Abstract
Aspergillus fumigatus is an important opportunistic fungal pathogen. This organism must be able to adapt to stress changes in the microenvironment during host invasion and systemic spread. The high-osmolarity-glycerol (HOG) mitogen-activated protein kinase (HOG-MAPK) signaling pathway plays an important role in regulating morphology, growth, and adaptation to stress and virulence in a number of fungal pathogens. The Sho1 adaptor protein is one important element of the two upstream branches of the HOG-MAPK pathway in Saccharomyces cerevisiae, a signal transduction cascade involved in adaptation to stress. We constructed a sho1 mutant of A. fumigatus, MA21. Both the growth and germination rates of the mutant were reduced, and the MA21 strain had an irregular hyphal morphology characterized by reduced production of phialides and conidia. This gene deletion mutant was sensitive to 2.5 mM hydrogen peroxide and 15 μM menadione, but it appeared to be minimally sensitive to diamide compared to the wild-type strain. In an immunosuppressed mouse model, the mutant was as virulent as the wild-type or complemented strains. These data support the idea that the loss of sho1, a highly conserved gene among fungi, regulates radial hyphal growth and delays germination of A. fumigatus conidia. In addition, the sho1 gene has a visible effect in the adaptation to oxidative stress in A. fumigatus similar to that in S. cerevisiae.
Aspergillus fumigatus is a saprophytic, filamentous fungus that ensures its survival and dispersion by producing large numbers of conidia. Inhalation of these conidia by severely immunosuppressed patients can lead to serious, life-threatening infections (14). Due to the increasing numbers of immunosuppressed bone marrow or organ transplant and cancer chemotherapy patients, the number of individuals developing invasive aspergillosis is increasing. In spite of advances in early diagnosis and development of new antifungal therapies, the rate of mortality due to invasive aspergillosis is still very high, approaching 80 to 95% (6, 11). A. fumigatus is found throughout the environment and is exposed highly variable conditions in terms of the availability and quality of nutrients, temperature, pH, and especially oxidative and osmotic stress. Some of the stress conditions are similar to those that an organism might encounter in vivo (9). Therefore, adaptive mechanisms that confer resistance to environmental stress may contribute to the efficient colonization and persistence of this organism in the human host. Given the role of signal transduction pathways as sensing mechanisms, it is important to understand the adaptation of fungal pathogens to the host. The well-characterized high-osmolarity-glycerol (HOG)-mitogen-activated protein kinase (MAPK) pathway is essential for regulating stress adaptation in several fungi, as it triggers adaptation through intracellular accumulation of glycerol as the adaptive osmolyte (3). The HOG-MAPK is one of a group of stress-activated protein kinases that specifically transmit environmental stress signals (1). Two different types of proteins, Sln1p (a histidine kinase) and Sho1p, have been described as sensors of the two upstream branches controlling the HOG-MAPK pathway. Sho1p, through Ste11p, activates the Pbs2p and Hog1p pathway to regulate glycerol synthesis and other adaptive responses (20). sho1 homologues have also been studied in Candida albicans and Saccharomyces cerevisiae. In S. cerevisiae, a sho1 null mutation not only is responsible for adaptation to hyperosmotic stress but also contributes to hydrogen peroxide adaptation (25). Recent studies demonstrated that Sho1p plays only a minor role in osmotic stress adaptation in C. albicans. Nevertheless, Sho1p is important for growth under oxidative stress conditions, and it mediates phosphorylation of the Cek1 MAPK in exponentially growing cells, so the sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen C. albicans (23). However, the primary sensor(s) that triggers the signaling pathway(s) in A. fumigatus is not completely understood. In this study, we described the functions of the A. fumigatus sho1 gene in oxidative stress adaptation, growth, and sporulation. We also investigated the role of sho1 in the pathogenesis of A. fumigatus in a murine model of invasive pulmonary aspergillosis.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study are listed in Table 1. A. fumigatus AF293.1 was used to replace the sho1 gene (resulting in a Δsho1 strain, designated MA21). AF293.1 is a uracil/uridine auxotroph (pyrG) mutant of A. fumigatus strain AF293 (17). AF293 was used as the wild-type strain for all in vitro and animal model experiments. All A. fumigatus cultures were grown in glucose minimal media with or without uracil/uridine as previously described (31) at 37°C unless otherwise specified. Escherichia coli DH10B (Invitrogen) was used for routine cloning and was grown in Luria-Bertani broth at 37°C. Agrobacterium tumefaciens strains were grown either in Luria-Bertani broth supplemented or not supplemented with kanamycin (50 μg/ml) or in induction medium (4) supplemented with 0.2 mM acetosyringone (IMAS). A. fumigatus transformants were selected on IMAS lacking uracil/uridine but containing 200 μg of cefotaxime per ml.
