Abstract
Staphylococcus aureus biofilm formation is induced in iron-restricted growth conditions in vitro. In this study, we showed that Emp and Eap play important roles in low-iron-induced biofilm formation of S. aureus Newman. Eap and Emp are secreted proteins which are non-covalently attached to the S. aureus cell surface and have previously been implicated in a number of aspects of S. aureus pathogenesis. We showed here that the transcription of these important virulence factors is induced by growth in low-iron medium, reflective of the in vivo environment. Our results show that iron regulation of Eap and Emp is Fur independent. However, Fur is required for full induction of eap and emp expression in low-iron conditions. In this study, we demonstrated that in addition to Fur, low-iron-induced biofilm formation requires Sae, Agr, and SarA. In iron-restricted growth conditions, Sae and Agr are essential for Emp and Eap expression and hence for biofilm formation, whereas SarA appears to have a less-significant role. We also showed that expression of the ica operon is required for biofilm formation in iron-restricted growth conditions. We demonstrated that in fact, ica is required for the expression of the important multifunctional virulence determinants eap and emp.
Staphylococcus aureus is one of the major pathogens associated with nosocomial infections and bacteremia. Patients particularly at risk of developing staphylococcal infections are those with indwelling medical devices, which S. aureus may colonize as a biofilm. Formation of a biofilm is a major factor in the organism's pathogenicity and is a complex, multifactored process. Initially, bacterial cells adhere either directly to the surface of the implanted device or to the host matrix components that coat the device upon insertion. This step involves both nonspecific interactions mediated via hydrophobicity and cell surface charge and specific interactions between host and bacterial proteins. Subsequently, adherent cells form a multilayered biofilm covered by a protective “slime” or glycocalyx from which individual cells or multicellular clusters are released, facilitating the systemic dissemination and infection of secondary sites (18, 38).
Regulation of biofilm formation is extremely complex and involves an array of coordinated regulatory mechanisms which have yet to be fully elucidated. Biofilm-specific transcriptional regulators include Rbf, which mediates the induction of biofilm formation at the cell-cell interaction stage in response to glucose and osmotic stress (30), and IcaR and TcaR, both of which negatively regulate biofilm formation (25). Global regulators include SarA, which is required for biofilm formation, as the mutation of sarA results in a reduced capacity to form a biofilm (2, 49), and the two-component regulator ArlRS, a repressor of biofilm formation (48). The role of the Agr quorum-sensing system appears to vary depending on the strain or growth conditions, as disruption of agr can inhibit, enhance, or have no effect on biofilm formation (2, 39, 51).
Staphylococcal biofilm formation is influenced by many environmental factors, including anaerobic growth (11), osmotic stress (43), and the availability of glucose (30). A major environmental stress encountered by bacteria in vivo is severe iron restriction. We have previously shown that iron plays a major role in the regulation of S. aureus biofilm formation (26). We demonstrated that biofilm formation in the Newman strain was induced in a low-iron environment and repressed by the addition of iron. S. aureus iron-responsive gene regulation is usually mediated by the global regulator Fur (ferric uptake regulator) (22, 50). We found that S. aureus biofilm formation was positively regulated in low-iron conditions by Fur and negatively regulated by iron via a Fur-independent mechanism (26). The factors involved in this iron-mediated biofilm formation have yet to be identified.
The best-understood factor implicated in successful biofilm formation is the polysaccharide PNAG (polymeric N-acetylglucosamine) (31), which is synthesized by products of the ica operon (10). There is, however, some controversy as to the role of PNAG. Cramton et al. (10) have shown that PNAG is involved in bacterial cell-to-cell adhesion and is critical for biofilm formation in vitro. Deletion of the ica operon results in a biofilm-negative phenotype. Other groups argue that under different conditions, S. aureus is capable of forming a biofilm in the absence of PNAG (2, 48). A number of clinical isolates from infected implants were found to be biofilm negative in vitro despite testing positive for icaA (16). This finding suggests that S. aureus biofilm production in vivo may be influenced by factors other than or in addition to PNAG, and in fact, evidence for ica-independent biofilm mechanisms is accumulating (reviewed in reference 38).
In this study, we demonstrated that biofilm formation in iron-restricted growth conditions involves the secreted proteins Emp and Eap. We also showed that the expression of these proteins is dependent on the ica operon and the global regulators Fur, Agr, SarA, and Sae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table 1 and were stored at −80°C in 10% (vol/vol) glycerol Trypticase soy broth (TSB; BBL). The strains were plated out fresh from frozen stocks onto 6% defibrinated horse blood agar for each biofilm assay. The strains for the biofilm assays and protein extraction were cultured either in TSB or under iron-restricted conditions in CRPMI (RPMI 1640 tissue culture medium [Sigma Ltd.] that had been depleted of iron by batch incubation with 6% [wt/vol] Chelex 100 [Sigma Ltd.] and then supplemented with 10% RPMI 1640 to provide the trace elements required for growth) (35). All cultures were incubated statically for 24 h at 37°C in 5% CO2 in air; where indicated, the medium was supplemented with 50 μM Fe2(SO4)3 to produce iron-replete growth conditions and supplemented with the appropriate antibiotics where required. Escherichia coli TOP10 was cultured in LB (Luria broth).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| S. aureus | ||
| RN4220 | Restriction negative; 8325 derivative | Laboratory stock |
| Newman | Wild type | 13 |
| Newman fur | Newman fur::tet | 26 |
| Newman ica | Newman Δica::tet | 28 |
| Newman Δica complemented | Newman Δica::tet complemented | 28 |
| Newman sae | Newman sae::Tn917 (AS3) | 17 |
| ALC1342 | RN6390 ΔsarA::ermC | 8 |
| Newman sarA | Newman ΔsarA::ermC | This work |
| RN6911 | RN6390 Δagr::tetM | 37 |
| Newman agr | Newman Δagr::tetM | This work |
| Newman eap | Newman Δeap::erm (AH12) | 24 |
| Newman emp | Newman Δemp::erm | This work |
| E. coli | ||
| TOP10 | Invitrogen | |
| Plasmids | ||
| pMK4 | E. coli/S. aureus shuttle vector | 47 |
| pEmp | PMK4 with emp promoter and gene | This work |
| pEap | PMK4 with eap promoter and gene | This work |
Biofilm formation assay.
