a negative reaction with anticapsular antibody from the Iatron Crypto kit, which is used for serotyping of C. neoformans cells. Our array study validates the previous findings by showing the profound differences between the encapsulated and acapsular strains in regulating maturation and activation of DC.
It is well known that mature DC express high levels of MHC and costimulatory molecules such as CD86 and CD83 (23) and the up-regulation of these genes observed after stimulation with acapsular strain reflects the capacity of the fungus to promote a series of events involved in DC maturation and activation.
These results are consistent with previous observations demonstrating that the acapsular strain, differently from the encapsulated one, promotes the up-regulation of several costimulatory molecules (44); however, in this study, we provide evidence for the existence of other genes that participate in the final biological effect. As a matter of fact, besides the increase in CD86, CD83, and MHC-II-related genes, other genes, such as Relb, encoding a transcription factor associated most directly with differentiation and function of DC (5, 22, 46), as well as the CD1d2 and ICAM1 genes, which are involved in DC activation and maturation, are also activated. In both cases, up-regulation was exclusively ascribed to the acapsular strain, while the encapsulated strain failed to stimulate any activation.
There is compelling evidence that encapsulation of C. neoformans is a potent obstacle for the induction of proinflammatory cytokines such as IL-12 and TNF-α (19). Although our results validate previously reported data on the capacity of acapsular strain to induce IL-12 and TNF-α (45) and the inability of the encapsulated virulent strain to induce TNF-α (19), this is the first demonstration that an acapsular strain induces the chemokine genes in DC, while the encapsulated strain produced the opposite results either by down-regulating or not modulating the expression of some of these genes.
Exposure to the acapsular strain resulted in a prompt and intense response by DC, which resulted in the induction of a large set of cytokine genes, including those encoding TNF-α, IL-1, and interferon, as well as the genes for chemokines and cell adhesion molecules. Several cytokine-induced genes appeared to persist in an activated state even after 18 h of incubation. Thus, the DC were highly responsive to acapsular cells, and the nature of the response had the typical features of an immunostimulatory reaction. Conversely, the expression of only seven genes was modulated in 4 h of DC incubation with the encapsulated strain. Among these genes, Tgfb3 and Ifnra2 were up-regulated. The former is involved in inducing TGF-β and is considered an inhibitor of DC maturation (10), and the latter is involved in the production of alpha interferon and suppresses IL-12 production (7, 32), evidencing for the first time that not only was the encapsulated strain unable to induce activation but that it was capable of activating the gene that regulated the production of alpha interferon and could be involved in the suppression of IL-12 production (7). Thus, the inability of DC to produce IL-12 following stimulation with encapsulated yeast could be related to the induction level of this gene and the activation of other signaling pathways that suppress IL-12. These results reveal a complex network of gene expression that contributes to highlight the evasive maneuvers of the encapsulated strain. Another piece of evidence for this assertion is related to the capacity of the encapsulated fungus to induce down-regulation of five genes, including those encoding the E74-like factor 1 (Elf1) and sequestosome 1 (Sqstm1). The Elf1 gene is a transcription factor that regulates the expression of cytokines such as granulocyte-macrophage colony-stimulating factor and IL-3 (27, 35), and the Sqstm1 gene is involved in induction of NF-κB signaling (16, 48). NF-κB is a ubiquitous transcription factor that is activated by a variety of stimuli and is a key regulator of the production of proinflammatory cytokines such as TNF-α, IL-1β, and IL-6 and chemokines such as CXCL8 (41). Therefore, it is conceivable that capsular material inhibits NF-κB activation and the consequent cytokine release via down-regulation of these genes.
The effect of capsular material as an immune inhibitor has been shown previously (42). Glucuronoxylomannan is able to induce an atypical response of peripheral blood mononuclear cells, allowing activation of NF-κB without stimulating TNF-α secretion (38, 42). Indeed, our data show that DC stimulated for 4 or 18 h with encapsulated strain are for the most part unresponsive and the nature of their minimal early response resembles an immune-inhibitory reaction. The only DC gene that was significantly up-regulated after 18 h of incubation with the encapsulated strain was IIf2, which is involved in IL-2 mRNA induction (14, 25). This suggests that the induction of IL-2 expression by DC could be regulated by the presence of capsular material.