TABLE 1.
Strains and plasmids used in this study
Construction of the MA21 and MA22 complemented strains.
The deletion vector was constructed by cloning a 2.9-kb sequence, including a 1, 040-bp fragment upstream and a 733-bp fragment downstream of the coding region of the A. fumigatus sho1 gene, into pDHt/SK to produce plasmid A using primers P1 and P2 (Table 2). Then a 1.4-kb PCR product of Aspergillus nidulans pyrG was amplified from pALX223 (33) using primers P3 and P4 (Table 2), to which NcoI (5′ end) and PstI (3′ end) restriction sites were added. The pyrG gene, digested with NcoI and PstI, was ligated into plasmid A digested with NcoI and PstI to create plasmid B. Plasmid B was transformed into competent A. tumefaciens EHA105 by using the freeze-thaw method (22). The resulting strain of A. tumefaciens was designated the At sho1 strain. A sho1 mutant was constructed by A. tumefaciens-mediated transformation as described previously (27, 28, 29). To obtain a sho1 mutant, MA21, the At sho1 strain was cocultured with strain AF293.1 at 24°C for 48 h. Cell mixtures were grown on IMAS agar plates and then transferred to 37°C and incubated for 48 h. Transformants were scraped from plates and transferred onto glucose minimal medium agar plates supplemented with cefotaxime (200 μg/ml). Single colonies of transformants were consecutively transferred three times on this medium to kill A. tumefaciens. The MA21 strain was initially screened by PCR with primers designed to amplify the deleted regions of sho1 (primers P5 andP6 [Table 2]), which should have been absent in the MA21 strain. An additional PCR screen was done to amplify the junctions of sho1::pyrG to indicate replacement and homologous recombination (primers P7 and P8 [Table 2]). To ensure that a mutant phenotype was attributable to the specific deletion, the MA21 strain was reconstituted by integration of the AF293 sho1 allele to create a reconstituted strain, MA22. The MA21 strain was complemented by transformation with a 2,950-bp DNA fragment composed of the hph gene that confers resistance to hygromycin B (primers P9 and P10 [Table 2]) and the AF293 sho1 gene (a 2.9-kb fragment including a 1.0-kb region upstream and 720-bp region downstream of the sho1 coding regions) (primers P11 and P12 [Table 2]). These two fragments were transformed into the MA21 mutant protoplasts by using the method described previously (16). Transformants were selected on glucose minimal medium agar plates (supplemented with hygromycin). An analysis to confirm the constructed strains was performed by Southern analysis with SacI-digested genomic DNA according to the manufacturer's instructions (DIG High Prime DNA labeling and detection starter kit I; Roche). A 600-bp fragment of the sho1 gene was used as a probe for Southern hybridization (primers P13 and P14 [Table 2]).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Restriction enzyme |
|---|---|---|
| P1 | 5′-TATAGGGCCCAAACGGTACCTGTCTTTA-3′ | ApaI |
| P2 | 5′-CTCCCAGGGCTTGTATGGTCTATCTAGA-3′ | XbaI |
| P3 | 5′-ACTCCATGGGTCAATACCGTTACACATTTCCA-3′ | NcoI |
| P4 | 5′-TCACTGCAGCCGCAGACAATGCTCTCTATC-3 ′ | PstI |
| P5 | 5′-ATAAGCAGACAAGCCCAGGA-3′ | |
| P6 | 5′-GGGTTCATCCTCATGTTCCA-3′ | |
| P7 | 5′-AGTCCGTGCAGCCTTGTAGT-3′ | |
| P8 | 5′-AAAACCAACGTCACCGTCTC-3′ | |
| P9 | 5′-ATATGGGCCCTTCTATTTGTGTTTGATCGAGACC-3′ | ApaI |
| P10 | 5′-AGAACTCGAGCCTCTAAACAAGTGTACCTGTGC-3′ | XhoI |
| P11 | 5′-TCTGGGCCCATGGTCTATCTAGATCCCATA-3′ | ApaI |
| P12 | 5′-TCTAGATGCGCCCAAACGGTACCTGTCTTT-3′ | KpnI |
| P13 | 5′-TTTGCGCATTTACATTTCCA-3′ | |
| P14 | 5′-CGATACATGCGATCAACCAC-3′ |
Underlining indicates restriction enzyme sites.
Radial growth and germination.
