Abstract
Aeromonas salmonicida subsp. salmonicida, a bacterial pathogen of Atlantic salmon, has no visible pili, yet its genome contains genes for three type IV pilus systems. One system, Tap, is similar to the Pseudomonas aeruginosa Pil system, and a second, Flp, resembles the Actinobacillus actinomycetemcomitans Flp pilus, while the third has homology to the mannose-sensitive hemagglutinin pilus of Vibrio cholerae. The latter system is likely nonfunctional since eight genes, including the gene encoding the main pilin subunit, are deleted compared with the orthologous V. cholerae locus. The first two systems were characterized to investigate their expression and role in pathogenesis. The pili of A. salmonicida subsp. salmonicida were imaged using atomic force microscopy and Tap- and Flp-overexpressing strains. The Tap pili appeared to be polar, while the Flp pili appeared to be peritrichous. Strains deficient in tap and/or flp were used in live bacterial challenges of Atlantic salmon, which showed that the Tap pilus made a moderate contribution to virulence, while the Flp pilus made little or no contribution. Delivery of the tap mutant by immersion resulted in reduced cumulative morbidity compared with the cumulative morbidity observed with the wild-type strain; however, delivery by intraperitoneal injection resulted in cumulative morbidity similar to that of the wild type. Unlike the pili of other piliated bacterial pathogens, A. salmonicida subsp. salmonicida type IV pili are not absolutely required for virulence in Atlantic salmon. Significant differences in the behavior of the two mutant strains indicated that the two pilus systems are not redundant.
Aeromonas salmonicida subsp. salmonicida is a gram-negative, nonmotile, rod-shaped bacterium that is the etiologic agent of an infectious bacteremia-septicemia of salmonids known as furunculosis. Furunculosis is a complex disease that exists in different forms depending on the health, age, and species of fish. Many cell-associated and secreted factors have been implicated as virulence determinants in this bacterium (for reviews, see references 5 and 7). Despite this, much of the pathogenesis of A. salmonicida subsp. salmonicida remains poorly understood, and no single characteristic or phenotype was found only in virulent strains (16, 17, 30, 42) until the recent description of an A. salmonicida subsp. salmonicida type III secretion system (6, 11).
In order to better understand the virulence strategies employed by A. salmonicida subsp. salmonicida, we have focused on the initial stages of infection: adherence and invasion. Bacteria use complex intercellular mechanisms and specific and nonspecific adhesins to achieve these aims (34). The most well-studied A. salmonicida subsp. salmonicida adhesin is the surface layer or S-layer, sometimes referred to as the A-layer (additional layer) (33). This layer is a nonspecific but important factor for adherence due to its hydrophobic nature.
Pili allow bacteria to attach to solid surfaces, including host tissues, and are considered important virulence factors in many pathogenic bacteria (36). Pili are filamentous, extracellular organelles that may be peritrichous or polar and may be present singly or in bundles. There are currently four recognized types of pili that have been found on gram-negative bacteria, and type IV pili have often been implicated in host attachment and virulence. Type IV pili are subdivided into types IVa and IVb on genetic and morphological grounds. Type IVa (non-bundle-forming) pili are important in a number of bacterial processes, including flagellum-independent “twitching” motility, DNA uptake, biofilm formation, and adherence to substrates, including host cells (27). The Tap pilus, a type IVa pilus that is encoded in part by the tapABCD operon, has been described for multiple aeromonad species (2, 32). It has been implicated in A. salmonicida subsp. salmonicida virulence in a rainbow trout model (Oncorhynchus mykiss) on the basis of an increase in the 50% lethal dose (LD50) for a tapA mutant as assessed by intraperitoneal injection (26).
Analysis of the A. salmonicida subsp. salmonicida A449 genome sequence has revealed the presence of two type IV pilus systems in addition to the Tap system. In this study, Flp-like and mannose-sensitive hemagglutinin (MSHA)-like type IV pilus systems not previously identified in this species are described. The contribution to virulence of two of the pilus systems was assessed using a live animal model and isogenic knockout mutant strains. In addition, pili of A. salmonicida subsp. salmonicida were visualized for the first time using atomic force microscopy (AFM) and pilin-overexpressing strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacteria and plasmids used in this study are listed in Table 1. The parental strain for all knockouts was A. salmonicida subsp. salmonicida strain A449, which was originally isolated from a natural furunculosis epizootic. All Aeromonas strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) (Difco) for 3 days at 17°C. Escherichia coli strains were grown in Luria-Bertani broth (LB) or on LB agar at 37°C. Antibiotics were used at the following concentrations: for E. coli, 100 μg/ml ampicillin, 25 μg/ml kanamycin, and 20 μg/ml chloramphenicol; and for A. salmonicida subsp. salmonicida, 50 μg/ml ampicillin, 200 μg/ml kanamycin for selection, 50 μg/ml kanamycin for maintenance, and 20 μg/ml chloramphenicol. Iron concentrations were reduced by addition of 120 μM 2,2′-dipyridyl (Sigma).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| A. salmonicida subsp. salmonicida strains | ||
| A449 | Virulent, Cmr | 29 |
| 02-02 | A449 flpA::Km, Cmr Kmr | This study |
| 02-06 | Lab-derived mutant of A449, O side chain negative, surface layer secretor, Cmr | 43 |
| 02-10 | A449 tapA::KO, Cmr Apr | This study |
| 03-01 | A449 flpA::Km tapA::KO, Cmr Kmr Apr | This study |
| P. aeruginosa PAK | Wild type | D. Bradley |
| E. coli strains | ||
| TOP10 | K-12 | Invitrogen |
| DH10B | K-12 for large plasmids | Invitrogen |
| EC100D pir-116 | K-12 pir-116 | Epicenter |
| BW20767 | K-12 pir+, conjugation positive | 28 |
| Plasmids | ||
| pUTkm1 | Cloning vector, Apr Kmr | 13 |
| pCRScript-Amp | oriE1, Apr | Stratagene |
| pAH34 | oriR6Kγ mobRP4, Apr | 28 |
| pKO | pAH34 with tmRNA tag | This study |
| pKO-tapA | Internal fragment of tapA in pKO, in frame with tmRNA tag | This study |
| pGP704 | oriR6Kγ mobRP4, Apr | 21 |
| pJW109-pir | pGP704 with flpA-flanking regions surrounding kanamycin resistance cassette, Kmr Apr | This study |
| pMMB67EH | Low-copy-number, broad-host-range expression vector, mobRSF1010, Apr | 20 |
| pMMB67EH.Km | pMMB67EH with Apr replaced by Kmr | This study |
| ptapA.K | pMMB67EH.Km carrying complete tapA gene | This study |
| pflp1.K | pMMB67EH.Km carrying complete flp-1 gene | This study |
| pBACe3.6 | Large insert vector, Cmr | 19 |
| pAs01a04 | Derivative of pBACe3.6 with 71.2-kb insert carrying the flp locus | This study |
DNA techniques.