Quantitative measurement of staphylococcal adherence to polystyrene, indicative of biofilm formation, was done in a microtiter assay as described previously (10), with the following modifications. Bacteria were grown overnight in CRPMI at 37°C in 5% CO2 in air, diluted to an optical density at 595 nm of 0.1 in fresh CRPMI, inoculated into quadruple wells of 96-well flat-bottomed tissue culture plates (Nunc), and incubated statically at 37°C in 5% CO2 in air for 24 h. After being incubated, the plates were read at 595 nm in a plate reader (Bio-Rad) to monitor growth. The wells were then washed three times with sterile phosphate-buffered saline (PBS) to remove nonadherent cells, dried in a hot-air oven, and stained with 1% safranine. After being stained, the plates were read in a plate reader at 490 nm to determine the extent of biofilm formation. The means and standard deviations shown in the figures were calculated from at least four independent experiments. A two-tailed Student t test was used to determine the differences in biofilm formation between mutant and wild-type strains. The differences were considered statistically significant when P was <0.05.
Disruption of the emp gene.
Primer pairs Emp-E and EmpK-F and EmpK-R and Emp-B (Table 2) were used to amplify emp sequences from Newman genomic DNA, using rTth DNA polymerase (Roche). The amplified PCR product was digested with KpnI, ligated, and reamplified to generate a single product which was then A-tailed with Taq DNA polymerase per the manufacturer's instructions (Promega), ligated into pGEM-T (Promega) to generate pEmp-1, and transformed into E. coli TOP10 electrocompetent cells (Invitrogen). The ermB erythromycin resistance cassette was amplified from pEC4 with rTth DNA polymerase and primers KpnF and KpnR (Table 2). The resulting ∼1.2-kb fragment was digested with KpnI, ligated into pEmp-1 digested with KpnI to generate plasmid pEmp-2, and transformed into E. coli TOP10. pEmp2 was digested with BamHI and EcoRI, and the resulting ∼2-kb fragment was ligated into similarly digested pTS1 and transformed into E. coli TOP10. Recombinants were selected on Luria agar plates containing erythromycin (150 μg/ml) at 30°C to generate plasmid pEmp.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Primer sequencea | Description |
|---|---|---|
| Emp-E | GATGAATTTAAGGAATTCGCCAAAG | Emp mutant |
| EmpK-F | AAAGGTACCATGTGTATATCTAGGCGATTCTG | Emp mutant |
| EmpK-R | AAAGGTACCTTACACCAGGGCATGCTAGC | Emp mutant |
| Emp-B | AAAGGATCCTTTATACTCGTGGTGCTGGTAAGC | Emp mutant |
| KpnF | AAAAGGTACCTCTTGCGTATGGTTAACCC | Erythromycin |
| KpnR | AAAGGTACCAACTTCCAAATTTACAAAGCG | Erythromycin |
| SAR-F | GAGTTGTTATCAATGGTC | SarA probe |
| SAR-R | GTTTGCTTCAGTGATTCG | SarA probe |
| TETM-F | CGTCTGCCCTCATTATTGG | Tetracycline |
| TETM-R | CGATTTAGAAATCCCTTTGAG | Tetracycline |
| EapF | CTCGGATCCTTCAGTTAATTCAAAAAATAG | Eap complement |
| EapPF | CTCGGATCCAAAAGGAGAGATAATTTATGA | Eap probe |
| EapR | CTCCTGCAGTTAAAATTTAATTTCAATGTCTACTTTTTTAATGTC | Eap complement |
| EmpF | CTCGGATCCTGTTAAGACAACGTTTACT | Emp complement |
| EmpPF | CTCGGATCCAAGGAGAAATAACAGATGAAAAG | Emp probe |
| EmpR | CTCCTGCAGTTATACTCGTGGTGCTGGTAAGC | Emp complement |
| 16S-F | GATCCTGGGTCAGGATG | 16S probe |
| 16S-R | CTAGAGTTGTCAAAGGATG | 16S probe |
Underlined characters represent restriction sites.
pEmp was transformed into S. aureus RN4220 as previously described (27), with selection on LB agar plus 5 μg/ml erythromycin at 30°C. A single transformant was grown overnight at 30°C in LB plus 5 μg/ml erythromycin and plated onto LB agar plus 5 μg/ml erythromycin overnight at 42°C. The resulting colonies were screened for chloramphenicol sensitivity by growing them overnight on LB agar plus 10 μg/ml chloramphenicol. Genomic DNA from chloramphenicol-sensitive, erythromycin-resistant colonies was used as a template for PCR, using primer pair Emp-E/Emp-B to check for the loss of the wild-type emp gene and KpnF/KpnR for the presence of ermB.
Transduction of mutations into S. aureus strain Newman.
The emp::ermB, ΔsarA::ermC, and Δagr::tetM mutations were transferred to S. aureus strain Newman by transduction with phage phi11 (7) from RN4220 emp::empB, ALC1342, and RN6911, respectively. Transductants were selected on LK agar (1% tryptone, 0.5% yeast extract, 0.7% KCl, and 1.5% agar) plates containing 10 μg/ml of the appropriate antibiotic and 0.05% sodium citrate. Colonies containing the relevant mutation were confirmed by PCR, using primers Emp-E/Emp-B, SAR-F, SAR-R and TETM-F, and TETM-R, respectively (Table 2).
Construction of eap- and emp-complementing plasmids.
The eap and emp genes were PCR amplified, using TripleMaster mix (Eppendorf), from Newman wild-type DNA, using primer pairs EapF/EapR and EmpF/EmpR, respectively. The PCR products were A-tailed with Taq DNA polymerase per the manufacturer's instructions (Applied Biosystems), ligated into pGEM T Easy cloning vectors (Invitrogen), and transformed into electrocompetent E. coli TOP10 cells (Invitrogen). Plasmids containing eap and emp inserts were confirmed by PCR with the above primers and digestion with BamHI and PstI. The DNA fragments resulting from restriction digestion with BamHI and PstI were ligated into similarly digested pMK4, creating plasmids pEap and pEmp, respectively, and transformed into E. coli TOP10. Plasmids containing the relevant inserts were transformed into electrocompetent restriction-deficient S. aureus RN4220. The complementing plasmids were then transduced into Newman eap::erm and Newman emp::erm as described previously, using chloramphenicol at 10 μg/ml for selection.