Radial growth of the AF293 strain and strains MA21 and MA22 on glucose minimal medium agar was measured daily over a period of 72 h. Plates were inoculated centrally with 500 conidia (5 μl of a suspension containing 1 × 105 conidia/ml) and grown at 37°C in triplicate. The means ± standard deviations of the colony diameters for each 24-h period were determined. Data were analyzed using the repeated measures of SPSS 13.0. To measure germination, 100 μl of a conidial suspension of each strain (107 conidia/ml) was inoculated into 10 ml of YG (0.5% yeast extract, 2% glucose) liquid medium. All cultures were incubated at 37°C with shaking at 200 rpm. The germination percentage was assessed microscopically every 20 min beginning at approximately 4 h postinoculation (the swollen conidial stage) until 12 h postinoculation. For each time point, 100 conidia of each strain were counted and the germination percentage was determined. All experiments were repeated at least three times.
Microscopy procedures and image analysis.
CZA slide cultures (0.2% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, 0.001% FeSO4, 3% sucrose, 1.5% agar) for microscopic analysis were inoculated and incubated for 1 week at 37°C. Microscopy was performed with an Olympus DP71 microscope equipped with a digital camera. Digital images were analyzed using Adobe Photoshop 8.0 software.
Analysis of sensitivity to oxidative stress.
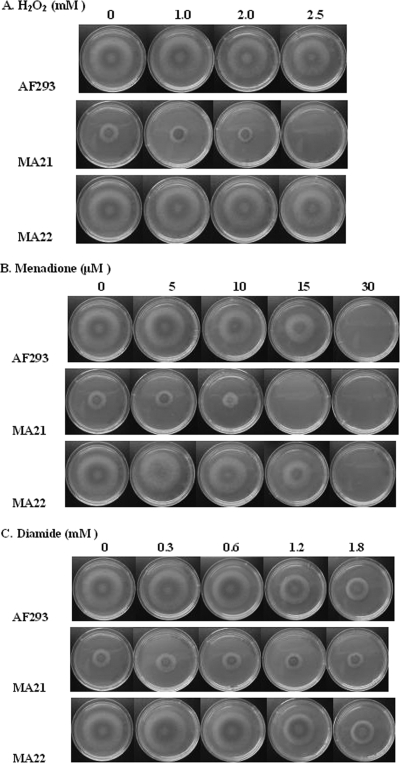
Conidia of strain AF293 and strains MA21 and MA22 were harvested with sterile 0.85% saline-0.1% Tween 20 from 5-day-old glucose minimal medium agar slants (37°C), filtered through four layers of Miracloth, and counted with a hemacytometer. Drop plates were prepared by spotting 5 μl from each conidial stock suspension containing 1 × 105 conidia/ml onto glucose minimal medium plates supplemented with 2.5 mM hydrogen peroxide (30% stock), 15 μM menadione (Sigma-Aldrich), or 1.8 mM diamide (Sigma-Aldrich). The plates were incubated at 37°C for 72 h and photographed. In addition, all strains were evaluated similarly to determine their sensitivity to 1.5 to 3.0 M NaCl or glycerol.
Murine inhalational model of invasive pulmonary aspergillosis.
All animal studies were approved by the institutional animal care and use committee. Six-week-old BALB/c male mice were immunosuppressed with cyclophosphamide (150 mg/kg of body weight; Sigma) on days −4, −1, and 3 of infection and with triamcinolone acetonide (40 mg/kg of body weight; Sigma) using a single intraperitoneal injection on the day of infection. Mice were housed under sterile conditions and provided with sterile drinking water containing tetracycline hydrochloride (500 μg/ml; Sigma). Four groups of 15 immunosuppressed mice, anesthetized with ether, were challenged with 20-μl portions of suspensions containing 2.5 × 105 conidia/ml of AF293, MA21, and MA22 and a diluent control (0.9% physiological saline). Mice were evaluated daily to determine morbidity and mortality. Survival was plotted on a Kaplan-Meier curve for each strain, and a log rank test was used for pairwise comparison of strains. Statistical significance was defined as a two-tailed P value of <0.05.
Histopathologic and CFU quantification of fungal burden.
To evaluate the histopathologic progression of disease, four groups of 13 additional mice were similarly infected with a sublethal dose (5 × 104 conidia in 20 μl) of each strain. Mice were sacrificed on days 0, 3, and 5. Lung tissues were removed for determination of the number of CFU. Lungs were harvested and stained with hematoxylin and eosin to characterize inflammation and with periodic acid-Schiff stain to document fungal invasion.
RESULTS
Deletion and complementation of sho1 in A. fumigatus.