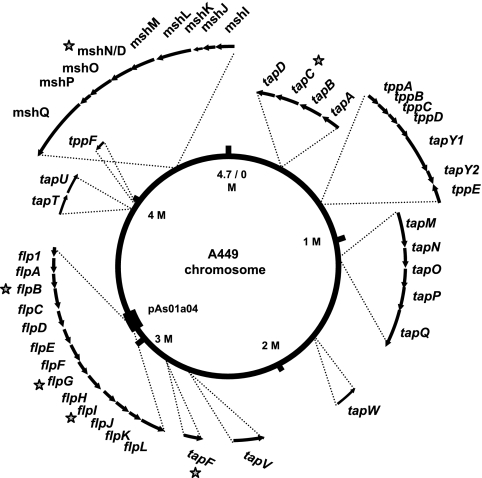
DNA manipulations were performed by using standard genetic and molecular techniques (1). Genomic DNA from A449 was isolated using a PureGene DNA isolation kit (Gentra Systems, Minneapolis, MN) and was used as the PCR template for construction of all mutant strains. Oligonucleotides were prepared by Integrated DNA Technologies Inc. (Coralville, IA). PCR was performed with either rTaq (Amersham) or Pfu (MBI Fermentas) by following the manufacturer's directions. During the A. salmonicida subsp. salmonicida genomic sequencing project, a BAC library of A. salmonicida subsp. salmonicida genomic DNA was made. Briefly, A. salmonicida subsp. salmonicida A449 genomic DNA was partially digested with EcoRI to generate fragments ranging from 50 to 150 kb long; these fragments were ligated into the vector pBACe3.6 (19) digested with the same enzyme. The library was transformed into E. coli DH10B. BAC pAs01a04 was one of the clones obtained and contained a 71-kb insert that corresponds to the chromosomal region between bp 3126400 and 3197600 (see Fig. 2).
FIG. 2.
Genome of A. salmonicida subsp. salmonicida showing the pilus system genes. The arrows indicate the approximate sizes and orientations of the genes. The numbering of the chromosome reflects the origin of replication. The thick line on the chromosome at 3.1 Mbp shows the extent of BAC pAs01a04 that carries the Flp locus. The stars indicate disrupted genes.
RT-PCR.
A. salmonicida subsp. salmonicida grown under a variety of conditions was harvested in RNAprotect bacterial reagent (Qiagen). Total RNA was extracted with RNeasy mini kits (Qiagen) used according to the manufacturer's directions. DNA was removed with DNA-free (Ambion). cDNA was made from the RNA using random decamers (Ambion) and SuperScript II reverse transcriptase (RT) (Invitrogen). The primers used for amplification of tapA and flpA are shown in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ → 3′)a | Description |
|---|---|---|
| tmRNA tag | GGCCGCAACTAGTTGCAGCAAACGACGAAAACTACGCACTAGCAGCTTAATAACAGAGCT | Coding strand of tmRNA tag |
| tmRNA tag | CTGTTATTAAGCTGCTAGTGCGTAGTTTTCGTCGTTTGCTGCAACTAGTTGC | Noncoding strand of tmRNA tag |
| tapA-1 | ATTGCACTTCCTGCATATCAGACC | Used to amplify internal fragment for knockout |
| tapA-2 | CGGGACCAGTATATATGTGTATGC | Used to amplify internal fragment for knockout |
| tapA-3 | AGGAGAATTCACATGAAGAAGCAATCAGGC | Used for RT-PCR of tap mRNA and to clone complete tapA gene |
| tapA-4 | TATTGGATCCAGAGGTCATGCGTTAGCAG | Used for RT-PCR of tap mRNA and to clone complete tapA gene |
| flpA-1 | CGGCCTTGCATGCCAGCAAGGTTGCG | Used to amplify 5′ flanking region of flpA for knockout |
| flpA-2 | CCAAGATCTGAGCGCTGTCACTTGCCCGCGGC | Used to amplify 5′ flanking region of flpA for knockout and to clone complete flp-1 gene |
| flpA-3 | GCGAGATCTCTGACCACTCCTCTATTTATGTAGTC | Used to amplify 3′ flanking region for knockout |
| flpA-4 | GCTTGCTGGTGAGGATCACATCCACCC | Used to amplify 3′ flanking region for knockout |
| flpA-F | CATATTGATGGCGTGCTGGT | Used for RT-PCR of flp mRNA |
| flpA-R | CACAAAATGGCGAATCCATA | Used for RT-PCR of flp mRNA |
| flpA-5 | GGACGAATTCATGAAGATGCGAGTTTGGG | Used to clone complete flp-1 gene |
| MMB01 | CATCATAACGGTTCTGGC | Used to sequence or amplify insert of pMMB plasmids |
| MMB02 | GAAAATCTTCTCTCATCCGC | Used to sequence or amplify insert of pMMB plasmids |
Restriction sites are underlined, and non-sequence-specific regions are indicated by bold type.
Bacterial conjugation.
E. coli BW20767 carrying the appropriate mobilizable plasmid was grown in LB medium with the appropriate antibiotic overnight at 37°C. A. salmonicida subsp. salmonicida was grown in TSB with chloramphenicol for 3 days at 17°C. One milliliter of a culture of each bacterial species was harvested and washed twice in fresh TSB without antibiotics. After the last wash, the bacterial pellet was resuspended in a small volume (15 to 30 μl) of TSB. The resulting thick suspensions of the bacteria were mixed together thoroughly and spotted in the center of a TSA plate with no antibiotic. After 2 days of incubation at 17°C, the spot was removed with a sterile toothpick and resuspended in 1 ml TSB. Appropriate dilutions were plated on TSA with chloramphenicol (to select against E. coli) and the appropriate antibiotic to select for the transformed colonies. Large colonies were picked, streaked onto fresh plates, and passaged two more times.
Construction of mutant strains. (i) tapA::KO strain.
The integrative knockout vector pKO was created so that the complete plasmid could be inserted into the gene of interest in such a way that the 3′ fragment of the affected gene would make an in-frame fusion to a transfer-messenger (tmRNA) tag so that any potentially translated peptide would be recognized by Clp proteases and degraded. The tmRNA tag was created using complementary oligonucleotides based on the sequence of A. salmonicida subsp. salmonicida tmRNA (45) with NotI and SstI sites at either end (Table 2). The tmRNA tag was inserted into the NotI and SstI sites of the polylinker of pCRScript-Amp (Stratagene) in such a way that the lacZα fragment was not disrupted to create pCRscript-tmRNA. The new polylinker and lacZα region of pCRScript-tmRNA was used to replace the polylinker of pAH34 (28) using PvuII sites to create pKO. Plasmid pAH34 and its derivative pKO are mobilizable, pir-dependent, ampicillin-resistant plasmids that allow blue/white selection of cloned inserts and cannot replicate in A. salmonicida subsp. salmonicida.
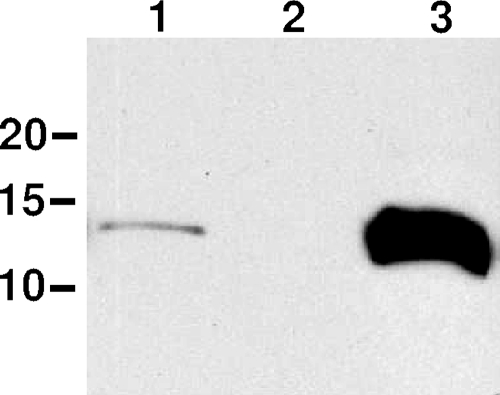
An internal fragment of the tapA gene was amplified from A449 genomic DNA using Pfu polymerase and primers tapA-1 and tapA-2. The fragment was blunt end ligated into the SrfI site of vector pKO to make an in-frame fusion to the tmRNA tag. The pKO-tapA plasmid was conjugated into A449 from E. coli pir+ mating strain BW20767 as described above, and single-crossover integrants (in which the tapA gene was interrupted by the pKO plasmid) were selected by growth on ampicillin and passaged in fresh medium three times. Proper integration was checked by PCR amplification of genomic DNA using primers within the plasmid and flanking the insertion. As determined by immunoblotting (Fig. 1), no intact TapA subunits were produced in this strain.
FIG. 1.