RNA extraction and Northern blotting.
Exponentially growing cultures in CRPMI were pelleted and resuspended in RNAlater as directed by the manufacturer (Ambion). The total RNA was then extracted, using the hot-phenol method as described by Schmitt et al. (45) but with 300 μg/ml lysostaphin added to the initial cell lysis step. The RNA (10 μg) was electrophoresed on a 1.5% denaturing formaldehyde gel, and the RNA was transferred to a nylon filter by Northern blotting overnight. The filters were hybridized overnight at 65°C in Church & Gilbert's buffer (0.5 M Na2HPO4, 0.5 M NaH2PO4, 7% sodium dodecyl sulfate [SDS], 1 mM EDTA) with probes labeled with [α-32P]dCTP (Perkin Elmer), using the Klenow fragment of DNA polymerase I and random hexamers as primers. The probes were constructed by PCR, using the EapPF and EapR and EmpPF and EmpR primers described in Table 2. The filters were washed with 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) SDS and exposed to X-ray film at −80°C. The blots were then stripped of the probes by washing the filters with two to three changes of boiling 0.1% SDS and rinsing them with 2× SSC. The blots were then hybridized with [α-32P]dCTP-labeled 16S RNA probes as a loading control.
SDS-PAGE.
All protein fractions were prepared from aliquots of the same 20-ml cultures used as inocula for the biofilm assays (see above). SDS cell surface extracts were prepared as described by Hussain et al. (23) from 10-ml, 24-h CRMPI cultures, except that dialysis to remove SDS was omitted and an equal volume of 2× Laemmli sample buffer (29) was added to the cell suspensions prior to boiling them for 3 min and separating them by SDS-10% polyacrylamide gel electrophoresis (PAGE). The culture supernatants from the 10-ml, 24-h cultures were retained and concentrated, using Amicon ultra columns with a nominal molecular weight limit of 30,000 (Millipore). Cell wall, cell membrane, and cytoplasmic proteins were extracted essentially as described by Morrissey et al. (35). Briefly, the cells from the 10-ml, 24-h CRPMI cultures were harvested by centrifugation. The cell pellets were resuspended in 250 μl per 10 mg of cells of PBS containing 1 mg/ml benzamidine, 30% (wt/vol) raffinose, and 80 μg/ml lysostaphin and incubated for 15 min at 37°C to release the cell wall proteins. After being incubated, the cell suspension was centrifuged, and the supernatant containing the cell wall proteins was removed. The remaining protoplast pellet was resuspended in 250 μl of PBS per 10 mg of cells as described previously and sonicated on ice in a Biorupter sonicating water bath (Diagenode) on high for 30 seconds on and 30 seconds off for a total of 10 min. The membrane proteins were isolated by centrifugation for 10 min at 11,000 × g. The supernatant containing the cytoplasmic proteins was removed, and the remaining pellet containing the membrane proteins was resuspended in 1× Laemmli sample buffer at 250 μl per 10 mg of cell pellets as previously described. All other fractions were mixed with an equal volume of 2× Laemmli sample buffer and boiled for 3 min prior to being separated by SDS-10% PAGE.
MALDI-TOF mass spectrometry analysis.
Bands excised from SDS polyacrylamide gels were subjected to in-gel trypsin digestion according to the standard procedures (46) of the PNACL Proteomics Facility, University of Leicester. The resulting digests were mixed 1:1 with a 10-mg/ml solution of α-cyano-4-hydroxycinnamic acid (Sigma, United Kingdom) and 0.5 μl was spotted onto a stainless steel target plate. Analysis of peptide digests was carried out on a Voyager-DE STR matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems, Warrington, United Kingdom) in positive-ion reflectron mode. Calibrated mass spectra were searched against a monthly updated copy of the NCBI nr protein database, using the PNACL Proteomics Facility version of the Mascot search tool (Matrix Science Ltd., United Kingdom) (41). The criteria for protein identification were based on the manufacturer's definitions. Basically, the candidate proteins with probability-based Mowse scores exceeding the threshold (P < 0.05), thus indicating significance, were referred to as “hits.”
RESULTS
ica is required for biofilm formation and expression of Emp and Eap in low-iron media.
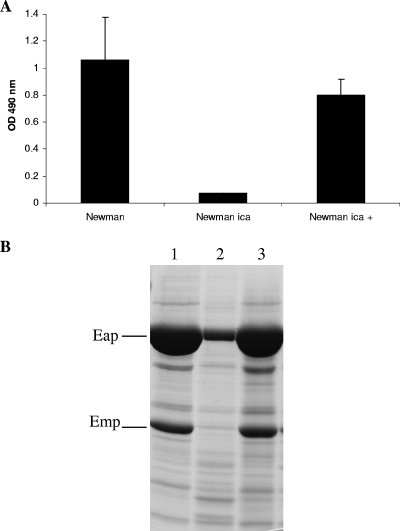
To investigate the role of PNAG in low-iron-induced biofilm formation, we compared the biofilm formation levels of the wild-type strain Newman and the isogenic Newman Δica::tet mutant, which is deficient in the production of PNAG (Fig. 1A), in iron-restricted CRPMI. The biofilm formation observed in the Newman Δica mutant was 93% less than that observed in the wild-type strain, suggesting that ica does play a role in biofilm formation in a low-iron environment.
FIG. 1.
Biofilm assays (A) and SDS extracts (B) showing the effect of the ica mutation on biofilm formation and expression of surface proteins of the S. aureus Newman (lane 1), Newman ica mutant (lane 2), and Newman ica-complemented mutant (Newman ica +; lane 3) strains grown for 24 h in CRPMI. The results shown in panel A represent the means and standard deviations of at least four independent experiments. OD, optical density.