Different from the encapsulated strain, the acapsular strain activates genes that regulate the activation of NF-κB family genes and the endothelin-1 (ET-1) gene, which promotes TNF-α production through its action on the ETA receptor, thus demonstrating its proinflammatory function (49). Since ET-1 is almost ubiquitous in both lymphoid and nonlymphoid tissues, it is reasonable to hypothesize that it may participate in DC activation. In addition, up-regulation of the ICAM1 gene suggests that the acapsular strain could favor the migration of DC through the endothelium, thereby promoting an inflammatory response that is considered essential for clearing C. neoformans from host tissues (9, 26, 29, 39). This is consistent with the observed rapid elimination of acapsular C. neoformans from infected animals (21).
The absence of DC stimulation observed with the encapsulated strain was not due to the lack of recognition by DC, because, in our experimental system, encapsulated C. neoformans cells interacted with DC and were even internalized. Previously, Kelly et al. reported that murine DC rarely phagocytized encapsulated C. neoformans (20). There were, however, several differences in the experimental systems that could account for the apparent discrepancy: (i) Kelly et al. used C. neoformans opsonized with 10% fetal bovine serum, whereas in our experiments, the fungus was opsonized with 20% of normal mouse serum; (ii) Kelly et al. used murine bone marrow-derived DC, while we used the D1 cell line; and (iii) Kelly et al. used the C. neoformans strain ATCC 62070, while we used the JEC21 strain. Several receptors present on our D1 cell line, such as CR3, FcγRII/FcγRIII (48), and CD205 (36), which is homologous to the mannose receptor (18), are known to be involved in C. neoformans phagocytosis (24, 40). These receptors could have been associated with the observed internalization of encapsulated yeast cells. Further evidence that supports the presence of interaction between encapsulated C. neoformans and D1 cells is that the encapsulated cells are able to modulate the expression of negative signals, such as TGF-β3 and Ifnra2.
The results of this study show for the first time the overall profile of gene expression in DC during their interaction with C. neoformans as well as the role of capsular material in negatively influencing the pattern of gene expression and molecules involved in driving the T cells. Our data suggest that the cell wall antigens, such as α- and β-glucans and mannoproteins, that in encapsulated C. neoformans are confined to the inner layer play a fundamental role in inducing a protective response. In contrast, the outermost capsular part is involved in evasion or even silencing of the DC response. The cryptococcal capsule is reminiscent of the extracellular polysaccharides of bacteria, such as the capsules of gram-negative species, including Streptococcus pneumoniae, Neisseria meningitidis, and Pseudomonas aeruginosa (47). Both bacterial and C. neoformans capsules are antiphagocytic and primarily composed of carbohydrates which are not readily removed from the cell wall. Moreover, the encapsulated bacteria S. pneumoniae, N. meningitidis, Haemophilus influenzae, and Klebsiella pneumoniae share common pattern recognition receptors with C. neoformans, such as TLR2 and TLR4 (31, 34). Although no bacterial extracellular glycan is structurally similar to glucuronoxylomannan or galactoxylomannan of C. neoformans capsule, these polymers provide a context for investigation not only of the synthesis of the capsule but also of the nature of immune-silencing properties of encapsulated microorganisms. Collectively, these results could help elucidate the nature of immune-regulation that determines survival of encapsulated microorganisms in different host environments.
Supplementary Material
Acknowledgments
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD.