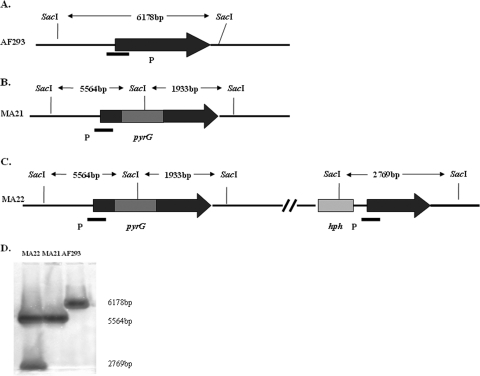
The sequence of the A. fumigatus sho1 gene was obtained from GenBank (accession no. afu5g08420), and an examination revealed a putative 1,124-bp open reading frame (Fig. 1A) encoding a putative 311-amino-acid protein homologous to the sho1 genes of S. cerevisiae (71% identity), Aspergillus clavatus (83% identity), and A. nidulans (71% identity). SMART analysis (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/) revealed that Sho1p contained four putative transmembrane domains (amino acids 36 to 58, 68 to 87, 92 to 114, and 124 to 146) near its N terminus, a linker domain between these domains, and an SH3 domain at the C terminus (amino acids 255 to 311) that is believed to interact with the Pbs2p MAPK kinase (21). As expected, sequence conservation is highest in the transmembrane region. To examine the functions of Sho1p of A. fumigatus, we constructed a sho1 deletion mutant by replacing part of the transmembrane domains of the A. fumigatus sho1 gene with the A. nidulans pyrG gene (Fig. 1B). Transformation of the uracil/uridine-auxotrophic strain A. fumigatus AF293.1 with the sho1 replacement construct yielded 20 transformants when A. tumefaciens-mediated transformation was performed. Seven transformants had both PCR amplicons when primers designed to amplify the predicted sho1 replacement locus after homologous recombination were used and no detectable PCR amplicon when primers designed to amplify the wild-type sho1 locus were used. To complement the sho1 defect in the MA21 strain, a PCR product containing the entire AF293 sho1 locus plus 1.0 kb of additional 5′ and 3′ sequence and a 3.0-kb hph gene were transformed into the recipient MA21 strain (Fig. 1C). The complemented strain was designated MA22. Transformants were selected on glucose minimal medium containing hygromycin B. The results of Southern hybridization of each strain are shown in Fig. 1D.
FIG. 1.
Deletion and complementation of the sho1 gene. (A) AF293 sho1 locus. (B) Coding region for the transmembrane domains of A. fumigatus sho1 was replaced with the A. nidulans pyrG gene by homologous recombination to produce the sho1 mutant (MA21). (C) AF293 sho1 gene, flanked by the hph gene that confers hygromycin B resistance, was integrated into the genome in the sho1 mutant to complement the deletion strain (MA22). (D) Southern analyses. Genomic DNA from each of the strains was cut with SacI and probed (as indicated by filled boxes in panels A to C) to detect the wild-type (6.2-kb), deletion (5.6-kb), and recombinant (2.8-kb) bands.
A. fumigatus sho1 contributes to radial growth and germination.
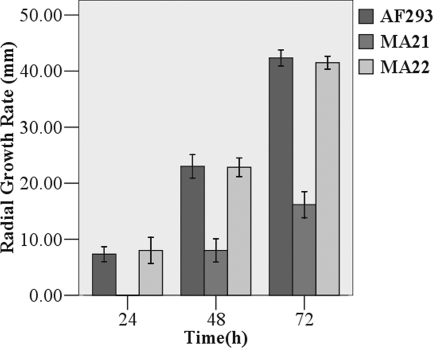
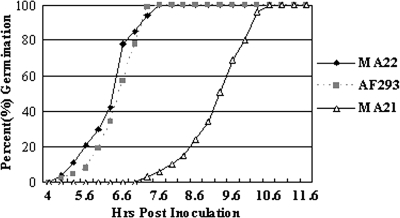
Strain MA21 had profound defects in growth and filamentation compared to strain AF293 and the MA22 strain with the gene reconstituted. Point inoculation of each strain onto glucose minimal medium plates, followed by incubation at 37°C, showed that the radial growth rate of the MA21 strain was decreased compared to those of AF293 and MA22 (Fig. 2). The radial growth rate of the mutant was approximately one-half that of the AF293 strain. This phenotype of strain MA21 was apparent regardless of the initial inoculum concentration and incubation time. In YG liquid medium, germination of MA21 was delayed and was characterized by an approximately 2.3-h lag in initiation. After the initial lag, the rates of germination for all strains appeared to be similar. Both AF293 and MA22 began to produce germ tubes shortly after 5 h, and the percentage of germinating conidia increased rapidly over the next 2 h to 100% by 7.5 h, while germination of MA21 began after 7.3 h and the maximum level was reached over the next 3.5 h (Fig. 3). The smaller-colony phenotype thus could have been due to slower or delayed germination, to a decrease in the rate of apical extension, or to a combination of factors.