Our previous studies have shown that PNAG levels do not differ between strong and poor biofilm-producing strains in low-iron growth conditions (26). Therefore, to further investigate the effect of the ica mutation on the Newman strain, we compared SDS cell surface extracts containing non-covalently bound cell surface proteins from the Newman wild type and its isogenic mutant, Newman Δica::tet, grown under the same iron-restricted conditions as used in the biofilm assay. In the SDS cell surface extracts prepared from wild-type Newman, several proteins of various molecular weights were detected, two of which were particularly abundant (Fig. 1B). Interestingly, there was a significant reduction in the levels of these two proteins in the surface protein extracts prepared from the Newman Δica mutant (Fig. 1B). The two proteins were identified by MALDI-TOF mass spectrometry as Eap, a 70,000-Da secreted protein (40), and Emp, a 35,000-Da secreted protein (23), both of which are non-covalently attached to the S. aureus cell surface (Table 3). Eap is a virulence factor implicated in many aspects of S. aureus disease that can promote adhesion to itself and a broad spectrum of host proteins, such as fibronectin, fibrinogen, vitronectin, and collagen (40). Though studied in less detail than Eap, Emp has also been found to bind to a range of host proteins (23) and is implicated in disease causation (6). Complementation of the Newman Δica mutant (28) restored biofilm formation to wild-type levels and also restored the cell surface expression of Eap and Emp (Fig. 1B).
TABLE 3.
S. aureus Newman proteins identified by MALDI-TOF mass spectrometry
| Protein | Cell fraction | Scorea |
|---|---|---|
| Eap | Cytoplasmic | 336 |
| Membrane | 656 | |
| Cell wall | 761 | |
| SDS surface extract | 822 | |
| Supernatant | 1,445 | |
| Emp | Cytoplasmic | 258 |
| Membrane | 686 | |
| Cell wall | 91 | |
| SDS surface extract | 697 | |
| Supernatant | 220 |
The percent sequence coverage is factored into the score. A score higher than 76 is deemed significant.
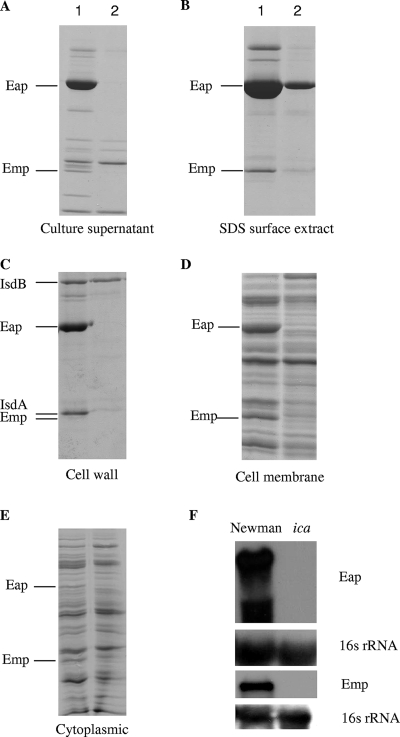
To determine the global effect of the ica mutation on staphylococcal proteins and to establish if there is any abnormal accumulation of proteins in any cell fraction, culture supernatant, SDS surface, cell wall, membrane, and cytoplasmic extracts were prepared from the same 24-h cultures for both the Newman wild-type and Newman Δica mutant strains grown in CRPMI (Fig. 2A to E). In the ica mutant cell extracts, the levels of the majority of the proteins were unaffected; however, Eap and Emp were not detectable in any of the protein fractions except at a low level in the SDS surface extract. This finding is in contrast to that for the wild-type Newman strain, where Eap and Emp polypeptides were identified by MALDI-TOF mass spectrometry in all fractions (Table 3). In addition to Eap and Emp, a small number of other proteins showed markedly altered levels in the Newman Δica mutant (Fig. 2). One of these proteins in the cell wall fraction was identified by MALDI-TOF mass spectrometry as FrpA, also known as IsdA, which is an iron-regulated multifunctional protein with a characteristic LPXTG motif that covalently anchors it to the cell surface (9, 33, 36). There was no obvious accumulation of proteins in any of the fractions to account for the loss of proteins in the ica mutant.
FIG. 2.
Culture supernatant (A), SDS surface extract (B), cell wall (C), cell membrane (D), and cytoplasmic (E) fractions obtained from the same cultures of the S. aureus Newman (lane 1) and Newman ica mutant (lane 2) strains grown for 24 h in CRPMI. All lanes were equivalently loaded. Eap and Emp polypeptides were identified in each fraction by MALDI-TOF mass spectrometry. (F) Northern blot analysis of the S. aureus Newman and Newman ica mutant strains. Total RNA (10 μg) prepared from S. aureus cells growing exponentially in CRPMI was resolved by agarose gel electrophoresis and hybridized with emp or eap DNA probes. The blots were then stripped and rehybridized with the 16S rRNA control probe.
To investigate how the ica mutation affected Eap and Emp protein expression, total RNA was extracted from exponentially growing Newman wild-type and Newman Δica mutant strains grown in CRPMI. Northern blot analysis with eap and emp DNA probes showed that ica was in fact required for eap and emp transcription (Fig. 2F), as no eap and emp transcripts were detected in the ica mutant, although some protein was still detected in the SDS cell surface extracts (Fig. 2B). Therefore, these data show that the ica operon is necessary for the expression of the Eap and Emp virulence factors in iron-restricted growth conditions.
Emp is essential for biofilm formation in iron-restricted growth conditions.
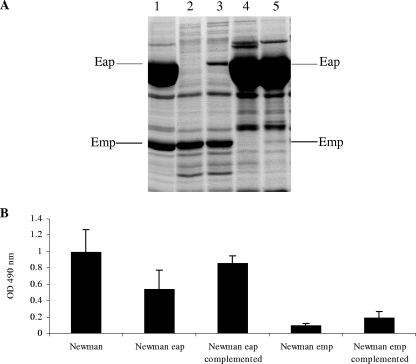
As the loss of ability to form biofilm in the Newman Δica mutant correlated with the loss of Eap and Emp on the cell surface, we investigated the effect on biofilm formation of deleting eap and emp individually. SDS surface extracts prepared from the Newman Δeap::erm (24) and Newman emp::erm mutant strains grown in CRPMI confirmed the absence of the 70,000-Da and 35,000-Da Eap and Emp proteins in the respective strains (Fig. 3A). The Newman wild type and its isogenic eap and emp mutants were then assessed for biofilm formation under low-iron conditions. Disruption of the eap gene resulted in a reduction in biofilm formation of 45% (P = 0.007), whereas disruption of the emp gene resulted in a reduction in biofilm formation of 90% compared to that observed for Newman wild type (Fig. 3B). The reduction in biofilm observed for the emp mutant strain was equal to that observed for the Newman Δica mutant, suggesting that Emp is essential for biofilm formation under low-iron growth conditions.
