Abstract
Our previous studies demonstrated that Mycobacterium bovis bacillus Calmette-Guérin (BCG) can directly interact with human NK cells and induce the proliferation, gamma interferon production, and cytotoxic activity of such cells without the need for accessory cells. Thus, the aim of the present study was to identify the putative receptor(s) responsible for the recognition of BCG by human NK cells and potentially involved in the activation of NK cells. To this end, we first investigated the surface expression of three NK cell-activating receptors belonging to the natural cytoxicity receptor (NCR) family on highly purified human NK cells upon in vitro direct stimulation with BCG. An induction of the surface expression of NKp44, but not of NKp30 or NKp46, was observed after 3 and 4 days of in vitro stimulation with live BCG. The NKp44 induction involved mainly a particular NK cell subset expressing the CD56 marker at high density, CD56bright. In order to establish whether NKp44 could directly bind to BCG, whole BCG cells were stained with soluble forms of the three NCRs chimeric for the human immunoglobulin G (IgG) Fc fragment (NKp30-Fc, NKp44-Fc, NKp46-Fc), followed by incubation with a phycoerythrin (PE)-conjugated goat anti-human IgG antibody. Analysis by flow cytometry of the complexes revealed a higher PE fluorescence intensity for BCG incubated with NKp44-Fc than for BCG incubated with NKp30-Fc, NKp46-Fc, or negative controls. The binding of NKp44-Fc to the BCG surface was confirmed with immunogold labeling using transmission electron microscopy, suggesting the presence of a putative ligand(s) for human NKp44 on the BCG cell wall. Similar binding assays performed on a number of gram-positive and gram-negative bacteria revealed a pattern of NKp44-Fc binding restricted to members of the genus Mycobacterium, to the mycobacterium-related species Nocardia farcinica, and to Pseudomonas aeruginosa. Altogether, the results obtained indicate, for the first time, that at least one member of the NCR family (NKp44) may be involved in the direct recognition of bacterial pathogens by human NK cells.
Natural killer (NK) cells are an important component of the innate immune system and have the ability to both lyse target cells and provide an early source of immunoregulatory cytokines (21, 25). Although first identified by their cytotoxic activity against tumor and virally infected cells, there is now increasing evidence that NK cells are important mediators of the innate resistance to a variety of pathogenic microorganisms, including intracellular bacteria (18, 23). Human NK cells represent approximately 10% of peripheral blood lymphocytes and are phenotypically defined as CD56+ and CD3− cells. On the basis of the surface density of the CD56 marker, two main subpopulations of NK cells can be identified (11). The majority (∼90%) of peripheral NK cells have low-density expression of CD56 (CD56dim) and express high levels of the FcγIII receptor (CD16). In contrast, about 10% of NK cells express CD56 at high levels (CD56bright), while they lack or express low levels of the CD16 marker.
One of the most important discoveries of NK cell biology over the last few years is the finding that NK cell activity results from the sum of signals deriving from inhibitory and activatory receptors expressed on the surfaces of NK cells. Inhibitory receptors include killer immunoglobulinlike receptors (KIR), which inhibit NK cytotoxic functions following the recognition of major histocompatibility complex class I molecules on target cells (8, 22, 25). Natural cytotoxicity receptors (NCRs) that include three molecules (i.e., NKp30 [CD337], NKp44 [CD336], and NKp46 [CD335]) are instead among the activator receptors (8, 22, 25). Cellular ligands of NCRs are still poorly characterized, while more information is available about the microbial ligands of such receptors expressed on the surfaces of infected cells. For example, viral hemagglutinins have been identified as ligands that bind NKp46 and NKp44 (1, 19), while the main tegument protein (pp65) of human cytomegalovirus has been reported as a ligand of NKp30 (2).
It is well known that macrophages and dendritic cells recognize molecular structures formed by repeated motifs and expressed on the surfaces of pathogens that are shared among various microorganisms (pathogen-associated molecular patterns [PAMPs]) (20). Although the dominant pathway of NK cell activation is accessory cell dependent, there is increasing evidence to suggest that, in some cases, the direct interaction of NK cells with pathogen-derived ligands may also activate NK cells (23). For example, Chalifour et al. demonstrated that human NK cells directly recognize PAMPs expressed on bacterial surfaces (i.e., OmpA of Klebsiella pneumoniae and flagellin of Escherichia coli) by Toll-like receptor 2 (TLR-2) and TLR-5, respectively, and that such recognition triggers gamma interferon (IFN-γ) and α-defensin production (10). It has also been reported that the infective forms of Leishmania species (live promastigotes) are able to directly activate NK cells to secrete IFN-γ in the absence of accessory cells (7, 24). Finally, evidence for a direct contact between bovine NK cells and the protozoan Neospora caninum has recently been reported (9).
Previous studies from our laboratory have demonstrated that Mycobacterium bovis bacillus Calmette-Guérin (BCG) can directly interact with human NK cells in the absence of monocytes/macrophages or interleukin 12 (IL-12) and can induce the proliferation, IFN-γ production, and cytotoxic activity of such cells (13). The effector functions of human NK cells were also induced upon stimulation with killed BCG or mycobacterial cell wall preparations and were totally abrogated when NK cells and bacteria were separated by a 0.2-μm membrane which inhibits cell-bacterium contact but not the passage of soluble factors (13). Altogether, these results have suggested a direct interaction between BCG surface components and human NK cells that promotes their activation, proliferation, IFN-γ production, and cytotoxic activity. Interestingly, we also demonstrated that following direct stimulation with BCG, CD56bright cells were those mainly involved in IFN-γ production and proliferation but that the CD56dim subset had a major role in the cytotoxic activity (5).