FIG. 2.
Radial growth rates. Conidia from each strain were inoculated onto glucose minimal medium agar and incubated for 24, 48, and 72 h at 37°C. The sho1 mutant grew less than AF293 and MA22 (statistical data were analyzed using repeated measures of SPSS 13.0).
FIG. 3.
Germination of AF293, MA21, and MA22. A total of 105 conidia of each strain were inoculated into YG medium at 37°C, and the percent germination was determined at the indicated times.
A. fumigatus sho1 regulates hyphal development.
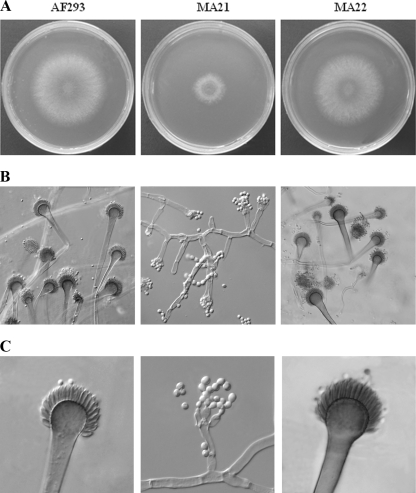
Colonies of all strains are shown in Fig. 4A. Using slide culture, we observed that MA21 had fewer phialides, and vesicle formation was attenuated (Fig. 4B). The conidial heads appeared to be on shorter stalks, while hyphae of AF293 and the MA22 strain formed long, regularly branched, and dispersed filaments. With identical concentrations of spores and with identical incubation times, MA21 exhibited a severely restricted growth pattern with appreciably shorter, hyperbranching filaments and limited aerial growth (Fig. 4B and 4C). These phenotypes are different from those of the wild type and the strains with the gene reconstituted.
FIG. 4.
Morphology of MA21 compared with that of AF293 and MA22. (A) A. fumigatus conidia (5 × 102 conidia) were incubated on glucose minimal medium agar at 37°C for 72 h. Reduced growth was observed with MA21. (B) CZA slide cultures incubated at 37°C were examined microscopically at 72 h postinoculation after staining with lactophenol cotton blue. Shorter, hyperbranching filaments and small conidial heads of MA21 were observed. (C) Vesicle formation by MA21 is attenuated, and phialides appear to be inflated, sparse, and scattered compared those of AF293 and MA22.
sho1 gene deletion mutant is sensitive to oxidative stress.
Conidia were prepared from 3-day-old cultures of all strains and suspended in Tween 20-NaCl saline. The concentration of conidia was adjusted to 105 conidia/ml, and 5 μl of conidia from each strain was spotted on media containing 0.5 to 2.5 mM hydrogen peroxide, 15 to 30 μM menadione, and 0.3 to 1.8 mM diamide. All cultures were incubated at 37°C. Figure 5A shows that strain MA21 was sensitive to 2.5 mM hydrogen peroxide. Figure 5B shows that MA21 was also sensitive to 15 μM menadione compared to the other strains. However, strain MA21 was not inhibited by (or was slightly sensitive to) diamide compared to growth of the strain in the absence of diamide, but it did grow less with and without diamide than the other strains (Fig. 5C). These results indicate that Sho1p protects cells from oxidative damage due to hydrogen peroxide and menadione but not from oxidative damage due to diamide.
FIG. 5.
Sensitivity of AF293, MA21, and MA22 to various stress conditions. (A) Hydrogen peroxide. (B) Menadione. (C) Diamide. A total of 1 × 105 conidia (5 μl) were inoculated onto all media. Cultures were incubated for 72 h at 37°C.
sho1 is not a virulence factor in a murine model of invasive pulmonary aspergillosis.