FIG. 3.
SDS surface extracts (A) and biofilm assays (B) showing the effects of the eap and emp mutations on biofilm formation and expression of the surface proteins of the S. aureus Newman wild-type (lane 1), Newman eap mutant (lane 2), Newman eap-complemented mutant (lane 3), Newman emp mutant (lane 4), and Newman emp-complemented mutant (lane 5) strains grown for 24 h in CRPMI. The results shown in panel B represent means and standard deviations from at least four independent experiments. OD, optical density.
To confirm the role of Eap and Emp in biofilm formation in low-iron growth conditions, the mutant strains were complemented with the respective wild-type copies of the eap and emp genes carried on a plasmid (pEap and pEmp). SDS cell surface extracts showed that expression of the proteins in the eap and emp mutants was partially restored in the complemented strains (Fig. 3A). It has been previously noted that there is a low level of expression of Eap in complemented strains; however, eap mutant phenotypes were still complemented, confirming the role of Eap (24). Our biofilm assays demonstrated that although there is only a low level of Eap and Emp expression in the complemented mutants, there was an increase in biofilm formation in comparison to that of the respective mutant strains, thus confirming the role of the Eap and Emp proteins in low-iron-induced biofilm formation (Fig. 3B).
Emp and Eap are transcriptionally regulated by a low-iron environment and Fur.
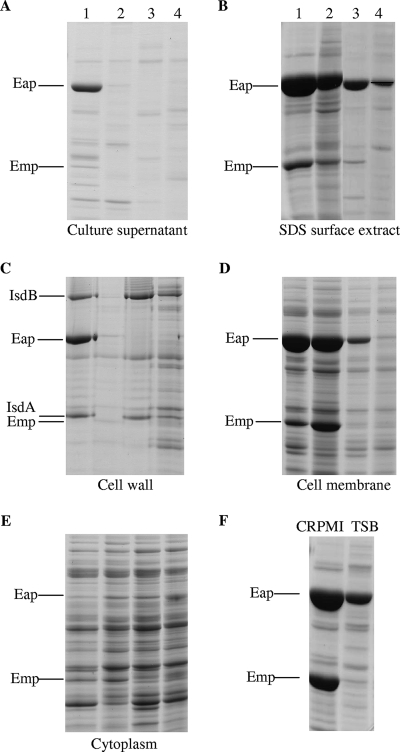
We have previously demonstrated that low-iron-induced biofilm formation is regulated by iron and Fur (26). Therefore, to determine the effect of iron and Fur on Emp and Eap expression, cell fractions from wild-type Newman and an isogenic Newman Δfur::tet mutant grown under iron-restricted (CRPMI) and iron-replete (CRPMI with iron added or TSB) conditions were compared. In the wild type, surface expression of Emp and Eap is partially repressed in high-iron growth conditions (Fig. 4B, C, and F), which correlates with our observation that biofilm formation is repressed by high-iron conditions. Protein profiles of the fur mutant revealed that Eap and Emp are actually positively regulated by Fur, as expression of Eap and Emp by the fur mutant is reduced in all extracts compared to that for the wild type. However, surface levels of Eap and Emp continued to be iron regulated in the absence of Fur, suggesting that Eap and Emp are iron regulated via a Fur-independent mechanism. These findings agree with our previous observation that under low-iron growth conditions, biofilm formation was positively Fur regulated and iron regulated independently of Fur (26).
FIG. 4.
S. aureus Newman (lanes 1 and 2) and Newman fur mutant (lanes 3 and 4) culture supernatant (A), SDS surface extract (B), cell wall (C), cell membrane (D), and cytoplasmic (E) fractions obtained from the same cultures grown for 24 h in iron-restricted CRPMI (lanes 1 and 3) or CRPMI with 50 μM Fe2(SO4)3 added (lanes 2 and 4) to give iron-replete growth conditions. (F) SDS extracts of S. aureus Newman grown in CRPMI and TSB. All lanes were equivalently loaded. Eap, Emp, IsdA, and IsdB polypeptides were identified in each fraction by MALDI-TOF mass spectrometry.
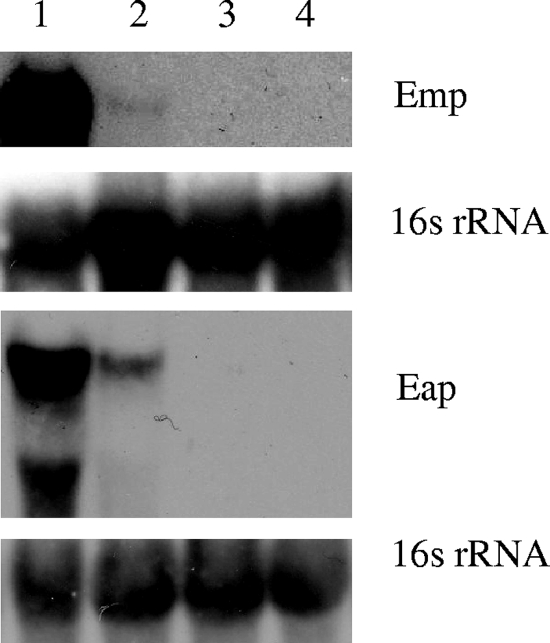
To further investigate how low-iron conditions and Fur regulate Eap and Emp protein expression, total RNA was extracted from exponentially growing Newman wild-type and Newman Δfur mutant strains grown in CRPMI. Northern blot analysis showed that regulation of Eap and Emp expression in response to low-iron conditions was in fact occurring at the transcriptional level, as there are reduced levels of both eap and emp transcripts in high-iron conditions (Fig. 5). Northern blot analysis also confirmed that Fur is required for expression of eap and emp in low-iron conditions, as no eap and emp transcripts were detected in the fur mutant (Fig. 5), although some Eap and Emp proteins were still present in the cell extracts (Fig. 4). Therefore, these results demonstrate that a low-iron environment and the global regulator Fur have pivotal roles in the expression of these important virulence factors.