Immunoblot to confirm deletion and overexpression of tapA. Lane 1, A449(pMMB67EH.K); lane 2, tapA::KO(pMMB67EH.K); lane 3, A449(ptapA.K). All strains were grown in TSB with 50 μg/ml kanamycin and 0.05 mM IPTG. The positions of molecular mass standards (in kDa) are indicated on the left.
(ii) flpA::Km strain.
The flpA gene, encoding the putative prepilin peptidase, was interrupted by insertion of a kanamycin resistance gene cassette. Fragments flanking the flpA gene were amplified from genomic DNA using the primers shown in Table 2. Primers flpA-2 and flpA-3 included BglII sites that were used subsequently to introduce the 1.5-kb BamHI fragment carrying the kanamycin resistance cassette from pUTKm1. In a multistep process, the three fragments were cloned into the mobilizable, ampicillin-resistant, pir-dependent vector pGP704 to generate pJW109-pir. pJW109-pir was conjugated into A449 from BW20767 as described above, and single-crossover integrants were selected by growth on ampicillin. Double-crossover segregants were selected by isolation of ampicillin-sensitive, kanamycin-resistant colonies. Proper integration was checked by PCR amplification of genomic DNA using primers flanking the insertion.
(iii) flpA::Km/tapA::KO strain.
The double flpA::Km/tapA::KO mutant strain was constructed by introducing the tapA mutation plasmid, pKO-tapA, into the flpA::Km strain.
Construction of pilin-overproducing strains.
The pilin genes were amplified from A449 genomic DNA using primers flpA-2 and flpA-5 for flp-1 and primers tapA-3 and tapA-4 for tapA (Table 2). The fragments were cloned into the broad-host-range expression vector pMMB67EH.Km using the EcoRI and BglII or BamHI sites provided by the amplification primers. pMMB67EH.Km was created by cloning the 1.7-kb BamHI fragment carrying the kanamycin resistance gene from plasmid pUTkm1 into the β-lactamase gene of pMMB67EH. The plasmids were conjugated into A449 and an O-side-chain-deficient, S-layer-deficient derivative of A449, IMB 02-06, as described above. Transformation was confirmed by PCR amplification of total genomic DNA with primers complementary to the vector flanking the cloned insert (MMB01 and MMB02).
Immunoblotting.
Interruption of the tapA gene and overexpression of the TapA pilin in A449 (ptapA.K) were confirmed by immunoblotting with anti-TapA antibody (kindly donated by Mark Strom, National Oceanic & Atmospheric Administration, Seattle, WA) (Fig. 1). Briefly, whole-cell lysates were separated on a 15% Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (37), transferred to a polyvinylidene difluoride membrane, and visualized with anti-TapA horseradish-peroxidase conjugated secondary antibody and chemiluminescence (1). Overexpression of TapA in 02-06 and 02-10 was confirmed in the same way (data not shown). Antiserum to the Flp1 pilin is currently unavailable.
AFM.
For AFM imaging, A. salmonicida subsp. salmonicida cells were grown in TSB with chloramphenicol (20 μg/ml), kanamycin (50 μg/ml), 2,2′-dipyridyl (120 μM), and isopropyl-β-d-thiogalactopyranoside (IPTG) (50 μM) for 3 days at 17°C. E. coli cells were grown in LB medium with chloramphenicol (20 μg/ml) and 2,2′-dipyridyl (120 μM) overnight at 37°C. After the bacteria were washed in phosphate-buffered saline (pH 7.4) and resuspended in the same buffer, 1 drop of bacterial suspension was placed on a freshly cleaved mica surface. Excess fluid was drawn off, and the bacteria were allowed to adsorb for 15 min, after which the sample was rinsed with deionized water and air dried at room temperature for 30 min. Images were recorded using the contact mode at room temperature with a Molecular Imaging AFM microscope (Tempe, AZ). Images were recorded in both height and deflection modes. While height images provided quantitative information on sample surface topography, deflection images often had higher contrast of morphological details. V-shaped cantilevers with oxide-sharpened Si3N4 tips were used with spring constants of 0.01 N/m. High-resolution images were recorded for dried preparations with optimized feedback parameters at a scan frequency of 4 Hz. The diameters of the pilus filaments were measured by using AFM cross sections.
Animal care.
All relevant animal care committees approved the animal procedures, which were conducted under Canadian Council on Animal Care guidelines. As “death as an endpoint” is not considered acceptable under these guidelines (14; www.ccac.ca), morbid animals were euthanized when a set of previously agreed limiting clinical signs were reached. Thus, the challenge data are reported below as morbidity data rather than mortality data to reflect this.
Juvenile Saint John River or Sackville River stock Atlantic salmon were obtained from Nova Scotia hatcheries certified under Canadian Fish Health Protection regulations. They were stocked in 100-liter fiberglass resident tanks at a stocking density of ca 2.4 kg/tank and maintained at 14 ± 2°C in flowthrough dechlorinated municipal water under subdued lighting. They were fed a maintenance ration of a commercially available extruded feed (Signature Salmon Ration; Shurgain, Truro, Nova Scotia, Canada) daily which was equivalent to 1% of the body weight. Feeding was suspended for 1 day before manipulation and for 1 day after manipulation.
In vivo culture of A. salmonicida subsp. salmonicida.
In vivo culture was performed as described previously (10). Briefly, sections of dialysis tubing filled with A. salmonicida subsp. salmonicida cultures were surgically implanted into the peritoneum of Atlantic salmon. After 22 h the implants were recovered from the fish, and the bacteria were transferred to RNAprotect bacterial reagent (Qiagen) for RT-PCR analysis.
Challenge.
Both intraperitoneal injection and immersion bacterial challenges of Atlantic salmon were conducted as described elsewhere (11). The parental, tapA::KO, and flp::Km strains were tested by immersion challenge in Saint John River Atlantic salmon. The flp::Km/tapA::KO strain was tested by immersion challenge in Sackville River Atlantic salmon. All three pilin mutant strains were tested by intraperitoneal injection into Sackville River Atlantic salmon.
For immersion challenges, two tanks per group with 40 fish per tank were used; the weight of the fish was ca. 60 g each. For intraperitoneal challenge, two tanks per group with ca. 25 fish per group were used; the weight of the fish was ca. 200 g each.
Morbid animals were euthanized with an overdose of tricaine methanosulfonate (Syndel Laboratories). Posterior kidney samples were placed onto TSA supplemented with 20 μg/ml chloramphenicol, as recommended for detection of A. salmonicida subsp. salmonicida in clinical infections (38). Bacteria cultivated from the posterior kidney were subsequently cultured on TSA supplemented with kanamycin or ampicillin.
For assessment of the competitive index (CI), Sackville River stock Atlantic salmon were exposed by immersion to 106 CFU/ml of A449 and one of the three pilus mutant strains (tapA::KO, flpA::Km, or tapA::KO/flpA::Km). The posterior kidney was sterilely dissected from moribund animals after the fish were euthanized. After the posterior kidney was weighed, it was homogenized in 1 ml of sterile phosphate-buffered saline, and the bacterial titer was determined by direct colony counting on TSA supplemented with chloramphenicol, ampicillin, or kanamycin as described above. Counts were normalized to obtain values expressed in CFU per milligram of kidney. The CI was calculated as follows: CFU of mutant mg−1/CFU of parent mg−1.
Statistical differences in cumulative morbidity between groups were assessed by the G test (a modified χ2 test). Mean times to death were compared by an unpaired t test using GraphPad Prism 3.0 (GraphPad Software, San Diego, CA).
Nucleotide sequence accession numbers.