Editor: A. Casadevall
Footnotes
Published ahead of print on 4 February 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alexander, W. S. 2002. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2410-416. [DOI] [PubMed] [Google Scholar]
- 2.Bacci, A., C. Montagnoli, K. Perruccio, S. Bozza, R. Gaziano, L. Pitzurra, A. Velardi, C. F. d'Ostiani, J. E. Cutler, and L. Romani. 2002. Dendritic cells pulsed with fungal RNA induce protective immunity to Candida albicans in hematopoietic transplantation. J. Immunol. 1682904-2913. [DOI] [PubMed] [Google Scholar]
- 3.Bulmer, G. S., and M. D. Sans. 1968. Cryptococcus neoformans. 3. Inhibition of phagocytosis. J. Bacteriol. 955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhardt, C., M. Muller, A. Badde, C. C. Garner, E. D. Gundelfinger, and A. W. Puschel. 2005. Semaphorin 4B interacts with the post-synaptic density protein PSD-95/SAP90 and is recruited to synapses through a C-terminal PDZ-binding motif. FEBS Lett. 5793821-3828. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco, D., R. P. Ryseck, and R. Bravo. 1993. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development 1181221-1231. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 144912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousens, L. P., J. S. Orange, H. C. Su, and C. A. Biron. 1997. Interferon-alpha/beta inhibition of interleukin 12 and interferon-gamma production in vitro and endogenously during viral infection. Proc. Natl. Acad. Sci. USA 94634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d'Ostiani, C. F., G. Del Sero, A. Bacci, C. Montagnoli, A. Spreca, A. Mencacci, P. Ricciardi-Castagnoli, and L. Romani. 2000. Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans. Implications for initiation of T helper cell immunity in vitro and in vivo. J. Exp. Med. 1911661-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelhardt, B., and H. Wolburg. 2004. Transendothelial migration of leukocytes: through the front door or around the side of the house? Eur. J. Immunol. 342955-2963. [DOI] [PubMed] [Google Scholar]
- 10.Fainaru, O., T. Shay, S. Hantisteanu, D. Goldenberg, E. Domany, and Y. Groner. 2007. TGFbeta-dependent gene expression profile during maturation of dendritic cells. Genes Immun. 8239-244. [DOI] [PubMed] [Google Scholar]
- 11.Feldmesser, M., and A. Casadevall. 1998. Mechanism of action of antibody to capsular polysaccharide in Cryptococcus neoformans infection. Front. Biosci. 3d136-d151. [DOI] [PubMed] [Google Scholar]
- 12.Feldmesser, M., A. Mednick, and A. Casadevall. 2002. Antibody-mediated protection in murine Cryptococcus neoformans infection is associated with pleotrophic effects on cytokine and leukocyte responses. Infect. Immun. 701571-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Förster, R., A. Schubel, D. Breitfeld, E. Kremmer, I. Renner-Muller, E. Wolf, and M. Lipp. 1999. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 9923-33. [DOI] [PubMed] [Google Scholar]
- 14.Geetha, T., and M. W. Wooten. 2002. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 51219-24. [DOI] [PubMed] [Google Scholar]
- 15.Gunn, M. D., S. Kyuwa, C. Tam, T. Kakiuchi, A. Matsuzawa, L. T. Williams, and H. Nakano. 1999. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 189451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havard, L., S. Rahmouni, J. Boniver, and P. Delvenne. 2005. High levels of p105 (NFKB1) and p100 (NFKB2) proteins in HPV16-transformed keratinocytes: role of E6 and E7 oncoproteins. Virology 331357-366. [DOI] [PubMed] [Google Scholar]