The aim of the present study was to identify the putative receptor(s) responsible for the recognition of BCG by human NK cells and potentially involved in the activation of NK cells. To this end, we investigated the surface expression of NCRs on highly purified human NK cells upon in vitro stimulation with BCG in the absence of monocytes/macrophages. We observed an induction of the surface expression of NKp44, but not of NKp30 or NKp46, after stimulation with live BCG. In addition, the staining of BCG by soluble forms of the three NCRs demonstrated that NKp44 was the only NCR able to bind the bacterium. NKp44 was also able to bind other species within the Mycobacterium genus, including M. tuberculosis, the mycobacterium-related species Nocardia farcinica, and Pseudomonas aeruginosa. The binding of BCG by soluble NKp44 was confirmed, at the single-cell level, by immunogold labeling using transmission electron microscopy, suggesting the presence of putative ligands for human NKp44 on the mycobacterial cell wall. To our knowledge, this is the first evidence that a receptor belonging to the NCR family is involved in the direct recognition of bacterial pathogens by human NK cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Mycobacterium bovis BCG (Pasteur Merieux, Lyon, France), Mycobacterium avium (strain NBL112/87) (14), Mycobacterium smegmatis (strain mc2 155) (4), and Mycobacterium tuberculosis H37Rv (ATCC) were grown in rolling bottles in Middlebrook 7H9 medium supplemented with 0.5% bovine serum albumin (BSA), 0.2% glucose, and 0.085% NaCl. Clinical isolates of Salmonella enterica serovar Enteritidis, Escherichia coli, Streptococcus pyogenes, Enterococcus faecium, Pseudomonas aeruginosa, and Actinomyces meyeri were grown in Trypticase soy yeast extract medium under optimal culture conditions for each species. Cellulomonas denverensis (DSM 15764) and Nocardia farcinica (DSM 43665) were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and grown in Trypticase soy yeast extract medium and Middlebrook 7H9 medium (see above), respectively, according to the instructions of the DSMZ. Bacteria were harvested during the logarithmic growth phase, washed by centrifugation, and resuspended in phosphate-buffered saline (PBS) at 5 × 107 CFU/ml. Aliquots were kept frozen at −80°C for future use.
Cell populations.
Heparinized venous blood was obtained from nine healthy volunteers. Six subjects had been vaccinated with BCG at least 8 years prior to donation, while three subjects had no previous history of BCG vaccination. Informed consent was obtained, and the protocol was approved by the local ethics committee. Blood was diluted in PBS containing 10% (vol/vol) sodium citrate and layered on a standard density gradient (Lymphoprep, Cedarlane, Canada). After centrifugation at 160 × g for 20 min at room temperature, supernatants were removed, without disturbing the lymphocyte layer at the interface, to eliminate platelets. The gradient was further centrifuged at 800 × g for 20 min, and peripheral blood mononuclear cells (PBMC) were collected from the interface. Cells were washed three times with PBS containing 0.1% (wt/vol) BSA and 10% sodium citrate and enriched for NK cells via a magnetic cell sorter by using NK cell isolation kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's protocol. The negatively isolated fraction, containing untouched NK cells, was collected, and purity was assessed by fluorescence-activated cell sorter analysis. The purified cell population consistently contained more than 97% CD56+ CD3− NK cells. Contaminant CD3+ cells were less than 3% of the population, and CD15+ and CD14+ cells were undetectable. Negatively selected NK cells were resuspended in RPMI 1640 supplemented with 2 mM l-glutamine and 10% heat-inactivated autologous serum and seeded in 96-well plates at a density of 3 × 105 NK cells/well.
NK cell stimulation.
For each experiment, an aliquot of BCG was thawed and inoculated in liquid growth medium. After 8 days of rolling culture at 37°C, bacteria were harvested, washed twice in PBS, and vortexed for 20 min with glass beads to dissolve clumps. The bacterial suspension was incubated in the dark for 20 min at 1 × g to allow sedimentation of residual clumps. The density of the suspension was determined spectrophotometrically at 600 nm. After centrifugation, the bacterial pellet was diluted in RPMI 1640 supplemented with 2 mM l-glutamine (HyClone Europe, Ltd., Cramlington, United Kingdom) and 10% heat-inactivated autologous serum to achieve approximately 1 × 106 CFU/ml. Immediately before use, the bacterial suspension was subjected to three consecutive 10-s pulses (45 W) in a water bath sonicator to obtain a predominantly single-bacterial-cell suspension. The number of BCG organisms per well was assessed by plating 10-fold dilutions of the bacterial suspension, in duplicate, on Middlebrook 7H11 agar enriched with oleic acid, albumin, dextrose, and catalase (Becton Dickinson Microbiology Systems, Cockeysville, MD). Isolated NK cells (3 × 105) were directly cultured with live BCG at (1.8 ± 0.5) × 104 (mean value ± standard error of the mean [SEM]) CFU per well (96-well plate) and at 37°C in humidified air containing 5% CO2. Antigen-free cultures were established as negative controls. The expression of NCRs (NKp30, NKp44, and NKp46) on CD56bright and CD56dim NK cell subsets was evaluated upon 3 and 4 days of in vitro stimulation. To this aim, cells were harvested from wells, washed twice, and stained with appropriate monoclonal antibodies (MAbs) (see below).
In three neutralization experiments, NK cells were pretreated with FcR blocking reagent (Miltenyi Biotec, Bergisch Gladbach, Germany) to avoid nonspecific capture of MAbs by NK cell Fc receptors. After being washed twice with RPMI 1640, NK cells were preincubated with 5 μg/ml of anti-NKp30 (Z25), anti-NKp44 (Z231), or anti-NKp46 (BAB281) (Beckman Coulter, Fullerton, CA) MAbs for 1 h at 4°C. NK cells were then cultured with BCG or left unstimulated as described above. Following a 48-h culture, 1.5 μg/ml of each anti-NCR MAb was added to corresponding wells. The percentage of CD69+ NK cells was evaluated after 3 days.
Immunofluorescence staining for surface markers.
Cells were resuspended in PBS and incubated with saturating amounts of antibodies for 30 min at 4°C. Two- or three-color immunofluorescence staining was performed as previously described (13). The following MAbs were used for staining: fluorescein isothiocyanate (FITC)-conjugated anti-CD3, anti-CD8, and anti-CD15 (Ancell Corp., Bayport, MN); phycoerythrin (PE)-conjugated anti-CD14 and anti-CD20 (Ancell Corp., Bayport, MN); anti-CD69 (Miltenyi Biotec, Bergisch Gladbach, Germany); anti-NKp30 (Z25, CD337); anti-NKp44 (Z231, CD336); anti-NKp46 (BAB281, CD335); rhodamine-PE-cyanin 5.1-conjugated anti-CD56 (N901) (Beckman Coulter, Fullerton, CA); and isotype-matched mouse immunoglobulin G (IgG) (as negative controls) (BD Biosciences, Mountain View, CA). A nonconjugated anti-KIR2DL4 MAb (R&D Systems, Minneapolis, MN) and an FITC-conjugated anti-mouse IgG F(ab′)2 antibody (DakoCytomation, Glostrup, Denmark) were used for indirect immunofluorescence staining.