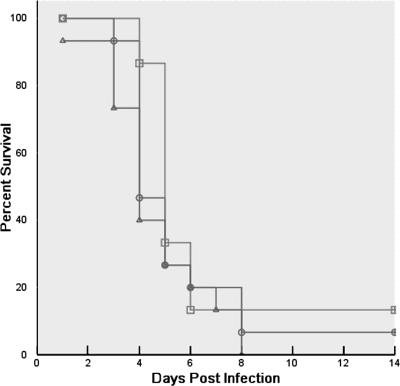
In order to determine whether sho1 was required for virulence, a murine inhalation model of invasive pulmonary aspergillosis was utilized to mimic human disease. Survival curves for mice infected with each strain over a 14-day period are shown in Fig. 6. The percentages of mice infected with AF293, MA21, and MA22 that survived were similar. Infection with the AF293 strain resulted in 90% mortality by 14 days after infection, while infection with the MA21 mutant led to 100% mortality (P > 0.05). Animals infected with each strain displayed progressive and severe signs of invasive disease that have been well described previously, including ruffled fur, hunched posture, and an increased respiratory rate (26). No significant differences in symptoms among strains were observed. Additionally, on days 0, 3, and 5 postinfection, lung sections (data not shown) and the numbers of CFU of the strains obtained from lung homogenates (Table 3) were also similar. We concluded that sho1 is not a virulence factor in this murine model of pulmonary invasive aspergillosis.
FIG. 6.
Experimental invasive pulmonary aspergillosis. Kaplan-Meier analysis showed that survival of infected mice was not significantly different for the three treatment groups (AF293 [▴], MA21 [□], and MA22 [○]). BALB/c mice (in cohorts containing 15 mice each) were immunosuppressed by intraperitoneal injection of cyclophosphamide (150 mg/kg of body weight) on days −4 and −1 prior to infection and on day 3 postinfection and of a single dose of triamcinolone acetonide (40 mg/kg) just prior to infection (day 0).
TABLE 3.
Lung tissue burdens of AF293, MA21, and MA22 in infected micea
| Strain | Log10 CFU/g lung tissue (mean ± SE)
|
||
|---|---|---|---|
| Day 0 | Day 3 postinfection | Day 5 postinfection | |
| AF293 | 5.06 ± 0.13 | 5.22 ± 0.15 | 4.88 ± 0.16 |
| MA21 | 5.47 ± 0.19 | 5.08 ± 0.08 | 4.63 ± 0.29 |
| MA22 | 5.57 ± 0.20 | 4.83 ± 0.23 | 4.73 ± 0.32 |
The fungal burden of lung tissues from infected mice was determined by plating lung tissue homogenates onto Sabouraud dextrose agar plates (35°C). No significant difference in the number of CFU was observed for each group (P > 0.05).
DISCUSSION
The aim of this study was to characterize the function of the Sho1 adaptor protein in A. fumigatus. We obtained evidence that this protein (i) plays an essential role in morphogenesis and radial growth, (ii) has an oxidative stress adaptation role, and (iii) is not a virulence factor. Our initial hypothesis was that sho1 is a stress sensor since deletion of SHO1 in S. cerevisiae and C. albicans demonstrated its functions in osmotic and/or oxidative stress via the Hog1p MAPK pathway. However, the response to oxidative stress is limited to specific oxidants because the sho1 null mutant of S. cerevisiae was only slightly sensitive to hydrogen peroxide but was not sensitive to other oxidants (25). Furthermore, the sho1 mutant was also found to be sensitive to hydrogen peroxide and menadione. In addition, Sho1p plays an essential role in control of the Cek1 MAPK but only a minor role in the activation of Hog1 in C. albicans (23). In this study, we showed that the sho1 gene participates in the morphogenesis of A. fumigatus. In C. albicans, deletion of SHO1 also abolished pseudohyphal growth under nitrogen starvation conditions on solid media (23). The sho1 mutant of A. fumigatus displays significant physiological defects that critically affect the growth of the fungus on minimal medium. The sho1 deletion mutant was phenotypically different from AF293 in both macroscopic and microscopic features, as the MA21 strain had extremely stunted growth, suggesting that sho1 plays a regulatory role in hyphal growth and/or at least in conidial germination. These changes may demonstrate that Sho1p interacts with Ste11p, which acts in the filamentous growth pathway. Thus, the sho1 mutant may affect signal transduction via the Ste11p protein in A. fumigatus.