The nucleotide sequences for the loci described in this study have been deposited in the GenBank database under the following accession numbers: tapABCD, DQ396478; tppABCD tapY1Y2 tppE, DQ396479; tppF, DQ396480; tapMNOPQ, DQ396481; tapTU, DQ396482; tapW, DQ396483; tapF, DQ396484; tapV, DQ396485; MSHA gene, DQ396486; and flp, DQ396487. The accession number for the complete A. salmonicida subsp. salmonicida A449 genome is NC_009348.
RESULTS
Analysis of the genome sequence of A. salmonicida subsp. salmonicida A449 revealed the presence of genes encoding three type IV pilus complexes: Tap, Flp, and MSHA (Fig. 2). The tapABCD operon encoding the Tap pilin and three accessory genes were described previously for A. salmonicida subsp. salmonicida and some other Aeromonas species (2, 24), while the Flp pilus, which exhibits homology to pili of Actinobacillus actinomycetemcomitans and Caulobacter crescentus, has not been described previously for Aeromonas species. The third system is very similar to the Vibrio cholerae MSHA pilus. This system has been previously described for other aeromonad species (25) but not for A. salmonicida subsp. salmonicida.
Pilin genes and operons.
The tapABCD locus of A. salmonicida subsp. salmonicida and other Aeromonas species (3, 32) is very similar to the pilABCD locus of Pseudomonas aeruginosa. In P. aeruginosa, an additional 18 genes that are scattered throughout the chromosome in several loci are required for pilus biogenesis (27). Homologues of these additional genes have been identified in the A. salmonicida subsp. salmonicida genome (Fig. 2). Table 3 shows the predicted gene products of the A. salmonicida subsp. salmonicida Tap system based on their similarity to the P. aeruginosa Pil system. When possible, the P. aeruginosa nomenclature is used (e.g., tapA is the homologue of pilA); the only exception is the pseudopilin genes, which are designated tpp (tap pseudopilin).
TABLE 3.
Predicted gene products of the A. salmonicida subsp. salmonicida Tap pilus system based on similarity to the P. aeruginosa Pil system
| A. salmonicida subsp. salmonicida gene product | Homologue in P. aeruginosa | % Amino acid identitya | Predicted function, location, and/or Gsp homologueb |
|---|---|---|---|
| TapA | PilA | 32 | Pilin subunit |
| TapB | PilB | 56 | Cytoplasm, traffic ATPase, GspE |
| TapC | PilC | 50 | Inner membrane, GspF |
| TapD | PilD | 58 | Inner membrane, prepilin peptidase |
| TapM | PilM | 39 | Inner membrane, MreB homologue |
| TapN | PilN | 34 | Inner membrane, GspL |
| TapO | PilO | 40 | Inner membrane |
| TapP | PilP | 32 | Outer membrane, pilotin, lipoprotein |
| TapQ | PilQ | 38 | Outer membrane, secretin |
| TapT | PilT | 75 | Cytoplasm, twitching ATPase |
| TapU | PilU | 56 | Cytoplasm, twitching ATPase |
| TapW | PilU | 52 | Cytoplasm, twitching ATPase |
| TppA | PilE | 37 | Pseudopilin |
| TppB | PilV | 12 | Pseudopilin |
| TppC | PilW | 9 | Pseudopilin |
| TppD | PilX | 12 | Pseudopilin |
| TppE | FimT | 19 | Pseudopilin |
| TppF | FimU | 12 | Pseudopilin |
| TapY1 | PilY1 | 23 | Outer membrane |
| TapY2 | PilY2 | 18 | Periplasm |
| TapV | FimV | 26 | Inner membrane, regulator |
| TapF | PilF | 33 | Outer membrane, tetratricopeptide repeat lipoprotein |
The percent amino acid identity of frameshifted gene products was calculated by reading through the frameshifts.
The cellular location was predicted using the PSORTb program (http://www.psort.org/psortb/) and the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/).
An A. salmonicida subsp. salmonicida locus with similarity to flp gene clusters of A. actinomycetemcomitans and Haemophilus ducreyi was also identified (Fig. 2 and Table 4). This gene cluster encodes the biosynthetic proteins for a type IV pilus, and the genes are designated flp, rcp, cpa, or tad depending on the system. For clarity, the A. salmonicida subsp. salmonicida genes are designated flpA through flpL. Some of these pilus systems have more than one major pilin subunit. A. actinomycetemcomitans has two subunits, encoded by flp-1 and flp-2, and H. ducreyi has three subunits, encoded by flp-1, flp-2, and flp-3. Even though A. salmonicida subsp. salmonicida has only one major pilin gene, in keeping with the nomenclature used for these organisms, a numerical suffix is used for the pilin subunit, Flp1.
TABLE 4.
Predicted gene products of the A. salmonicida subsp. salmonicida Flp pilus system based on similarity to the A. actinomycetemcomitans Flp system
| A. salmonicida subsp. salmonicida gene product | Homologue in A. actinomycetemcomitans | % Amino acid identity | Predicted function, location, and/or Gsp homologue |
|---|---|---|---|
| Flp1 | Flp1 | 27 | Pilin subunit |
| FlpA | TadV | 27 | Inner membrane, prepilin peptidase |
| FlpB | RcpC | 19 | Periplasm, unknown function |
| FlpC | RcpA | 27 | Outer membrane, secretin |
| FlpD | RcpB | 18 | Periplasm, unknown function |
| FlpE | TadZ | 17 | Cytoplasm, ATPase, GspA |
| FlpF | TadA | 45 | Cytoplasm, traffic ATPase, GspE |
| FlpG | TadB | 22 | Inner membrane, GspF |
| FlpH | TadC | 22 | Inner membrane, GspF |
| FlpI | TadD | 22 | Outer membrane, tetratricopeptide repeat lipoprotein |
| FlpJ | TadE | 8 | Pseudopilin |
| FlpK | TadF | 10 | Pseudopilin |
| FlpL | TadG | 7 | Pseudopilin |
A cluster of genes with similarity to the genes encoding MSHA pili of V. cholerae was also identified in A449 (Fig. 2 and Table 5). However, analysis of the A. salmonicida subsp. salmonicida A449 locus showed that 8 of 16 genes, including the gene encoding the major pilin subunit, are missing or partially deleted. The deletion creates a fused gene with the 5′ end of mshN and the 3′ end of mshD. The absence of mshA, encoding the major pilin subunit, as well as several critical genes involved in pilin assembly, led us to expect that the MSHA-like locus is not expressed in A. salmonicida subsp. salmonicida A449.
TABLE 5.
Predicted gene products of the A. salmonicida subsp. salmonicida MSHA-type cluster based on similarity to the V. cholerae MSHA system
| A. salmonicida subsp. salmonicida gene product | Homologue in V. cholerae | % Amino acid identity | Predicted function, location, and/or Gsp homologue |
|---|---|---|---|
| MshI | MshI | 25 | Inner membrane, GspL |
| MshJ | MshJ | 35 | Inner membrane, GspM |
| MshK | MshK | 33 | Periplasm, unknown function |
| MshL | MshL | 49 | Outer membrane, secretin |
| MshM | MshM | 50 | Cytoplasm, ATPase, GspA |
| MshN/D | MshN N terminus and MshD C terminus | Fusion created by deletion | |
| Partially deleted | MshN | Outer membrane, tetratricopeptide repeat lipoprotein | |
| Absent | MshE | Cytoplasm, traffic ATPase, GspE | |
| Absent | MshG | Inner membrane, GspF | |
| Absent | MshF | Unknown function | |
| Absent | MshB | Pseudopilin | |
| Absent | MshA | Pilin subunit | |
| Absent | MshC | Pseudopilin | |
| Partially deleted | MshD | Pseudopilin | |
| MshO | MshO | 31 | Pseudopilin |
| MshP | MshP | 24 | Pseudopilin |
| MshQ | MshQ | 39 | Outer membrane, similar to PilY1 |
Six of the identified pilus genes have authentic frameshift errors compared with their closest orthologues that may affect their translation. Two genes of the Tap system show frameshifts; tapC has a 1-bp deletion that is predicted to cause premature termination of the protein, and tapF has a 7-bp duplication at the 5′ end, leading to a very short predicted protein. There are three genes in the Flp locus with frameshift errors; flpB and flpG have 1-bp changes that introduce stop codons, while flpI has a 4-bp duplication that moves the gene out of frame. Compared with the V. cholerae gene, mshI is either frameshifted or expressed as two separate genes. The disrupted genes are indicated in Fig. 2.