Following staining, 20,000 events were acquired ungated in a FACSort flow cytometer (BD Biosciences). CellQuest software (BD Biosciences) was used for computer-assisted analyses. For analyses of the two (i.e., CD56bright and CD56dim) NK cell subsets, first all viable cells were selected by a widely set gate on a two-parameter plot of side scatter versus forward-angle scatter. Among these cells, a second gate was set, on a two-color (CD3-FITC versus CD56-rhodamine-PE-cyanin) fluorescence intensity plot, to include all CD56+ CD3− cells. Finally, among CD56+ CD3− cells, CD56bright and CD56dim subsets were identified according to the fluorescence intensity of the CD56 marker. The levels of NCRs expression were evaluated by mean fluorescence intensity (MFI) analyses.
Determination of IL-2 in culture supernatants.
The levels of IL-2 in culture supernatants were quantified by using a commercially available enzyme-linked immunosorbent assay (Ready-SET-Go! human IL-2 enzyme-linked immunosorbent assay; eBioscience, San Diego, CA) according to the manufacturer's instructions. Supernatants were collected at different time points (1 and 3 days) from in vitro cultures of NK cells stimulated with BCG or left unstimulated and stored in aliquots at −80°C for future use. Recombinant IL-2 was used as a standard in the assay. The detection limit for the assay was 4 pg/ml.
Staining of bacteria with soluble NCR-Fc.
Staining of bacteria with recombinant human NKp30-, NKp44-, and NKp46-IgG1 (Fc) chimeras (R&D Systems, Minneapolis, MN) was performed as follows: 300 μl of each bacterial suspension (optical density at 600 nm, 0.1) was incubated with 1.5 to 3 μg/ml of soluble NKp30-Fc, NKp44-Fc, NKp46-Fc, or human IgG for 45 min at 4°C. Following three washes in PBS containing 0.1% BSA, bacteria were incubated for 30 min at 4°C with a human IgG-Fc fragment-specific, PE-conjugated goat antibody (eBioscience, San Diego, CA). After three washes, the bacteria were resuspended in PBS and analyzed by flow cytometry. In preliminary experiments, no difference in levels of binding of soluble NCRs was observed between live BCG and BCG fixed with 1% paraformaldehyde overnight at 4°C. Therefore, staining of M. avium, M. smegmatis, M. tuberculosis H37Rv, S. enterica serovar Enteritidis, E. coli, S. pyogenes, E. faecium, C. denverensis, N. farcinica, and P. aeruginosa was performed with fixed bacteria, including fixed BCG as a staining control. The binding of each soluble NCR-Fc was evaluated by calculating the MFI using CellQuest software. In some experiments, BCG killed by a 1-h incubation at 80°C was used in the binding assay.
To evaluate whether anti-NKp44 was able to neutralize NKp44 receptor binding to mycobacteria, NKp44-Fc was preincubated with 1 to 5 μg/ml of anti-NKp44 (Beckman Coulter, Fullerton, CA) for 45 min at 4°C and then incubated with BCG according to the staining procedure described above.
Analysis of NCR-Fc binding to the surfaces of BCG and E. faecium by transmission electron microscopy.
Immunoelectron microscopic observations were made by using the three NCRs chimeric for the human IgG Fc fragment (NKp30-Fc, NKp44-Fc, NKp46-Fc). Bacterial suspensions (optical density at 600 nm, 1) were mildly fixed with 0.5% paraformaldehyde, harvested by centrifugation, washed twice with PBS, incubated with each NCR-Fc (0.3 to 3 μg/ml) for 1 h at room temperature, washed twice in Aurion BSA-c solution (Wageningen, The Netherlands), and labeled with anti-human IgG (γ-chain specific) 10-nm-diameter-gold particle-conjugated antibody (diluted 1:20) (Sigma) for 45 min at room temperature. After three washes in Aurion BSA-c solution, two washes in PBS, and a final wash in distilled water, a drop of suspension was placed onto a carbon-coated grid, partially dried, and directly observed without any further treatment. Samples were examined with a Philips 208 transmission electron microscope (FEI Company, Eindhoven, The Netherlands). This method ensured the best antigenic preservation, allowing labeling of only the surface components.
Statistical analysis.
The statistical significance of the data was determined by Student's t test for paired samples or by the nonparametric Wilcoxon matched-pair signed-rank test. A P value of <0.05 was considered significant.
RESULTS
Surface expression of NKp30, NKp44, and NKp46 on human NK cells upon direct stimulation with BCG.
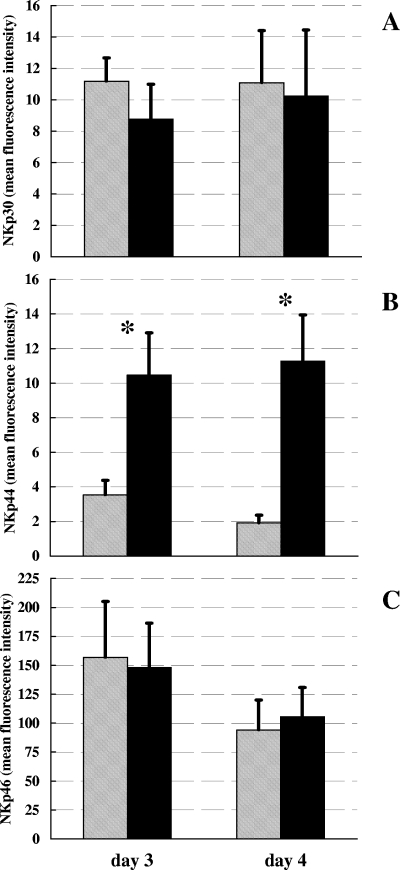
In order to evaluate whether the expression of the three NCRs (NKp30, NKp44, NKp46) on human NK cell surfaces was modified upon direct stimulation with BCG, isolated NK cells from seven healthy donors were incubated directly with live BCG or left unstimulated, and the surface expression of NCRs was assessed by flow cytometry as MFI after 3 and 4 days of culture. As depicted in Fig. 1, a significant induction of NKp44 expression was observed on BCG-stimulated NK cells after both 3 and 4 days of culture compared to expression in antigen-free cultures (P < 0.05, Student's t test for paired samples). There was no significant difference in the levels of expression of both NKp30 and NKp46 between NK cells stimulated with BCG and those left unstimulated.