In a recent study, Sho1p was shown to be required in an oxidative stress response in C. albicans (23). Previous work demonstrated that oxidative stress plays an important role in preventing invasive pulmonary aspergillosis (19). A. fumigatus conidia and mycelium are phagocytosed and killed by alveolar macrophages and neutrophils that produce powerful reactive oxygen species, such as hydrogen peroxide, hydroxyl radical (HO−), and superoxide anions (O2−) (5). We have been interested in how A. fumigatus responds to oxidative stress conditions and whether the responses may be involved in its pathogenicity, since as a pathogen A. fumigatus must survive in macrophages during the development of invasive aspergillosis (10). Therefore, we sought to compare the susceptibility to oxidants of a mutant that lacked the sho1 gene with that of the wild type and a complemented mutant. We used three different oxidants, hydrogen peroxide, menadione, and diamide, since no one oxidant is representative of oxidative stress conditions (30, 32). Using different oxidants should have given us a significantly better chance to detect the role of this gene than using a single agent. We found that deletion of sho1 resulted in sensitivity to hydrogen peroxide and menadione, but the mutant was relatively resistant to diamide, so we concluded that sho1 may play a role in protection against some oxidants, at least in vitro. The sho1 strain appeared to be functionally consistent with the sakA (hog1) mutant of A. fumigatus, which exhibited reduced growth, which was increased when H2O2 or menadione was present, compared to the growth of the wild-type strain. However, with the other stress sensor (tscB, sln1), there was no difference in hyphal growth, morphogenesis, or sensitivity to H2O2 or menadione compared to the wild-type strain (7). Thus, we presume that the sakA (hog1) gene may receive signals for growth regulation and oxidative stress adaptation via the sho1 branch pathway in A. fumigatus. Thus, it is possible that the sho1 protein is an important adaptor protein for oxidative stress adaptation in A. fumigatus in vitro. The drastic growth defect observed with MA21 and its oxidant sensitivity led us to believe that avirulence might be a consequence of the gene deletion. Other workers have established a direct correlation between in vitro growth defects and in vivo decreased virulence of A. fumigatus in an animal model (2, 8, 18), indicating that genes involved in growth of the fungus are likely important pathogenicity determinants of invasive disease. However, the sho1 deletion mutant was virulent in a murine infection model of invasive pulmonary aspergillosis. We presumed that the lack of an avirulent phenotype may have been due to the different nutritive conditions in vitro and in vivo, so radial growth was investigated under different nutritive and nitrogen conditions, such as brain heart infusion medium (high-protein medium), complete medium, and media containing NH4Cl, NaNO3, proline, and phenylalanine. However, with each medium, growth of MA21 was still reduced compared to growth of AF293 and MA22 (data not shown). We speculated that the virulent phenotype was related to the high numbers of conidia used in the animal experiments. It might also be true that the method used to immunosuppress animals with a combination of cyclophosphamide and triamcinolone acetonide may have contributed to virulence of the mutant (12, 29), even though this method has been used in many other similar studies (2, 8, 32). An independent study showed that a combination of these two drugs caused the immunosuppression to be so severe that even the uninfected control mice exhibited increased morbidity (24). Nonetheless, the lack of a virulence phenotype, despite the role in oxidative stress and growth, is consistent with what has been reported for Yap1 and Skn7 in A. fumigatus (13, 15). These studies indicated that increased sensitivity of A. fumigatus to peroxides in vitro is not correlated with modification of fungal virulence.
Our study indicated that the sho1 gene is an important gene for in vitro growth of A. fumigatus. However, sho1 is not a virulence factor of A. fumigatus in the murine model of invasive pulmonary aspergillosis.
Acknowledgments
We thank Dongmei Li of the Department of Microbiology & Immunology, Georgetown University Medical Center, Washington, DC, for her valuable advice and technical assistance. We thank Kyung J. Kwon-Chung for providing plasmid pDHt/SK and A. tumefaciens strain EHA105 and Alex Andrianopoulos for providing plasmid pALX223.
This investigation was supported by a grant from the John E. Fogarty International Center and by NIH NIAID award TW005926 to R.C. and R.L.