- 17.Herring, A. C., J. Lee, R. A. McDonald, G. B. Toews, and G. B. Huffnagle. 2002. Induction of interleukin-12 and gamma interferon requires tumor necrosis factor alpha for protective T1-cell-mediated immunity to pulmonary Cryptococcus neoformans infection. Infect. Immun. 702959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, W., W. J. Swiggard, C. Heufler, M. Peng, A. Mirza, R. M. Steinman, and M. C. Nussenzweig. 1995. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature 375151-155. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami, K., X. Qifeng, M. Tohyama, M. H. Qureshi, and A. Saito. 1996. Contribution of tumour necrosis factor-alpha (TNF-alpha) in host defence mechanism against Cryptococcus neoformans. Clin. Exp. Immunol. 106468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, R. M., J. Chen, L. E. Yauch, and S. M. Levitz. 2005. Opsonic requirements for dendritic cell-mediated responses to Cryptococcus neoformans. Infect. Immun. 73592-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lappin, M. B., J. M. Weiss, V. Delattre, B. Mai, H. Dittmar, C. Maier, K. Manke, S. Grabbe, S. Martin, and J. C. Simon. 1999. Analysis of mouse dendritic cell migration in vivo upon subcutaneous and intravenous injection. Immunology 98181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen, C. P., S. C. Ritchie, T. C. Pearson, P. S. Linsley, and R. P. Lowry. 1992. Functional expression of the costimulatory molecule, B7/BB1, on murine dendritic cell populations. J. Exp. Med. 1761215-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levitz, S. M., A. Tabuni, T. R. Kozel, R. S. MacGill, R. R. Ingalls, and D. T. Golenbock. 1997. Binding of Cryptococcus neoformans to heterologously expressed human complement receptors. Infect. Immun. 65931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, C., A. J. Lusis, R. Sparkes, A. Nirula, and R. Gaynor. 1992. Characterization and chromosomal mapping of the gene encoding the cellular DNA binding protein ILF. Genomics 13665-671. [DOI] [PubMed] [Google Scholar]
- 26.Lyck, R., Y. Reiss, N. Gerwin, J. Greenwood, P. Adamson, and B. Engelhardt. 2003. T-cell interaction with ICAM-1/ICAM-2 double-deficient brain endothelium in vitro: the cytoplasmic tail of endothelial ICAM-1 is necessary for transendothelial migration of T cells. Blood 1023675-3683. [DOI] [PubMed] [Google Scholar]
- 27.Lyons, R. M., and H. L. Moses. 1990. Transforming growth factors and the regulation of cell proliferation. Eur. J. Biochem. 187467-473. [DOI] [PubMed] [Google Scholar]
- 28.Martin, E., B. O'Sullivan, P. Low, and R. Thomas. 2003. Antigen-specific suppression of a primed immune response by dendritic cells mediated by regulatory T cells secreting interleukin-10. Immunity 18155-167. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin, F., B. P. Hayes, C. M. Horgan, J. E. Beesley, C. J. Campbell, and A. M. Randi. 1998. Tumor necrosis factor (TNF)-alpha and interleukin (IL)-1beta down-regulate intercellular adhesion molecule (ICAM)-2 expression on the endothelium. Cell Adhes. Commun. 6381-400. [DOI] [PubMed] [Google Scholar]
- 30.Mestas, J., and C. C. Hughes. 2004. Of mice and not men: differences between mouse and human immunology. J. Immunol. 1722731-2738. [DOI] [PubMed] [Google Scholar]
- 31.Mogensen, T. H., S. R. Paludan, M. Kilian, and L. Ostergaard. 2006. Live Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis activate the inflammatory response through Toll-like receptors 2, 4, and 9 in species-specific patterns. J. Leukoc. Biol. 80267-277. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, K. B., L. P. Cousens, L. A. Doughty, G. C. Pien, J. E. Durbin, and C. A. Biron. 2000. Interferon alpha/beta-mediated inhibition and promotion of interferon gamma: STAT1 resolves a paradox. Nat. Immunol. 170-76. [DOI] [PubMed] [Google Scholar]
- 33.Perfect, J. R., B. Wong, Y. C. Chang, K. J. Kwon-Chung, and P. R. Williamson. 1998. Cryptococcus neoformans: virulence and host defences. Med. Mycol. 36(Suppl. 1)79-86. [PubMed] [Google Scholar]