FIG. 1.
Surface density expression of NKp30 (A), NKp44 (B), and NKp46 (C) on human NK cells stimulated with live BCG (black bars) or left unstimulated (shaded bars) for 3 to 4 days in vitro. Levels of NCR expression by NK cells were evaluated by determining MFIs. Mean values from seven different blood donors ± SEM are reported. *, P < 0.05 (Student's t test for paired samples).
To assess whether NKp44 induction could be due to the presence of IL-2 released by a few contaminating cells, IL-2 levels were measured in culture supernatants from seven experiments following 1 and 3 days of in vitro stimulation. IL-2 was undetectable in all the supernatants tested. Furthermore, no induction of KIR2DL4—an NK cell surface molecule, the expression of which is known to be enhanced following IL-2 activation (17)—was observed on NK cells in two independent experiments (data not shown).
Evaluation of expression levels of NKp30, NKp44, and NKp46 by the CD56bright and CD56dim NK cell subsets in response to BCG.
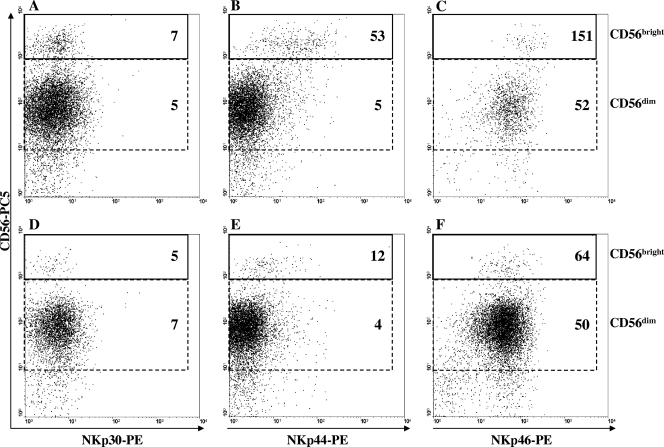
To investigate whether the induction of NKp44 expression on NK cells in response to BCG was preferentially attributable to one of the two NK subsets (i.e., CD56dim or CD56bright), the expression levels of the three NK-activating receptors (NKp30, NKp44, NKp46) were separately evaluated on CD56bright and CD56dim NK subsets stimulated with BCG for 3 and 4 days or left unstimulated. As shown in Fig. 2 (for a representative experiment) and in Table 1, while the levels of NKp44 expression did not differ between BCG-stimulated and unstimulated CD56dim cells, they were significantly induced on BCG-stimulated CD56bright NK cells compared to levels of expression on unstimulated cells. No differences were observed in any of the BCG-stimulated or unstimulated CD56bright or CD56dim NK cell subsets for the other two NCRs (Table 1).
FIG. 2.
Surface density expression of NKp30 (A, D) NKp44 (B, E), and NKp46 (C, F) on CD56bright and CD56dim cells after 4 days of in vitro stimulation with live BCG (A, B, C) or on cells left unstimulated (D, E, F). Plots from a representative experiment are illustrated. Numbers indicate the MFI levels of three NCRs on CD56bright and CD56dim NK cell subsets. Percentages of total CD56bright NK cells were as follows: for unstimulated NK cells, 2.4%, and for BCG-stimulated NK cells, 6.4%.
TABLE 1.
Expression levels of NKp30, NKp44, and NKp46 by CD56bright and CD56dim NK cell subsets in response to BCG upon 4 days of in vitro stimulationa
| NK cells | MFI (mean ± SEM)
|
|||||
|---|---|---|---|---|---|---|
| NKp30
|
NKp44
|
NKp46
|
||||
| CD56dim | CD56bright | CD56dim | CD56bright | CD56dim | CD56bright | |
| Unstimulated | 11.4 ± 3.4 | 8.3 ± 1.8 | 1.2 ± 0.6 | 21.6 ± 5.7 | 92.2 ± 26.3 | 160.1 ± 37.7 |
| BCG stimulated | 9.8 ± 4.5 | 10.5 ± 2.7 | 2.9 ± 0.5 | 58.6 ± 7.8b | 91.3 ± 25.6 | 189.1 ± 25.5 |
Percentages of total CD56bright NK cells were as follows: for unstimulated NK cells, 5.2% ± 1.2%, and for BCG-stimulated NK cells, 12.7% ± 2.8% (P < 0.05, Student's t test for paired samples). Values were derived from seven independent experiments.
P < 0.001 (Student's t test for paired samples).
Evaluation of soluble NCR-Fc binding to various mycobacterial species.
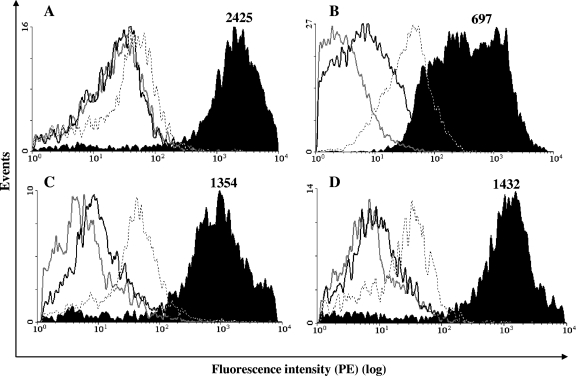
To evaluate whether the induction of NKp44 on BCG-stimulated NK cells involved a direct interaction of NKp44 with bacteria, soluble recombinant human NKp30-, NKp44-, and NKp46-IgG1 (Fc portion) chimeras were incubated with BCG, and the receptor-bacterium binding was revealed by staining with a PE-conjugated goat anti-human IgG antibody and flow cytometric analyses. The results obtained (Fig. 3A and Table 2) demonstrated a higher PE fluorescence intensity (MFI) for BCG incubated with NKp44-Fc than that for the negative control: 2,272 ± 298 (NKp44) versus 45.7 ± 7.6 (negative control) (P < 0.01 [eight experiments], Wilcoxon matched-pair signed-rank test). In contrast, no difference in MFI values was observed between BCG incubated with NKp30-Fc or NKp46-Fc and the negative control. In addition, no statistically significant difference in NKp44-Fc binding was observed among live, fixed, and heat-killed BCG organisms (Table 2). Similar binding assays demonstrated that NKp44-Fc was also able to bind other mycobacterial species, i.e., M. tuberculosis H37Rv (Fig. 3B), M. avium (Fig. 3C), and M. smegmatis (Fig. 3D), with MFI values comparable to that observed for BCG (Table 2). These results strongly suggest the presence of a mycobacterial ligand for NKp44.