FIG. 5.
Northern blot analysis of S. aureus Newman and the Newman fur mutant. Total RNA (10 μg) prepared from S. aureus cells growing exponentially in CRPMI (lanes 1 and 3) or CRPMI with 50 μM Fe2(SO4)3 (lanes 2 and 4) was resolved by agarose gel electrophoresis and hybridized with emp or eap DNA probes. The blots were then stripped and rehybridized with the 16S rRNA control probe.
Sae and Agr are essential for low-iron-induced biofilm formation.
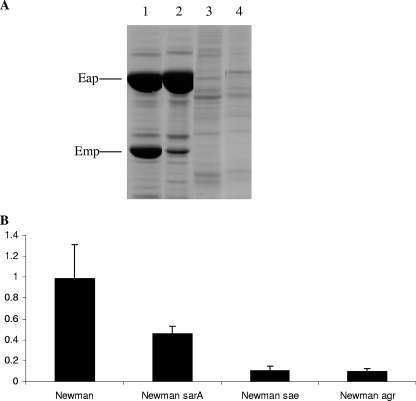
Eap and Emp expression is regulated at the transcriptional level by a number of global regulators, including SarA, Agr, and Sae (21). To investigate the roles of these transcriptional regulators in low-iron growth conditions, biofilm assays and cell surface protein extractions were performed, using wild-type Newman and its isogenic mutants Newman sae::Tn917 (AS3), Newman ΔsarA::erm, and Newman Δagr::tet grown in iron-restricted CRPMI. SDS surface protein extracts demonstrated that the levels of Eap and Emp were significantly reduced in the agr and sae mutants, but there were still significant levels of both proteins in the sarA mutant (Fig. 6A). These surface levels of Eap and Emp correlated with the biofilm assays, as there was only a 54% reduction in biofilm levels in the sarA mutant in comparison to that of the wild type (Fig. 6). In contrast, biofilm formation in the Newman sae and agr mutants was almost completely eliminated, reduced by 90% compared to that of the wild type (Fig. 6B), suggesting that in these iron-restricted, nutrient-poor growth conditions, which are reflective of in vivo environmental conditions, Sae and Agr are essential for biofilm formation.
FIG. 6.
SDS surface extracts (A) and biofilm assays (B) showing the effects of the sarA, saeRS, and agr mutations on biofilm formation and expression of the surface proteins of S. aureus Newman wild-type (lane 1), Newman sarA mutant (lane 2), Newman sae mutant (lane 3), and Newman agr mutant (lane 4) strains grown for 24 h in CRPMI. The results shown in panel B represent means and standard deviations from at least four independent experiments.
Proteins prepared from the cell wall, cell membrane, cytoplasm, and supernatants alongside the SDS protein extractions demonstrated that there was no increase in Eap or Emp in any of the other protein fractions of the sae, agr, and sarA mutant strains to account for their reduced levels of surface expression (data not shown). This finding is in agreement with sae, agr, and sarA being known transcriptional regulators of eap and emp expression (21), and indeed, our transcriptional analysis confirmed that there are reduced levels of eap and emp transcription in the mutant strains compared to that in the wild type (data not shown). Thus, these findings suggest that Sae, Agr, and, to a lesser degree, SarA all play an important role in the complex regulatory pathway involved in low-iron-induced biofilm formation, probably via the regulation of Eap and Emp expression.
DISCUSSION
Iron is an essential nutrient for virtually all organisms; however, free ferric iron is scarce in vivo, as the majority of iron is bound to high-affinity iron-binding proteins. Pathogenic bacteria have adapted well to the severe iron-restricted environment encountered in the host and have evolved to use the extremely low availability of iron in mammalian body fluids as a major environmental signal that they are in vivo. We have previously shown that S. aureus Newman biofilm formation is induced in iron-restricted growth conditions in vitro. In this study, we showed that the secreted proteins Eap and Emp are involved in low-iron-induced biofilm formation of S. aureus Newman and that there is a link between the ica-dependent and protein-dependent biofilm mechanisms that have been discussed in previous publications (38), as ica is required for expression of Eap and Emp proteins in low-iron growth conditions. Our complementation studies confirmed the roles of Eap and Emp in low-iron biofilm formation, although the expression of both proteins was very low, which was also noted in previous complementation studies (24). Nevertheless, we did see reproducible increases in Eap and Emp expression in the complemented strains which correlate with an increase in biofilm formation.
Eap and Emp are important virulence factors implicated in a number of aspects of S. aureus pathogenesis. Eap is involved in the inhibition of wound healing (1), evasion of the host immune system (5, 6), and the promotion of bacterial internalization into eukaryotic cells (20). Emp, though studied in less detail than Eap, has been shown to be associated with endovascular disease (6). Furthermore, Eap is known to be involved in cell-cell adhesion, and both proteins can promote adhesion to a broad spectrum of host proteins, such as fibronectin, fibrinogen, vitronectin, and collagen (23, 40), factors that may assist in biofilm formation in vivo, especially as biomaterials are rapidly coated by a conditioning film of host proteins. The identification of Eap and Emp as protein factors important for S. aureus biofilm formation correlates with a recent study which shows that proteins play an essential role in S. aureus biofilm formation, as biofilms are disintegrated by treatment with proteases (44). Furthermore, Eap was identified in 100% of S. aureus strains isolated from prosthetic joint infections after total knee arthroplasty, suggesting that Eap may be important for biofilm formation in vivo (44).
Previous studies have shown that S. aureus Newman forms only a low level of biofilm in iron-replete media like TSB and yet is able to form a substantial biofilm in vivo in a device-related animal infection model (15). This difference in the ability of Newman to form a biofilm may correlate with the levels of Eap and Emp that are expressed under different growth conditions. Our studies showed that Eap and, in particular, Emp expression levels are significantly lower when S. aureus Newman is grown in iron-replete, nutrient-rich media such as TSB than when grown in iron-restricted growth conditions. Indeed, we showed that maximal levels of Eap and Emp on the staphylococcal cell surface are obtained when S. aureus Newman is grown in a nutrient-poor, low-iron medium reflective of the in vivo environment. Furthermore, this increase of Eap and Emp expression in a low-iron environment is also seen in a number of other S. aureus strains, including 8325-4 and the clinical isolate MN8, although the levels of Eap and Emp in these strains are lower than in Newman, which correlates with our previous studies showing that 8325-4 and MN8 produce only low levels of biofilm in low-iron conditions compared to that of Newman (26; our unpublished data).