Regulation of pilus gene expression.
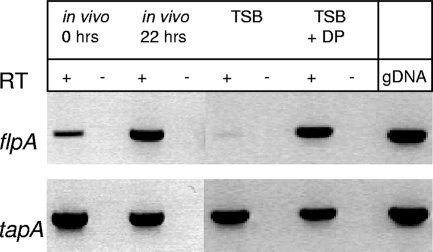
To investigate expression of the flp and tapABCD operons, which encode the pilin subunits, RT-PCR experiments were carried out with primers specific for the flpA and tapA genes and RNA from bacteria grown under a number of different conditions (Fig. 3). flpA was poorly transcribed in bacteria grown in TSB, but transcription was greatly increased when the bacteria were grown in vivo in implants in Atlantic salmon. mRNA induction was seen as early as 6 h after implantation (data not shown), and mRNA was highly expressed by 22 h (Fig. 3). Since most animals, including fish, maintain a low concentration of free iron in the serum and many pathogenic bacteria respond to low iron concentrations with increased expression of virulence factors, we asked whether low iron was the signal to which flp transcription responded. This was indeed the case as the expression of flp mRNA increased dramatically when the iron-specific chelator 2,2′-dipyridyl was added to TSB (Fig. 3).
FIG. 3.
RT-PCR of A. salmonicida subsp. salmonicida flpA and tapA genes in cultures grown under various conditions, including in vivo (bacteria from implants left in fish peritoneal cavities for 0 or 22 h), in TSB, and in TSB containing 120 μM 2,2′-dipyridyl (TSB + DP). Plus and minus signs indicate the presence and absence of the reverse transcription reaction. gDNA, genomic DNA.
In contrast to the regulated Flp pilus, the tapA gene was constitutively expressed under all conditions tested, including in vivo and in media with low iron concentrations (Fig. 3). Increased temperature, variations in the salt concentration, or the growth phase had no effect on the transcription of either pilus operon (data not shown).
Challenge experiments.
To investigate the role of pili in A. salmonicida subsp. salmonicida infection, isogenic mutants of strain A449 were created. Genes were inactivated using one of two methods. Plasmid pKO was introduced into the tapA gene in a single-crossover event to make an ampicillin-resistant, Tap-deficient strain. Alternatively, a double-crossover technique was used to introduce a kanamycin resistance cassette into flpA. Both methods produced stable mutants. The flpA::Km/tapA::KO double mutant was constructed as well. Reversion (the appearance of bacteria with inappropriate antibiotic sensitivities, indicating a loss of either pKO or the kanamycin cassette) was not seen. All pilus mutant strains had the same growth characteristics as the wild type in TSB at 17°C.
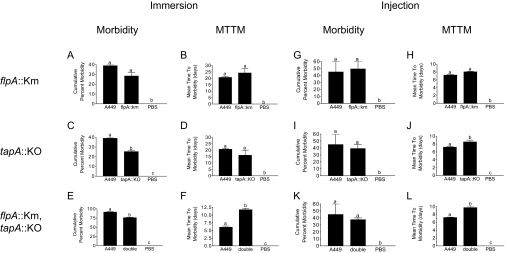
Two immersion challenge experiments were conducted. In the first experiment the tapA::KO and flpA::Km strains were used to challenge Saint John River Atlantic salmon (Fig. 4A to D). In the second experiment the tapA::KO/flpA::Km double mutant was used to challenge Sackville River Atlantic salmon (Fig. 4E to F). All three pilus mutant strains caused clinical disease that was visually indistinguishable from the disease caused by the parental strain. In the single-mutant challenges, the parental strain caused 40% cumulative morbidity with a mean time to morbidity (MTTM) of 20.8 days (Fig. 4A to D). In the group exposed to the flpA::Km strain, neither the cumulative morbidity (29%) nor the MTTM (24.3 days) was significantly different from the value obtained for the parent (Fig. 4A and B). However, the cumulative morbidity was significantly lower in the group exposed to the tapA::KO strain (25%) (Fig. 4C). The MTTM was also shorter in this group (16.2 days), but it was not significantly shorter (Fig. 4D). The virulence of the tapA::KO/flpA::Km double mutant, tested in a separate challenge, was similar that of the tapA::KO strain. In this challenge both the cumulative morbidity (76%) and the MTTM (11.8 days) were significantly different for the group exposed to the double mutant than for the group exposed to the parental strain (91% and 6.1 days) (Fig. 4E and F).
FIG. 4.
Effects of single and double type IV pilus mutants used to challenge Atlantic salmon by immersion or intraperitoneal injection. The data are the cumulative morbidity and MTTM obtained using duplicate tanks. Different letters above the bars indicate statistical significance; cumulative morbidity values were compared by using the G test, and MTTM values were compared by using the t test. A P value of <0.05 was considered significant for both tests. PBS, phosphate-buffered saline.
The virulence of all three mutant strains and the virulence of the parental strain were also tested by intraperitoneal injection into Sackville River Atlantic salmon (Fig. 4G to L). With this route of infection there were no significant differences in cumulative morbidity between the parental strain (45%) and the flpA::Km, tapA::KO, or tapA::KO/flpA::Km strain (50, 39, and 37%, respectively). There was also no difference in MTTM between the parental strain (7.3 days) and the flpA::Km strain (8.1 days). However, the MTTM was significantly longer for the tapA::KO strain (8.7 days) and the tapA::KO/flpA::Km strain (9.8 days).
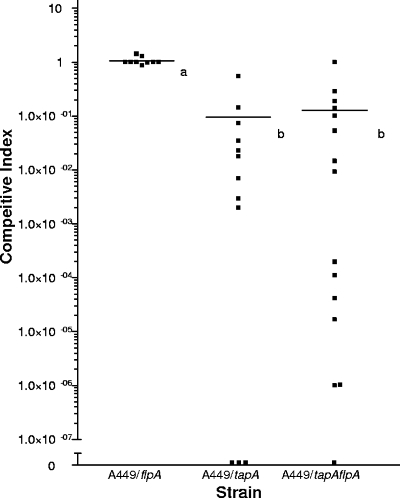
In order to more accurately assess any attenuation in virulence caused by a lack of pili, CIs of the mutants and the wild type were determined. In a separate challenge experiment, groups of fish were exposed by immersion to a mixture of equal amounts of the parental strain and one of the mutant strains. When the animals became moribund, they were euthanized, their posterior (renal) kidneys were sterilely dissected as described previously (38), and the bacteria were counted. Unlike other organs, the posterior kidney can be accessed sterilely, making it the preferred site for microbiological testing. The results are shown in Fig. 5. There were nine morbid animals following challenge with A449 and the flpA::Km strain, and the CI was 0.96 ± 0.38 (mean ± standard deviation), indicating that the flpA-deficient and wild-type strains were equally able to invade and survive within the posterior kidney. In marked contrast, however, the two tapA mutants had CIs of much less than 1, showing that they had decreased virulence. In the group exposed to A449 and the tapA::KO strain there were 12 moribund fish, and the CI was 0.07 ± 0.16 (mean ± standard deviation). In the group exposed to A449 and the double mutant there were 15 moribund fish, and the CI was 0.12 ± 0.26 (mean ± standard deviation). These results indicate that a lack of tapA leads to a reduced ability to invade and survive within the host.