FIG. 3.
Binding of soluble NCR-human IgG Fc chimeras to mycobacterial surfaces. (A) M. bovis BCG; (B) M. tuberculosis (H37Rv); (C) M. avium; (D) M. smegmatis. Binding was measured by determining MFIs. Dotted lines indicate staining with whole human IgG (negative control). Gray lines, NKp30-Fc; black lines, NKp46-Fc; black-filled histogram, NKp44-Fc. Numbers indicate MFI values for NKp44-Fc. Binding assays were repeated at least three times with each bacterial species. A representative histogram for each mycobacterial species is depicted.
TABLE 2.
Binding of soluble NCRs NKp30-Fc, NKp44-Fc, and NKp46-Fc to various bacteria
| Speciesa | MFI (mean ± SEM)b
|
|||
|---|---|---|---|---|
| NKp30-Fc | NKp44-Fc | NKp46-Fc | Negative control | |
| Mycobacteria | ||||
| Live M. bovis BCG | 37.6 ± 6.5 | 2,337.7 ± 766.5 | 133.7 ± 40.1 | 209.4 ± 59.5 |
| Heat-killed M. bovis BCG | 43.1 ± 2.5 | 1,852.6 ± 508.2 | 128.7 ± 38.9 | 166.8 ± 35.8 |
| M. bovis BCG | 35.7 ± 12.9 | 2,272.2 ± 298.1 | 110.1 ± 10 | 45.7 ± 7.6 |
| M. tuberculosis H37Rv | 14.2 ± 6.6 | 951.3 ± 480.8 | 27.8 ± 3.4 | 39.3 ± 5 |
| M. avium | 22.9 ± 2.2 | 1,426.5 ± 380.5 | 47.5 ± 4.2 | 47.2 ± 11.7 |
| M. smegmatis | 24 ± 3.3 | 1,812.1 ± 557.8 | 40 ± 4.5 | 35.3 ± 5.1 |
| Gram-positive bacteria | ||||
| Nocardia farcinica | 14.9 ± 5 | 868.7 ± 223.5 | 31.3 ± 3.3 | 23.8 ± 7.3 |
| Cellulomonas denverensis | 8 ± 2.5 | 65.8 ± 8.8 | 12.2 ± 1.9 | 18.7 ± 7.5 |
| Actinomyces meyeric | 5.0 | 35.7 | 8.2 | 21.2 |
| Streptococcus pyogenes | 8.9 ± 3.9 | 85.7 ± 9.9 | 68.3 ± 0.3 | 219.4 ± 67.6 |
| Enterococcus faecium | 3.4 ± 1.6 | 46.7 ± 14.5 | 27.1 ± 6.1 | 7.7 ± 3.9 |
| Gram-negative bacteria | ||||
| Pseudomonas aeruginosa | 46.8 ± 7.4 | 935.3 ± 204.4 | 112.9 ± 5.1 | 55.2 ± 18.3 |
| Salmonella enterica serovar Enteritidis | 9.4 ± 4.9 | 255.8 ± 145.1 | 42.6 ± 7.8 | 14.5 ± 8.0 |
| Escherichia coli | 9.3 ± 5.1 | 190.5 ± 70.4 | 14.6 ± 3.6 | 14.8 ± 4.8 |
| Acinetobacter baumannii | 15.3 ± 3.6 | 152.9 ± 53.5 | 36.2 ± 5 | 24.5 ± 12.8 |
All bacteria, except live and heat-killed M. bovis BCG, were fixed with 1% paraformaldehyde in PBS overnight at +4°C.
Values were derived from at least three independent experiments.
One observation only.
Analysis of NCR-Fc binding to various gram-positive or gram-negative bacteria.
To investigate whether the putative ligand for NKp44 was conserved only within the genus Mycobacterium or was more widely distributed among other bacteria, NCR-Fc binding assays were performed on various bacteria other than mycobacteria. The results obtained demonstrated that, among bacterial species tested, only N. farcinica (gram positive) and P. aeruginosa (gram negative) were able to bind NKp44-Fc at MFI levels comparable to those observed for mycobacteria (Table 2). None of the bacterial species tested exhibited evident binding of either NKp30-Fc or NKp46-Fc (Table 2).
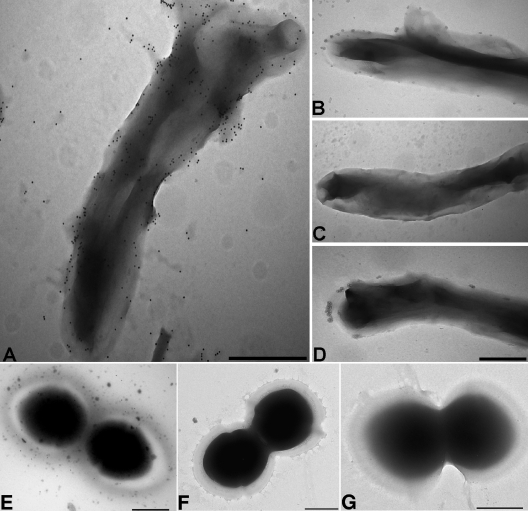
Evaluation of NKp44 binding to BCG and E. faecium surfaces by transmission electron microscopy.
In order to confirm the binding of NKp44-Fc to BCG at the single-cell level, bacteria were incubated with the soluble forms of the three NCRs, followed by incubation with a goat anti-human IgG (Fc) antibody conjugated with 10-nm colloidal-gold particles and examined by transmission electron microscopy. The observations revealed numerous gold particles distributed on the surface of a BCG organism incubated with NKp44-Fc (Fig. 4A). In contrast, very few particles or a complete absence of particles was observed on BCG incubated with the other two NCR-Fc chimeras or with the negative-control antibody (Fig. 4B, C, and D, respectively). Moreover, no discernible binding of the three NCR-Fc chimeras to the surface of E. faecium (Fig. 4E, F, and G), a species that had also revealed low levels of NKp44 binding by flow cytometry, was observed.
FIG. 4.