- 34.Pons, J., J. Sauleda, V. Regueiro, C. Santos, M. Lopez, J. Ferrer, A. G. Agusti, and J. A. Bengoechea. 2006. Expression of Toll-like receptor 2 is up-regulated in monocytes from patients with chronic obstructive pulmonary disease. Respir. Res. 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao, G. S., M. V. Ramachandran, and J. S. Bajaj. 2006. In silico structure-based design of a potent and selective small peptide inhibitor of protein tyrosine phosphatase 1B, a novel therapeutic target for obesity and type 2 diabetes mellitus: a computer modeling approach. J. Biomol. Struct. Dyn. 23377-384. [DOI] [PubMed] [Google Scholar]
- 36.Rovere, P., A. A. Manfredi, C. Vallinoto, V. S. Zimmermann, U. Fascio, G. Balestrieri, P. Ricciardi-Castagnoli, C. Rugarli, A. Tincani, and M. G. Sabbadini. 1998. Dendritic cells preferentially internalize apoptotic cells opsonized by anti-beta2-glycoprotein I antibodies. J. Autoimmun. 11403-411. [DOI] [PubMed] [Google Scholar]
- 37.Schulz, O., A. D. Edwards, M. Schito, J. Aliberti, S. Manickasingham, A. Sher, and C. Reis e Sousa. 2000. CD40 triggering of heterodimeric IL-12 p70 production by dendritic cells in vivo requires a microbial priming signal. Immunity 13453-462. [DOI] [PubMed] [Google Scholar]
- 38.Shoham, S., C. Huang, J. M. Chen, D. T. Golenbock, and S. M. Levitz. 2001. Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J. Immunol. 1664620-4626. [DOI] [PubMed] [Google Scholar]
- 39.Springer, T. A. 1994. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76301-314. [DOI] [PubMed] [Google Scholar]
- 40.Syme, R. M., J. C. Spurrell, E. K. Amankwah, F. H. Green, and C. H. Mody. 2002. Primary dendritic cells phagocytose Cryptococcus neoformans via mannose receptors and Fcγ receptor II for presentation to T lymphocytes. Infect. Immun. 705972-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian, B., and A. R. Brasier. 2003. Identification of a nuclear factor kappa B-dependent gene network. Recent Prog. Horm. Res. 5895-130. [DOI] [PubMed] [Google Scholar]
- 42.Vecchiarelli, A. 2007. Fungal capsular polysaccharide and T-cell suppression: the hidden nature of poor immunogenicity. Crit. Rev. Immunol. 27547-557. [DOI] [PubMed] [Google Scholar]
- 43.Vecchiarelli, A. 2005. The cellular responses induced by the capsular polysaccharide of Cryptococcus neoformans differ depending on the presence or absence of specific protective antibodies. Curr. Mol. Med. 5413-420. [DOI] [PubMed] [Google Scholar]
- 44.Vecchiarelli, A., D. Pietrella, P. Lupo, F. Bistoni, D. C. McFadden, and A. Casadevall. 2003. The polysaccharide capsule of Cryptococcus neoformans interferes with human dendritic cell maturation and activation. J. Leukoc. Biol. 74370-378. [DOI] [PubMed] [Google Scholar]
- 45.Vecchiarelli, A., C. Retini, D. Pietrella, C. Monari, C. Tascini, T. Beccari, and T. R. Kozel. 1995. Downregulation by cryptococcal polysaccharide of tumor necrosis factor alpha and interleukin-1 beta secretion from human monocytes. Infect. Immun. 632919-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weih, F., G. Warr, H. Yang, and R. Bravo. 1997. Multifocal defects in immune responses in RelB-deficient mice. J. Immunol. 1585211-5218. [PubMed] [Google Scholar]
- 47.Wilson, J. W., M. J. Schurr, C. L. LeBlanc, R. Ramamurthy, K. L. Buchanan, and C. A. Nickerson. 2002. Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 78216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winzler, C., P. Rovere, M. Rescigno, F. Granucci, G. Penna, L. Adorini, V. S. Zimmermann, J. Davoust, and P. Ricciardi-Castagnoli. 1997. Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J. Exp. Med. 185317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanagisawa, M., H. Kurihara, S. Kimura, Y. Tomobe, M. Kobayashi, Y. Mitsui, Y. Yazaki, K. Goto, and T. Masaki. 1988. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 332411-415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.