Our data show that Eap and Emp expression is iron regulated at the transcriptional level. Our proteomics results show that this iron regulation of Eap and Emp is Fur independent, as there is still iron-dependent repression of Eap and Emp proteins in the fur mutant. Therefore, iron regulation of Eap and Emp is via an unknown transcriptional regulator. However, Fur is required for the induction of eap and emp expression in low-iron conditions, as there is no observed eap and emp transcription in the fur mutant. This finding is in contrast to classical Fur regulation, as Fur would normally act as a transcriptional repressor in response to a high-iron environment, resulting in constitutive expression of target genes in the fur mutant. Fur could act as an activator of eap and emp transcription by binding directly to the promoters and inducing transcription, although there are no obvious Fur box consensus sequences in these promoters. This has been shown to occur in Neisseria meningitidis, where Fur binds to operators upstream of the nitric oxide reductase (norB) gene promoter, thus inducing transcription (12). Alternatively, Fur could act indirectly by repressing the expression of another regulator of emp and eap transcription.
It is possible that Fur may be involved in posttranscriptional regulation of eap and emp expression, as there was still a substantial level of Eap and Emp polypeptide observed in the fur mutant extracts, although no transcripts were detected. This suggests that transcription occurred, allowing some translation, but that the mRNA was quickly degraded, resulting in reduced levels of protein. In many bacteria, positive regulation by Fur is often mediated via small, untranscribed regulatory RNAs (32). Many small RNAs have now been identified in a wide range of organisms. These small RNAs can act negatively by targeting mRNA for degradation or positively by binding to 5′-untranslated regions of mRNA to induce translation (32). Regulatory small RNAs have been identified in S. aureus, but their roles in gene expression have yet to be fully elucidated (42), with the exception of that of RNAIII, which is a regulatory RNA that has been shown to be involved in the positive and negative regulation of the gene expression of a number of virulence factors (34). Therefore, it is possible that small RNAs are involved in Fur-mediated regulation of eap and emp transcription. However, Fur normally binds DNA when complexed with iron, thus repressing small RNAs and consequently inducing target mRNA transcription in a high-iron environment, whereas eap and emp expression is positively regulated by Fur in a low-iron environment. It is possible that the S. aureus iron-free form of Fur can bind DNA either as an activator or as a repressor, as Fur has been shown to directly repress gene expression in low-iron conditions in Helicobacter pylori (3). Alternatively, Fur may control the expression of the first of a cascade of regulators involved in eap and emp expression, leading to the expression of these important virulence factors in a low-iron environment.
Indeed, in this study we demonstrated that in addition to Fur, low-iron-induced biofilm formation requires Sae, Agr, and SarA. Our results show that in iron-restricted growth conditions, Sae and Agr are essential for Emp and Eap expression and hence for biofilm formation, whereas SarA appears to have a less-significant role. This finding is surprising, as in other growth conditions SarA is considered to be essential for biofilm formation (2, 49), yet in low-iron growth conditions the sarA mutation does not have a major effect on biofilm formation. The different effects of these regulator mutants on biofilm formation may be accounted for by different strain backgrounds and/or the growth conditions used and the multifactored nature of S. aureus biofilm production. We propose that in different growth conditions, factors other than Emp and Eap are important for biofilm formation, whereas in low-iron growth conditions, Emp is an essential factor, and therefore, the regulators required for its expression, such as Fur, Sae, and Agr, are more important. This hypothesis is supported by our data, which show that the agr mutation has different effects on biofilm formation that are dependent on growth conditions, as in contrast to our biofilm assays in low-iron conditions, there is an increase in biofilm levels of the Newman agr mutant grown in an iron-rich medium such as TSB (data not shown).
The Eap- and Emp-positive regulators, Sae, Agr, and SarA, may induce transcription of eap and emp in response to low-iron conditions. Our preliminary data suggest that agr transcription is not regulated by iron, and previous studies have shown that Sae protein expression is not regulated by Fur and iron (14). However, the signal that Sae senses and responds to has not been determined, and there are a myriad of other regulators which interact with Sae, Agr, and SarA that may mediate the induction of biofilm formation in response to low-iron conditions (38).
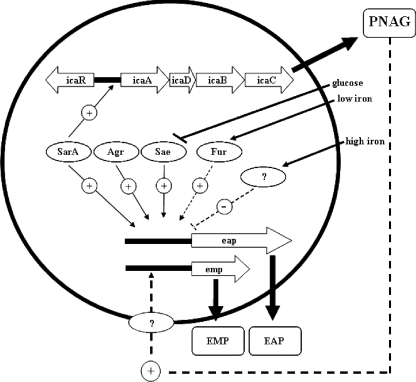
Our studies have shown that expression of the ica operon is required for biofilm formation in iron-restricted growth conditions. This result was surprising, as our previous studies had shown that PNAG levels did not differ between strong and poor biofilm-producing strains in these conditions (26). We showed here that in fact, ica is required for Eap and Emp expression, and it is the Eap and Emp protein levels which vary between the strains. We also showed that ica is required for the expression of other proteins, including the multifunctional FrpA or IsdA protein (9, 33, 36). However, FrpB/IsdB, which is also an iron-regulated protein with an LPXTG motif, is not affected by the ica mutation. This may not be surprising, as these two genes are not cotranscribed and are differentially regulated; for example, σB regulates isdA expression but not isdB expression (4). So why would the mutation of a polysaccharide biosynthetic pathway affect the transcription of at least three surface proteins? A possible explanation is that the loss of the hydrophobic polysaccharide on the surface in the ica mutant could result in localized environmental changes that are sensed by global transcriptional regulators which then repress eap and emp transcription. Many E. coli outer membrane proteins, including iron- and Fur-regulated proteins such as FepA, are regulated in response to different environmental conditions, often via small RNAs (19). Our preliminary evidence has shown that expression of Eap and Emp is repressed by osmotic stress (data not shown), suggesting that environmental conditions other than iron concentration do affect Eap and Emp expression. This regulation of eap and emp transcription in response to varying extracellular conditions could be mediated via any of the known or as-yet-unidentified regulators of eap and emp, as shown in the model in Fig. 7.