Immunoelectron microscopy of M. bovis BCG (A, B, C, D) and E. faecium (E, F, G) labeled with NCR-Fc. Bacteria labeled with NKp44-Fc (A, E), NKp30-Fc (B, F), NKp46-Fc (C, G), and a goat anti-human IgG antibody conjugated with 10-nm colloidal-gold particles. (D) Negative control. Bars = 0.5 μm.
Effects of anti-NKp30, anti-NKp44, and anti-NKp46 MAbs on the percentage of CD69+ NK cells and on NKp44-Fc binding to BCG.
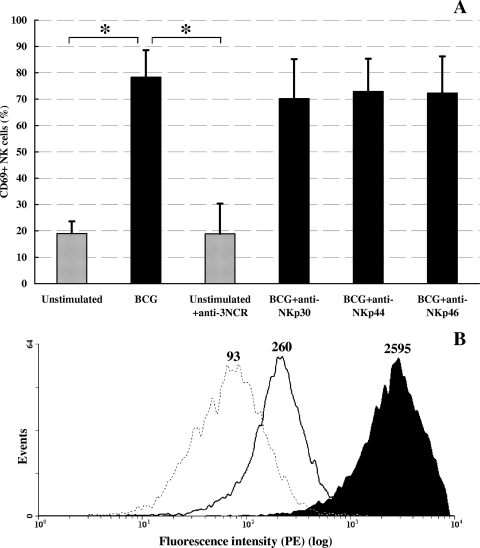
To determine whether BCG-induced activation of human NK cells could be mediated through NKp44 binding, NK cells from three healthy blood donors were stimulated with BCG in the presence of MAbs against NKp30, NKp44, or NKp46. As shown in Fig. 5A, although upon 3 days of stimulation with BCG a significant increase in the percentage of CD69+ NK cells was recorded compared to that of unstimulated NK cells (P < 0.05, Student's t test for paired samples), neither anti-NKp44 MAb nor MAbs against the other two NCRs could affect the percentage of CD69+ BCG-stimulated NK cells. Nevertheless, the same anti-NKp44 MAb was able to markedly reduce (90%) NKp44-Fc binding to the BCG surface, as assessed by flow cytometry (Fig. 5B).
FIG. 5.
Effects of anti-NKp44 antibody on BCG-induced NK cell activation and on NKp44-Fc binding to BCG surfaces. (A) Percentages of CD69+ human NK cells stimulated for 3 days in vitro with live BCG (black bars) or left unstimulated (shaded bars) in the presence or absence of antibodies against NKp30, NKp44, and NKp46. anti-3NCR, antibodies against NKp30, NKp44, and NKp46 mixed together. *, P < 0.05 (Student's t test for paired samples). (B) Binding of soluble NKp44-Fc to BCG's surface in the presence (black line) or absence (black-filled histogram) of anti-NKp44. The dotted line indicates staining with whole human IgG (negative control). Numbers indicate MFI values. A representative histogram is depicted.
DISCUSSION
The possibility that NK cells, similarly to other cells of innate immunity, can directly recognize pathogens via their surface receptors is a hypothesis that is being increasingly taken into consideration. However, to date there are only a few reports that have studied the effects of the direct interaction of human NK cells with microorganisms or with their products (23). This might be, at least partially, due to the inherent difficulty of obtaining a highly purified NK cell population and to the known ability of even a few contaminating monocytes/macrophages to activate NK cells, making it difficult to fully discriminate between direct and indirect NK cell activation signals. Despite these difficulties, what has become apparent over the last few years is that, although an optimal NK cell response requires the presence of PAMP-activated accessory cells, direct recognition of pathogens by NK cells can also occur with the corresponding activation of effector functions (23, 25).
In the present study, we investigated the expression of human NK cell-activating receptors (i.e., NCRs) potentially involved in the direct recognition of pathogens upon in vitro direct stimulation with BCG. By using a commercial NK cell-negative isolation kit, we obtained, in a highly reproducible manner, a virtually monocyte/macrophage-free NK cell population (>97% CD56+ CD3− cells), which was left untouched during the purification process. Compared to what occurred with antigen-free cultures, 3 and 4 days of in vitro stimulation with BCG induced a significant increase of NKp44 expression on highly purified human NK cells. NK cells are known to be activated to express NKp44 in the presence of IL-2 (our unpublished observation and reference 28); therefore, IL-2 was measured in culture supernatants. The lack of IL-2, together with the lack of KIR2DL4 induction (an NK cell receptor, the expression of which is known to be enhanced following IL-2 activation [17]), demonstrates that NKp44 induction on NK cells stimulated with BCG is not due to IL-2 release by putative contaminating cells.
Interestingly, the observed induction of NKp44 expression on BCG-stimulated human NK cells was predominantly attributable to the CD56bright subset, which was previously demonstrated to be involved mainly in the production of IFN-γ and immunoregulatory activities, rather than the cytotoxic activity, of NK cells in response to BCG (5). These observations may indicate a preferential direct interaction of mycobacteria with the CD56bright NK cell subset, as was recently also suggested by Schierloh and collaborators (26). Those authors demonstrated an enrichment of activated CD56bright NK cells in pleural fluids of tuberculosis patients due to an increased susceptibility to the apoptosis of resident CD56+ CD16− (CD56dim) cells mediated by heat-labile and stable soluble factors present in tuberculosis effusions and not in pleural fluids of other etiologies. In the same report, it was also demonstrated that IFN-γ production from pleural fluid CD56bright cells was induced by M. tuberculosis in the absence of accessory cells (i.e., CD3+, CD19+, and CD14+), suggesting that this activity was due to a direct interaction between M. tuberculosis and CD56bright cells (26).
The induction of NKp44 on BCG-stimulated NK cells might be merely secondary to the activation of NK cells by BCG or could be due to a direct interaction of the bacterium with the receptor. To investigate whether a ligand for NKp44 was present on the mycobacterial surface, we tested the ability of commercially available soluble forms of NKp30, NKp44, and NKp46 to bind BCG. These experiments demonstrated that NKp44-Fc, but not the other two NCRs (i.e., NKp30-Fc and NKp46-Fc), was able to form stable complexes with the bacterium. To exclude the possibility that the observed binding of NKp44-Fc to BCG was simply due to the trapping of the molecule as a consequence of the high tendency of mycobacterial cells to form aggregates, immunogold labeling experiments using transmission electron microscopy at the single-bacterium level were performed. The results obtained clearly demonstrated that NKp44-Fc did bind evenly to the BCG surface, suggesting the presence of a putative ligand for NKp44 on the mycobacterial cell wall. These findings are in agreement with our previous work that demonstrated the ability of mycobacterial-cell wall-enriched antigen preparations to directly interact with NK cells in the absence of macrophages or other accessory cell populations (6, 13).