FIG. 5.
CIs of single and double mutants. Atlantic salmon were challenged by immersion with equal amounts of the wild type strain and one of the mutant bacterial strains. Bacteria were isolated from the posterior kidney of moribund animals and counted. The symbols indicate the values for individual moribund fish. Different letters indicate statistical significance, as determined by the t test. A P value of <0.05 was considered significant.
Visualization of pili on the surface of A. salmonicida subsp. salmonicida.
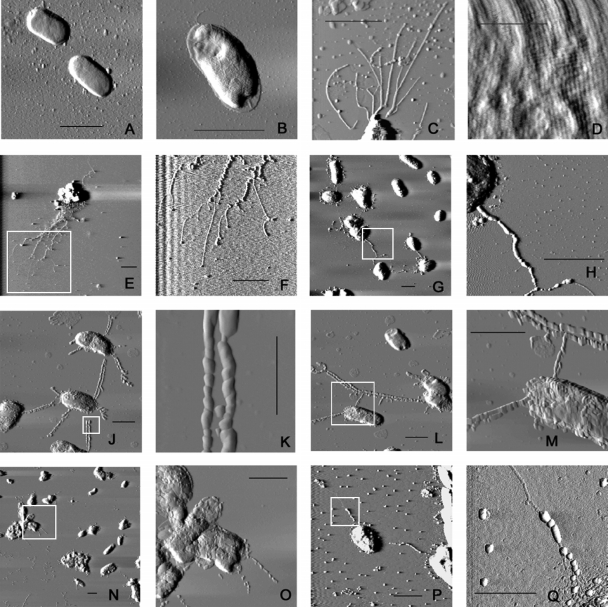
Despite several attempts by us and others, pili have never been visualized on the surface of A. salmonicida subsp. salmonicida using electron microscopy or immunofluorescence. We chose to use AFM to detect pili on A. salmonicida subsp. salmonicida (Fig. 6) because of our experience in detecting bacterial pili with this technique (40). The ability of AFM to visualize bacterial pili was confirmed by imaging P. aeruginosa strain PAK, which is known to express type IV pili. High-resolution AFM clearly demonstrated the presence of the pili on P. aeruginosa and also provided considerable structural detail, such as pilus width, length, and bundling characteristics (Fig. 6C). The pili in the images were between 6 and 8 nm wide and several thousand nanometers long, consistent with the size of P. aeruginosa pili measured by other techniques (5 to 8 nm wide and several thousand nanometers long) (9). Scans of the surface of A. salmonicida subsp. salmonicida revealed the closely packed tetragonal array of the surface layer (Fig. 6D). No pili or other extracellular structures were visible extending beyond this layer. In order to observe pili that might be hidden below the S-layer, a lipopolysaccharide O-side-chain-deficient (rough) mutant of A. salmonicida subsp. salmonicida, strain 02-06, was imaged. This strain was unable to secure the S-layer protein, VapA, to the cell surface (43), and consequently its surface was devoid of both the VapA protein and lipopolysaccharide (Fig. 6A). Pili were undetectable on this strain.
FIG. 6.
AFM images of A. salmonicida subsp. salmonicida 02-06 or E. coli DH10B carrying pilin-overexpressing plasmids. (A) 02-06(pMMB67EH.K); (B) E. coli DH10B(pBACe3.6); (C) P. aeruginosa PAK; (D) wild-type strain A449, showing tetragonal S-layer array; (E to H) 02-06(ptapA.K); (J to M) 02-06(pflp1.K); (N to Q) E. coli DH10B(pAs01a04). In panels E and F, G and H, J and K, L and M, N and O, and P and Q, the second image is an enlargement of the region indicated by the box in the first image. (A, C, E to J, and L to P) Scale bars = 2 μm; (B, K, and Q) scale bars = 1 μm; (D) scale bar = 0.2 μm.
In order to visualize pili that might be too short to be seen normally, the flp-1 and tapA pilin genes were overexpressed in A. salmonicida subsp. salmonicida by placing them under the control of the strong, inducible tac promoter and conjugating the overproducing plasmids into A. salmonicida subsp. salmonicida 02-06. Expression of the pilin subunits was induced by IPTG (50 μM). In order to induce expression of the other flp pilus assembly genes on the chromosome, the cultures were grown in low-iron medium using 2,2′-dipyridyl (120 μM). Under these conditions, AFM showed the appearance of long piluslike structures extending from the surface of the bacteria. Figures 6E to H show A. salmonicida subsp. salmonicida 02-06 overexpressing the tapA pilin gene. The pili seem to have a polar origin and have a ropelike appearance with many fibers wound together, although individual fibers were visible at the ends of the structures. The width of these bundles was between 15 and 35 nm, while the width of individual fibers varied from 3.5 to 4.6 nm, which is thinner than P. aeruginosa pili. Figures 6J to M show A. salmonicida subsp. salmonicida 02-06 overexpressing the flp-1 pilin gene. The pili appear to be peritrichous, and most often a pilus from one bacterium was in close, antiparallel contact with a pilus from another bacterium. The width of these pili was approximately 15 nm. We also imaged E. coli DH10B carrying the BAC clone pAs01a04 (Fig. 6N to Q), which contains the entire flp locus in its 71-kb insert (Fig. 2). This strain also produced pili that were similar but not identical to the Flp pili. These fibers were peritrichous and approximately 4.5 nm wide but did not bundle together and did not seem to interact with pili from adjacent bacteria. In all cases, 60 to 70% of the bacterial cells imaged had pili. Figure 6B shows E. coli DH10B carrying the empty BAC vector pBACe3.6, and no pili or other surface structures are visible on this bacterial strain. All of the pili, as well as the surface of the bacteria and, in some places, the mica surface were covered with an unidentified thick amorphous substance that may have led to an overestimate of the fiber size.
DISCUSSION
The A. salmonicida subsp. salmonicida genome contains genes for three type IV pilus systems: Tap, Flp, and an incomplete MSHA system. Since pili are well-established virulence factors for many bacteria (22, 36), our experiments were designed to characterize the expression and function of the two presumably functional pilus systems, Tap and Flp. A previous study (26) examined the role of the Tap pilus in A. salmonicida subsp. salmonicida virulence and concluded, on the basis of an increase in the LD50 for rainbow trout, that the Tap pilus is a virulence factor. In this study, the contributions to virulence of Tap and Flp were assessed by both injection and immersion exposure of Atlantic salmon to pilus-deficient mutant strains.
Pilin genes and expression.
The sequence, arrangement, and organization of the genes of the Tap, Flp, and MSHA pilus systems of A. salmonicida subsp. salmonicida are similar to those of the homologous systems of P. aeruginosa, A. actinomycetemcomitans, and V. cholerae, respectively. In addition to the previously characterized tapABCD cluster (26), 18 genes involved in the expression and assembly of the Tap pilus have been identified. The Tap genes are scattered widely throughout the genome, while the flp genes and the MSHA genes are each located in a single operon (Fig. 2). An eight-gene internal deletion in the MSHA operon encoding the pilin subunit, mshA, indicated that this system was inactivated and likely not expressed.