FIG. 7.
Model of the coordinated regulation of Eap and Emp expression. Positive and negative regulatory pathways are indicated with a plus (+) and a minus (−), respectively. Dotted lines indicate possible indirect regulatory pathways.
Our data suggesting that ica is required for expression of some proteins correlates with a recent study which showed that both ica and protein factors were required for biofilm formation in strains isolated from prosthetic hip and knee joint infections (44). The fact that the ica locus is so highly conserved in S. aureus may be explained if it is required for both PNAG-dependent biofilm formation and the expression of proteins which are involved in different aspects of S. aureus pathogenesis.
Acknowledgments
This work was supported by project grant 24681 from the Royal Society.
We thank Malcolm Horsburgh and Simon Foster for providing the 8325-4 fur mutant, Gerry Pier for providing Newman Δica::tet and Newman Δica::tet-complemented strains, Christiane Goerke for Newman sae::Tn917 (AS3), Ambrose Cheung for ALC1342, Richard Novick for RN6911, and Jan-Ingmar Flock for Newman Δeap::erm (AH12). We thank Peter Williams for his helpful comments on the manuscript and Andrew R. Bottrill and Shairbanu Ibrahim of the PNACL Proteomics Facility, University of Leicester, for the proteomic analyses.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 11 February 2008.
REFERENCES
- 1.Athanasopoulos, A. N., M. Economopoulou, V. V. Orlova, A. Sobke, D. Schneider, H. Weber, H. G. Augustin, S. A. Eming, U. Schubert, T. Linn, P. P. Nawroth, M. Hussain, H. P. Hammes, M. Herrmann, K. T. Preissner, and T. Chavakis. 2006. The extracellular adherence protein (Eap) of Staphylococcus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood 1072720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projen, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bereswill, S., S. Greiner, A. H. M. Van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 1825948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 1864085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavakis, T., M. Hussain, S. M. Kanse, G. Peters, R. G. Bretzel, J.-I. Flock, M. Herrmann, and K. T. Preissner. 2002. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 8687-693. [DOI] [PubMed] [Google Scholar]
- 6.Chavakis, T., K. Wiechmann, K. T. Preissner, and M. Herrmann. 2005. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb. Haemost. 94278-285. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 896462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 692448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, S. R., M. D. Wiltshire, and S. J. Foster. 2004. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol. Microbiol. 511509-1519. [DOI] [PubMed] [Google Scholar]
- 10.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 675427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 694079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 521081-1090. [DOI] [PubMed] [Google Scholar]
- 13.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 695-107. [DOI] [PubMed] [Google Scholar]
- 14.Freidman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 20777-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fluckiger, U., M. Ulrich, A. Steinhuber, G. Döring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 731811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler, V. G., P. D. Fey, L. B. Reller, A. L. Chamis, G. R. Corey, and M. E. Rupp. 2001. The intercellular adhesin locus ica is present in clinical isolates of Staphylococcus aureus from bacteremic patients with infected and uninfected prosthetic joints. Med. Microbiol. Immunol. 189127-131. [DOI] [PubMed] [Google Scholar]
- 17.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 401439-1448. [DOI] [PubMed] [Google Scholar]
- 18.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 431367-1378. [DOI] [PubMed] [Google Scholar]
- 19.Guillier, M., and S. Gottesman. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59231-247. [DOI] [PubMed] [Google Scholar]
- 20.Haggar, A., M. Hussain, H. Lonnies, M. Herrmann, A. Norrby-Teglund, and J. I. Flock. 2003. Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect. Immun. 712310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 1511789-1800. [DOI] [PubMed] [Google Scholar]
- 22.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 1836778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrman. 2002. Insertional inactivation of eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 702933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 1862449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, M., A. Cockayne, P. H. Williams, and J. A. Morrissey. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves Fur-dependent and Fur-independent mechanisms. J. Bacteriol. 1878211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21372-376. [Google Scholar]
- 28.Kropec, A., T. Maira-Litran, K. K. Jefferson, M. Grout, S. E. Cramton, F. Götz, D. A. Goldmann, and G. B. Pier. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 736868-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maira-Litrán, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 704433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 4093-113. [DOI] [PubMed] [Google Scholar]
- 33.Mazmanian, S. K., H. K. Su Ton-That, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 992293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 144569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 686281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrissey, J. A., A. Cockayne, J. Hammacott, K. Bishop, A. Denman-Johnson, P. J. Hill, and P. Williams. 2002. Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus. Infect. Immun. 702399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 1-37. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, Inc., New York, NY.
- 38.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270179-188. [DOI] [PubMed] [Google Scholar]
- 39.O'Neill, E., C. Pozzi, P. Houston, D. Smyth, H. Humphreys, D. A. Robinson, and J. P. O'Gara. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 451379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palma, M., A. Haggar, and J. L. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 1812840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins, D. N., D. D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 203551-3567. [DOI] [PubMed] [Google Scholar]
- 42.Pichon, C., and B. Felden. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA 10214249-14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 1826824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde, H., E. C. Burandt, N. Siemssen, L. Frommelt, C. Burdelski, S. Wurster, S. Scherpe, A. P. Davies, L. G. Harris, M. A. Horstkotte, J. K. Knobloch, C. Ragunath, J. B. Kaplan, and D. Mack. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 281711-1720. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speicher, K. D., O. Kolbas, S. Harper, and D. W. Speicher. 2000. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 1174-86. [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 2921-26. [DOI] [PubMed] [Google Scholar]
- 48.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Débarbouillé, J. R. Penandés, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 1875318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 481075-1108. [DOI] [PubMed] [Google Scholar]
- 50.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterization of the ferric regulator, Fur, from Staphylococcus aureus. Microbiology 146659-668. [DOI] [PubMed] [Google Scholar]
- 51.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 1861838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]