It is known that PAMPs are constituted of repeats of molecular motifs conserved among various classes of microorganisms, and this fact permits cells of innate immunity to be able to recognize a vast variety of pathogens by using a relatively limited receptor repertoire (20).
Therefore, to investigate the distribution of the putative NKp44 ligand on microorganisms other than BCG, NCR-Fc bacterial binding assays were performed using other mycobacteria (i.e., M. avium, M. smegmatis, and M. tuberculosis H37Rv) or other gram-positive and gram-negative species that are considered distantly related (i.e., S. enterica serovar Enteritidis, E. coli, S. pyogenes, E. faecium, P. aeruginosa, and A. baumannii) or closely related (i.e., members of the Actinobacteria class, such as Nocardia, Cellulomonas, and Actinomyces) to the genus Mycobacterium based on structural and taxonomic characteristics. The results obtained demonstrated that the putative ligand for NKp44 is highly conserved among species of the genus Mycobacterium. Although NKp44-Fc showed greater binding than NKp30-Fc and NKp46-Fc to all the bacterial species tested, the three NCR-Fcs bound equally to PBMC (data not shown). This may indicate that NKp44 is not more nonspecifically sticky than the other receptor preparations, at least when heterogeneous populations of eukaryotic cells such as PBMC are considered. Nevertheless, the binding levels of NKp44 to mycobacterial surfaces were consistently much higher than those observed with most of the other bacterial species. Analysis of NKp44-Fc binding to E. faecium by transmission electron microscopy confirmed the NCR-Fc binding levels assessed by flow cytometry, further supporting the specificity of NKp44-Fc's binding to the surfaces of mycobacteria. Moreover, a species belonging to the genus Nocardia also bound NKp44 at high MFI levels. Notably, members of the genus Nocardia share with mycobacteria the presence of mycolic acids in their cell walls (3). Interestingly, other mycobacterium-related species that do not contain mycolic acids (i.e., Cellulomonas denverensis and Actinomyces meyeri) did not bind NKp44-Fc, suggesting that such cell wall components are putative ligands for NKp44. Preliminary binding experiments performed by us using BCG previously digested with trypsin (12) demonstrated that this treatment did not abolish the ability of NKp44-Fc to bind the bacterial surface (data not shown), further suggesting a nonproteic nature of the putative mycobacterial ligand. In addition, the observation that NKp44-Fc equally binds to live BCG and to BCG killed by 1-h incubation at 80°C indicates that the putative NKp44 ligand might be a relatively heat-stable structural component.
Among the bacterial species distantly related to the mycobacteria tested, P. aeruginosa was the only one able to bind NKp44 at high MFI levels. Viral hemagglutinins (1) and, very recently, heparan sulfate epitope(s) (16) have been described as ligands/coligands of NKp44, suggesting that such a receptor, similarly to other members of the NCR family, may be involved in the recognition of various microbial/cellular ligands.
In order to assess whether the BCG-induced activation of human NK cells could be mediated through NKp44 binding, NK cells were stimulated with BCG in the presence of MAbs against NKp30, NKp44, or NKp46. None of the NCR-blocking antibodies could substantially alter the expression of the activation marker CD69 on NK cells stimulated with BCG. Nevertheless, the anti-NKp44 antibody was able to almost completely abolish NKp44 binding to the BCG surface. The data obtained, together with the observation that NKp44 is very poorly or not expressed on resting NK cells (28), suggest that NKp44 is not likely to be the main receptor involved in BCG-induced NK cell activation but rather may play a secondary role in the functional activities of NK cells, which has yet to be determined.
In conclusion, the results obtained herein demonstrate that (i) the interaction of BCG with highly purified NK cells induces on the surfaces of such cells the expression of NKp44, but not of NKp30 or NKp46, and (ii) NKp44-Fc, but not NKp30-Fc or NKp46-Fc, is able to bind mycobacteria and a few other gram-positive or gram-negative species, suggesting the existence of a bacterial ligand(s) for this receptor. A role for another member of the NCR family in immunity against mycobacteria has recently been reported (15, 27). Those authors demonstrated an involvement of the NKp46 receptor, but not of the NKp44 receptor, in the NK cell-mediated lysis of M. tuberculosis-infected macrophages via the recognition of vimentin on the surfaces of infected cells. This finding, together with the possible direct recognition of mycobacteria by the NKp44 receptor reported in this study, further stresses the hypothesis that the NK cell response to pathogens may result from the sum of indirect (i.e., via cytokines released by accessory cells) and direct stimuli.
Studies aimed at identifying the effects of the direct binding of mycobacteria to NKp44 on NK cell functions and the nature of the putative bacterial ligand(s) are in progress.
Acknowledgments
This work was supported by Progetti P.R.I.N. protocol no. 2004067822-001, Rome, Italy, and Fondazione Cassa di Risparmio di Verona, Vicenza, Belluno ed Ancona, Bando 2006. F.L.B. is supported by a scholarship from Fondazione Cassa di Risparmio di Verona, Vicenza, Belluno ed Ancona, Bando 2006.