While the A. salmonicida subsp. salmonicida Tap and P. aeruginosa Pil systems show overall high similarity, there are differences, primarily in the regulatory genes that control pilus function and biogenesis. Most notably, A. salmonicida subsp. salmonicida contains three homologues of the P. aeruginosa twitching motility ATPase gene pilT (8): tapT, tapU, and tapW. While tapT and tapU are adjacent to each other, tapW is situated on the opposite side of the chromosome. A. salmonicida subsp. salmonicida does not appear to have orthologues of the P. aeruginosa genes pilZ, fimX, and fimL, whose products regulate twitching motility, nor are there homologues of pilGHIJKL and chpABCDE, which control P. aeruginosa twitching motility chemotaxis (44), suggesting that the Tap system may not provide twitching motility to A. salmonicida subsp. salmonicida. The Tap prepilin methylase/peptidase, TapD, is likely also used in assembly of the two type II secretion systems, as well as the defunct MSHA pilus system (31).
Flp pili are relatively new members of the type IVb class, the first instance having been described in 2000 (23). It is now clear that these pili are widespread among bacteria (35), where they mediate phenotypes such as biofilm formation, autoaggregation, and rough colony morphology. The Flp pilins are very small (50 to 80 amino acids) and have larger leader peptides than other type IV pilins. After processing, A. salmonicida subsp. salmonicida Flp1 is predicted to be only 49 residues long and to contain only the hydrophobic α-helix that is conserved among all members of type IV pilin group. The putative Flp prepilin leader peptidase is also different from its paralogue, TapD, in that FlpA is missing the N-terminal one-third of the protein that carries the methylation active site. The remaining portion of the molecule is an aspartyl protease and is similar to the C terminus of other type IVa prepilin peptidases (12, 39).
Gene expression analysis of the pilus subunit operons by RT-PCR indicated that the Tap pilus was constitutively expressed. On the other hand, Flp expression was iron regulated and was expressed under in vivo or iron-depleted in vitro growth conditions. In contrast to what one might expect, the apparently unregulated system, Tap, makes a greater contribution to virulence than Flp.
Pilin visualization.
Since AFM easily detected the pili of P. aeruginosa but not those of A. salmonicida subsp. salmonicida, we reasoned that the A. salmonicida subsp. salmonicida pili are potentially too short for visualization. Consequently, strains overexpressing the TapA and Flp1 pilins were constructed. As the expression of accessory genes was not increased in these strains, only pilus length, not pilus number, was expected to increase. TapA-overexpressing strains of A. salmonicida subsp. salmonicida produced pili at their poles, the same location as the P. aeruginosa pili. The A. salmonicida subsp. salmonicida Tap pili appeared to be much more flexible than the pili of P. aeruginosa and bundled together. The overexpressed Flp pili, on the other hand, were peritrichous and appeared to form binary, antiparallel connections with pili from other cells. This is the first reported visualization of A. salmonicida subsp. salmonicida pili. The fact that pili were visible only under overexpressed conditions supported the hypothesis that A. salmonicida subsp. salmonicida pili are too short to be seen normally and also showed that the accessory genes responsible for pilus assembly are functional.
When the entire flp locus was expressed in a laboratory strain of E. coli, DH10B, pilus-like structures were again visible (Fig. 6N to Q). The pili were similar but not identical to the Flp pili produced by A. salmonicida subsp. salmonicida. The E. coli Flp pili were individual pili, while the A. salmonicida subsp. salmonicida pili were usually paired with a pilus from another bacterium. This morphological difference suggested that there may be Aeromonas-specific factors that are required for normal Flp pilus production that are not included on BAC pAs01a04.
Frameshifts.
The genome of A. salmonicida subsp. salmonicida is littered with frameshifted genes caused by small (1- to 14-bp) insertions or deletions, as well as point mutations (M. Reith, unpublished data). There are three such lesions in the Flp pilus system and two in Tap. It is unknown whether these lesions represent inactive pseudogenes or if the genes are somehow repaired during transcription or translation. As the data presented here demonstrate that both Flp and Tap pili can be expressed and assembled, the affected genes either are not required or are repaired during expression (4, 18). Intact genes may also be able to complement frameshifted genes as there are many sets of paralogous genes in the A449 genome, including the three type IV pilus systems, as well as two homologous type II secretion systems (one for general protein secretion and one dedicated to the S-layer). For example, the frameshifted genes tapC and flpG and the missing mshG gene are paralogous. However, there are intact paralogues of these genes in the Flp system (flpH) and in the type II secretion systems (exeF and spsF). It is possible that one or more of the uninterrupted, paralogous proteins (FlpH, ExeF, and SpsF) can substitute for the proteins with frameshifts.
Alternatively, the Flp and Tap pilins might be assembled by one of the complete type II secretion systems. This has been shown to be possible with the major pseudopilin of the P. aeruginosa type II secretion system, encoded by xcpT. Overexpression of xcpT generates long pilus-like structures (15, 41); however, these structures are not absolutely dependent on the Xcp secretion apparatus. When genes for this apparatus are deleted, pseudopili can still be assembled through the second type II secretion apparatus (hxc) or the type IV pilus (pil). This may happen with the Flp and Tap pilins in A. salmonicida subsp. salmonicida, but the assembly of the Flp pilin in E. coli (pAs01a04) cannot easily be explained by this kind of complementation.
Virulence.
Immersion and intraperitoneal challenges of Atlantic salmon using strains which were deficient in tapA and/or flpA were conducted to investigate the contribution of the pili to virulence. Two TapA-deficient strains were used in live bacterial challenges of Atlantic salmon, one strain with tapA inactivated and one strain with both tapA and flpA inactivated. When these strains were used to challenge Atlantic salmon by immersion, the cumulative morbidity with both the tapA::KO and tapA::KO/flpA::Km strains was slightly, but significantly reduced. When the strains were administered by intraperitoneal injection, however, there were no significant differences in cumulative morbidity between the tapA::KO, tapA::KO/flpA::Km, and parental strains, although the MTTM of the mutant strains was longer. However, the CIs measured by using the immersion route for both Tap-inactivated strains were much less than one indicating decreased virulence.
The study of Masada et al. (26) showed that there was a statistically insignificant reduction in the cumulative mortality of a tapA-deficient strain in a rainbow trout intraperitoneal injection model. However, calculation of the LD50s for the same assay showed that there was a 2.5-fold increase for the tapA-deficient strain compared with the wild type. These results are comparable to our results for Atlantic salmon, in that simple measurements of cumulative mortality after intraperitoneal injection did not reveal significant differences between the tapA mutant and the wild type; however, a closer examination of the data (mean time to death, LD50) or other challenge assays (CI, immersion challenge) did show that the Tap pili play a role, albeit a small role, in virulence.
In contrast to the Tap system, Flp made little contribution to virulence when either route of infection was used. The mean CI for the flpA::Km strain was also about 1, indicating that the flpA::Km strain was not attenuated and was able to adhere to and invade Atlantic salmon as effectively as the parental strain. The Flp pilus may therefore play another role in the life of A. salmonicida subsp. salmonicida, such as adherence to other vertebrate, invertebrate, or inanimate surfaces.
Summary.