Editor: A. Casadevall
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Alonso-Monge, R., F. Navarro-Garcia, E. Roman, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhabhra, R., M. D. Miley, E. Mylonakis, D. Boettner, J. Fortwendel, J. C. Panepinto, M. Postow, J. C. Rhodes, and D. S. Askew. 2004. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect. Immun. 724731-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 2591760-1763. [DOI] [PubMed] [Google Scholar]
- 4.Bundock, P., A. den Dulk-Ras, A. Beijersbergen, and P. J. Hooykaas. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 143206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chauhan, N., J. P. Latge, and R. Calderone. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4435-444. [DOI] [PubMed] [Google Scholar]
- 6.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26781-803. (Quiz, 26:804-805.) [DOI] [PubMed]
- 7.Du, C., J. Sarfati, J. P. Latge, and R. Calderone. 2006. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 44211-218. [DOI] [PubMed] [Google Scholar]
- 8.Fortwendel, J. R., W. Zhao, R. Bhabhra, S. Park, D. S. Perlin, D. S. Askew, and J. C. Rhodes. 2005. A fungus-specific ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell 41982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillardin, C. d'Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 471523-1543. [DOI] [PubMed] [Google Scholar]
- 10.Hospenthal, D. R., K. J. Kwon-Chung, and J. E. Bennett. 1998. Concentrations of airborne Aspergillus compared to the incidence of invasive aspergillosis: lack of correlation. Med. Mycol. 36165-168. [PubMed] [Google Scholar]
- 11.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21161-172. [DOI] [PubMed] [Google Scholar]
- 12.Kupfahl, C., T. Heinekamp, G. Geginat, T. Ruppert, A. Hartl, H. Hof, and A. A. Brakhage. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 62292-302. [DOI] [PubMed] [Google Scholar]
- 13.Lamarre, C., O. Ibrahim-Granet, C. Du, R. Calderone, and J. P. Latge. 2007. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44682-690. [DOI] [PubMed] [Google Scholar]
- 14.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lessing, F., O. Kniemeyer, I. Wozniok, J. Loeffler, O. Kurzai, A. Haertl, and A. A. Brakhage. 2007. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 62290-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, W., G. S. May, M. S. Lionakis, R. E. Lewis, and D. P. Kontoyiannis. 2004. Extra copies of the Aspergillus fumigatus squalene epoxidase gene confer resistance to terbinafine: genetic approach to studying gene dose-dependent resistance to antifungals in A. fumigatus. Antimicrob. Agents Chemother. 482490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osherov, N., D. P. Kontoyiannis, A. Romans, and G. S. May. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemother. 4875-81. [DOI] [PubMed] [Google Scholar]
- 18.Paisley, D., G. D. Robson, and D. W. Denning. 2005. Correlation between in vitro growth rate and in vivo virulence in Aspergillus fumigatus. Med. Mycol. 43397-401. [DOI] [PubMed] [Google Scholar]
- 19.Philippe, B., O. Ibrahim-Granet, M. C. Prevost, M. A. Gougerot-Pocidalo, M. Sanchez Perez, A. Van der Meeren, and J. P. Latge. 2003. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 713034-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86865-875. [DOI] [PubMed] [Google Scholar]
- 21.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 194623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramesh, S. A., B. N. Kaiser, T. Franks, G. Collins, and M. Sedgley. 2006. Improved methods in Agrobacterium-mediated transformation of almond using positive (mannose/pmi) or negative (kanamycin resistance) selection-based protocols. Plant Cell Rep. 25821-828. [DOI] [PubMed] [Google Scholar]
- 23.Roman, E., C. Nombela, and J. Pla. 2005. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 2510611-10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheppard, D. C., G. Rieg, L. Y. Chiang, S. G. Filler, J. E. Edwards, Jr., and A. S. Ibrahim. 2004. Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 481908-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh, K. K. 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 291043-1050. [DOI] [PubMed] [Google Scholar]
- 26.Steinbach, W. J., D. K. Benjamin, Jr., S. A. Trasi, J. L. Miller, W. A. Schell, A. K. Zaas, W. M. Foster, and J. R. Perfect. 2004. Value of an inhalational model of invasive aspergillosis. Med. Mycol. 42417-425. [DOI] [PubMed] [Google Scholar]
- 27.Sugui, J. A., Y. C. Chang, and K. J. Kwon-Chung. 2005. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 711798-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugui, J. A., J. Pardo, Y. C. Chang, A. Mullbacher, K. A. Zarember, E. M. Galvez, L. Brinster, P. Zerfas, J. I. Gallin, M. M. Simon, and K. J. Kwon-Chung. 2007. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 61552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugui, J. A., J. Pardo, Y. C. Chang, K. A. Zarember, G. Nardone, E. M. Galvez, A. Mullbacher, J. I. Gallin, M. M. Simon, and K. J. Kwon-Chung. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 61562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temple, M. D., G. G. Perrone, and I. W. Dawes. 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15319-326. [DOI] [PubMed] [Google Scholar]
- 31.Xue, T., C. K. Nguyen, A. Romans, D. P. Kontoyiannis, and G. S. May. 2004. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182346-353. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, W., J. C. Panepinto, J. R. Fortwendel, L. Fox, B. G. Oliver, D. S. Askew, and J. C. Rhodes. 2006. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 744865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuber, S., M. J. Hynes, and A. Andrianopoulos. 2002. G-protein signaling mediates asexual development at 25 degrees C but has no effect on yeast-like growth at 37 degrees C in the dimorphic fungus Penicillium mameffei. Eukaryot. Cell 1440-447. [DOI] [PMC free article] [PubMed] [Google Scholar]