Editor: F. C. Fang
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Arnon, T. I., M. Lev, G. Katz, Y. Chernobrov, A. Porgador, and O. Mandelboim. 2001. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 312680-2689. [DOI] [PubMed] [Google Scholar]
- 2.Arnon, T. I., H. Achdout, O. Levi, G. Markel, N. Saleh, G. Katz, R. Gazit, T. Gonen-Gross, J. Hanna, E. Nahari, A. Porgador, A. Honigman, B. Plachter, D. Mevorach, D. G. Wolf, and O. Mandelboim. 2005. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 6515-523. [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, R. E. Lee, K. Mdluli, A. E. Sampson, B. G. Schroeder, R. A. Slayden, and Y. Yuan. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37143-179. [DOI] [PubMed] [Google Scholar]
- 4.Batoni, G., D. Bottai, G. Maisetta, M. Pardini, A. Boschi, W. Florio, S. Esin, and M. Campa. 2001. Involvement of the Mycobacterium tuberculosis secreted antigen SA-5K in intracellular survival of recombinant Mycobacterium smegmatis. FEMS Microbiol. Lett. 205125-129. [DOI] [PubMed] [Google Scholar]
- 5.Batoni, G., S. Esin, F. Favilli, M. Pardini, D. Bottai, G. Maisetta, W. Florio, and M. Campa. 2005. Human CD56bright and CD56dim natural killer cell subsets respond differentially to direct stimulation with Mycobacterium bovis bacillus Calmette-Guérin. Scand. J. Immunol. 62498-506. [DOI] [PubMed] [Google Scholar]
- 6.Batoni, G., S. Esin, M. Pardini, D. Bottai, S. Senesi, H. Wigzell, and M. Campa. 2000. Identification of distinct lymphocyte subsets responding to subcellular fractions of Mycobacterium bovis bacille Calmette-Guerin (BCG). Clin. Exp. Immunol. 119270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, I., N. Salaiza, M. Aguirre, J. Delgado, N. Carrillo-Carrasco, L. G. Kobeh, A. Ruiz, R. Cervantes, A. P. Torres, N. Cabrera, A. Gonzalez, C. Maldonado, and A. Isibasi. 2003. Leishmania lipophosphoglycan (LPG) activates NK cells through Toll-like receptor-2. Mol. Biochem. Parasitol. 13065-74. [DOI] [PubMed] [Google Scholar]
- 8.Bottino, C., R. Castriconi, L. Moretta, and A. Moretta. 2005. Cellular ligands of activating NK receptors. Trends Immunol. 26221-226. [DOI] [PubMed] [Google Scholar]
- 9.Boysen, P., S. Klevar, I. Olsen, and A. K. Storset. 2006. The protozoan Neospora caninum directly triggers bovine NK cells to produce gamma interferon and to kill infected fibroblasts. Infect. Immun. 74953-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalifour, A., P. Jeannin, J. F. Gauchat, A. Blaecke, M. Malissard, T. N′Guyen, N. Thieblemont, and Y. Delneste. 2004. Direct bacterial protein PAMP recognition by human NK cells involves TLRs and triggers alpha-defensin production. Blood 1041778-1783. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, M. A., T. A. Fehniger, and M. A. Caligiuri. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22633-640. [DOI] [PubMed] [Google Scholar]
- 12.Delogu, G., C. Pusceddu, A. Bua, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52725-733. [DOI] [PubMed] [Google Scholar]
- 13.Esin, S., G. Batoni, M. Pardini, F. Favilli, D. Bottai, G. Maisetta, W. Florio, R. Vanacore, H. Wigzell, and M. Campa. 2004. Functional characterization of human natural killer cells responding to Mycobacterium bovis bacille Calmette-Guerin. Immunology 112143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esin, S., G. Batoni, G. Kallenius, H. Gaines, M. Campa, S. B. Svenson, R. Andersson, and H. Wigzell. 1996. Proliferation of distinct human T cell subsets in response to live, killed or soluble extracts of Mycobacterium tuberculosis and Myco. avium. Clin. Exp. Immunol. 104419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg, A., P. F. Barnes, A. Porgador, S. Roy, S. Wu, J. S. Nanda, D. E. Griffith, W. M. Girard, N. Rawal, S. Shetty, and R. Vankayalapati. 2006. Vimentin expressed on Mycobacterium tuberculosis-infected human monocytes is involved in binding to the NKp46 receptor. J. Immunol. 1776192-6198. [DOI] [PubMed] [Google Scholar]
- 16.Hershkovitz, O., S. Jivov, N. Bloushtain, A. Zilka, G. Landau, A. Bar-Ilan, R. G. Lichtenstein, K. S. Campbell, T. H. van Kuppevelt, and A. Porgador. 2007. Characterization of the recognition of tumor cells by the natural cytotoxicity receptor, NKp44. Biochemistry 467426-7436. [DOI] [PubMed] [Google Scholar]
- 17.Kikuchi-Maki, A., S. Yusa, T. L. Catina, and K. S. Campbell. 2003. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-γ production. J. Immunol. 1713415-3425. [DOI] [PubMed] [Google Scholar]
- 18.Lodoen, M. B., and L. L. Lanier. 2006. Natural killer cells as initial defense against pathogens. Curr. Opin. Immunol. 18391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandelboim, O., N. Lieberman, M. Lev, L. Paul, T. I. Arnon, Y. Bushkin, D. M. Davis, J. L. Strominger, J. W. Yewdell, and A. Porgador. 2001. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 4091055-1060. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1135-145. [DOI] [PubMed] [Google Scholar]
- 21.Moretta, A., C. Bottino, M. C. Mingari, R. Biassoni, and L. Moretta. 2002. What is a natural killer cell? Nat. Immunol. 36-8. [DOI] [PubMed] [Google Scholar]
- 22.Moretta, L., and A. Moretta. 2004. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 23255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman, K. C., and E. M. Riley. 2007. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat. Rev. Immunol. 7279-291. [DOI] [PubMed] [Google Scholar]
- 24.Nylén, S., K. Maasho, K. Söderström, T. Ilg, and H. Akuffo. 2003. Live Leishmania promastigotes can directly activate primary human natural killer cells to produce interferon-gamma. Clin. Exp. Immunol. 131457-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor, G. M., O. M. Hart, and C. M. Gardiner. 2005. Putting the natural killer cell in its place. Immunology 1171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schierloh, P., N. Yokobori, M. Aleman, R. M. Musella, M. Beigier-Bompadre, M. A. Saab, L. Alves, E. Abbate, S. S. de la Barrera, and M. C. Sasiain. 2005. Increased susceptibility to apoptosis of CD56dimCD16+ NK cells induces the enrichment of IFN-gamma-producing CD56bright cells in tuberculous pleurisy. J. Immunol. 1756852-6860. [DOI] [PubMed] [Google Scholar]
- 27.Vankayalapati, R., A. Garg, A. Porgador, D. E. Griffith, P. Klucar, H. Safi, W. M. Girard, D. Cosman, T. Spies, and P. F. Barnes. 2005. Role of NK cell-activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J. Immunol. 1754611-4617. [DOI] [PubMed] [Google Scholar]
- 28.Vitale, M., C. Bottino, S. Sivori, L. Sanseverino, R. Castriconi, E. Marcenaro, R. Augugliaro, L. Moretta, and A. Moretta. 1998. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1872065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]