A. salmonicida subsp. salmonicida has two functional type IV pilus systems, Tap and Flp, and one nonfunctional system, MSHA. The two functional systems are not redundant as they have different expression profiles, physical appearances, cellular locations, and abilities to cause clinical disease in Atlantic salmon. The Tap pili clearly contributed to virulence; however, tapA-deficient mutants retained much of their pathogenicity. On the other hand, the Flp pili did not contribute to virulence against Atlantic salmon. Type IV pili are well-known virulence factors for many bacterial species, including other aeromonads, but this work shows that they are not major virulence factors for A. salmonicida subsp. salmonicida infection of Atlantic salmon.
Acknowledgments
We thank William Kay for the kind gift of A. salmonicida subsp. salmonicida strain A449. We acknowledge the contributions of Hannah McKenzie (NRC-IMB and Dalhousie), Jason Williams and Ron Melanson (NRC-IMB), John Batt, and the Dalhousie University Aquatron team (Jerry Whynot, Steve Fowler, and Don Lawrence).
J.M.B., A.D., and L.K. were supported by the National Research Council of Canada's Genomics and Health Initiative.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 22 January 2008.
NRC publication 2007-42745.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Barnett, T. C., and S. M. Kirov. 1999. The type IV Aeromonas pilus (Tap) gene cluster is widely conserved in Aeromonas species. Microb. Pathog. 2677-84. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, T. C., S. M. Kirov, M. S. Strom, and K. Sanderson. 1997. Aeromonas spp. possess at least two distinct type IV pilus families. Microb. Pathog. 23241-247. [DOI] [PubMed] [Google Scholar]
- 4.Bayliss, C. D., K. M. Dixon, and E. R. Moxon. 2004. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis. A meeting review. FEMS Immunol. Med. Microbiol. 4011-19. [DOI] [PubMed] [Google Scholar]
- 5.Bernoth, E. M. 1997. Furunculosis: the history of the disease and of disease research, p. 1-20. In E. M. Bernoth, A. E. Ellis, P. J. Midtlyng, G. Olivier, and P. Smith (ed.), Furunculosis: multidisciplinary fish disease research. Academic Press, San Diego, CA.
- 6.Burr, S. E., K. Stuber, T. Wahli, and J. Frey. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 1845966-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cipriano, R. C., and G. Bullock. 2001. Furunculosis and other diseases caused by Aeromonas salmonicida. Fish disease leaflet 66. USGS/Leetown Science Center Fish Health Branch, Kearneysville, WV.
- 8.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 673625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2363-378. [DOI] [PubMed] [Google Scholar]
- 10.Dacanay, A., S. C. Johnson, R. Bjornsdottir, R. O. Ebanks, N. W. Ross, M. Reith, R. K. Singh, J. Hiu, and L. L. Brown. 2003. Molecular characterization and quantitative analysis of superoxide dismutases in virulent and avirulent strains of Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 1854336-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dacanay, A., L. Knickle, K. S. Solanky, J. M. Boyd, J. A. Walter, L. L. Brown, S. C. Johnson, and M. Reith. 2006. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 1521847-1856. [DOI] [PubMed] [Google Scholar]
- 12.de Bentzmann, S., M. Aurouze, G. Ball, and A. Filloux. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 1884851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demers, G., G. Griffin, G. De Vroey, J. R. Haywood, J. Zurlo, and M. Bedard. 2006. Animal research. Harmonization of animal care and use guidance. Science 312700-701. [DOI] [PubMed] [Google Scholar]
- 15.Durand, E., A. Bernadac, G. Ball, A. Lazdunski, J. N. Sturgis, and A. Filloux. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 1852749-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis, A. E., A. S. Burrows, and K. J. Stapleton. 1988. Lack of relationship between virulence of Aeromonas salmonicida and the putative virulence factors: A-layer, extracellular proteases and extracellular haemolysins. J. Fish Dis. 11309-323. [Google Scholar]
- 17.Fernandez, A. I., A. F. Fernandez, M. J. Perez, T. P. Nieto, and A. E. Ellis. 1998. Siderophore production by Aeromonas salmonicida subsp. salmonicida. Lack of strain specificity. Dis. Aquat. Organ. 3387-92. [DOI] [PubMed] [Google Scholar]
- 18.Field, D., M. O. Magnasco, E. R. Moxon, D. Metzgar, M. M. Tanaka, C. Wills, and D. S. Thaler. 1999. Contingency loci, mutator alleles, and their interactions. Synergistic strategies for microbial evolution and adaptation in pathogenesis. Ann. N. Y. Acad. Sci. 870378-382. [DOI] [PubMed] [Google Scholar]
- 19.Frengen, E., D. Weichenhan, B. Zhao, K. Osoegawa, M. van Geel, and P. J. de Jong. 1999. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58250-253. [DOI] [PubMed] [Google Scholar]
- 20.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 21.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonson, A. B., S. Normark, and M. Rhen. 2005. Fimbriae, pili, flagella and bacterial virulence. Contrib. Microbiol. 1267-89. [DOI] [PubMed] [Google Scholar]
- 23.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 1826169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirov, S. M., T. C. Barnett, C. M. Pepe, M. S. Strom, and M. J. Albert. 2000. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 684040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirov, S. M., L. A. O'Donovan, and K. Sanderson. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 675447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masada, C. L., S. E. LaPatra, A. W. Morton, and M. S. Strom. 2002. An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 5113-25. [DOI] [PubMed] [Google Scholar]
- 27.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 351-13. [DOI] [PubMed] [Google Scholar]
- 29.Michel, C. 1979. Furunculosis of salmonids: vaccination attempts in rainbow trout (Salmo gairdneri) by formalin-killed germs. Ann. Rech. Vet. 1033-40. [PubMed] [Google Scholar]
- 30.Olivier, G. 1990. Virulence of Aeromonas salmonicida: lack of relationship with phenotypic characteristics. J. Aquat. Anim. Health 2119-127. [Google Scholar]
- 31.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 1493051-3072. [DOI] [PubMed] [Google Scholar]
- 32.Pepe, C. M., M. W. Eklund, and M. S. Strom. 1996. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol. Microbiol. 19857-869. [DOI] [PubMed] [Google Scholar]
- 33.Phipps, B. M., T. J. Trust, E. E. Ishiguro, and W. W. Kay. 1983. Purification and characterization of the cell surface virulent A protein from Aeromonas salmonicida. Biochemistry 222934-2939. [DOI] [PubMed] [Google Scholar]
- 34.Pizarro-Cerda, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124715-727. [DOI] [PubMed] [Google Scholar]
- 35.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34193-198. [DOI] [PubMed] [Google Scholar]
- 36.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 365-72. [DOI] [PubMed] [Google Scholar]
- 37.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 38.Thoesen, J. C. (ed.). 1994. Suggested procedures for the detection and identification of certain finfish and shellfish pathogens, version 1, 4th ed., vol. 1. American Fisheries Society, Bethesda, MD.
- 39.Tomich, M., D. H. Fine, and D. H. Figurski. 2006. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 1886899-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touhami, A., M. H. Jericho, J. M. Boyd, and T. J. Beveridge. 2006. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vignon, G., R. Kohler, E. Larquet, S. Giroux, M. C. Prevost, P. Roux, and A. P. Pugsley. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 1853416-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vipond, R., I. R. Bricknell, E. Durant, T. J. Bowden, A. E. Ellis, M. Smith, and S. MacIntyre. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 661990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Z., J. Li, E. Vinogradov, and E. Altman. 2006. Structural studies of the core region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide. Carbohydr. Res. 341109-117. [DOI] [PubMed] [Google Scholar]
- 44.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52873-893. [DOI] [PubMed] [Google Scholar]
- 45.Williams, K. P., and D. P. Bartel. 1996. Phylogenetic analysis of tmRNA secondary structure. RNA 21306-1310. [PMC free article] [PubMed] [Google Scholar]