Abstract
The causative agent of tuberculosis, Mycobacterium tuberculosis, has two chaperonin (Cpn60) proteins and one cochaperonin (Cpn10) protein. We show here that cpn60.2 and cpn10, but not cpn60.1, are essential for cell survival. A mutant lacking Cpn60.1 was indistinguishable from the wild-type organism in plate and broth culture and within murine macrophages, although it showed increased sensitivity to high temperature (55°C). However, infection of mice with the Δcpn60.1 mutant revealed a major difference from the wild-type organism. In spite of having equal numbers of bacteria in infected sites, the Δcpn60.1 mutant failed to produce granulomatous inflammation in either mice or guinea pigs. This was associated with reduced cytokine expression in infected animals and macrophages. Cell wall lipid acid composition was not altered in the mutant strain. Thus, it appears that Cpn60.1 is an important agent in the regulation of the cytokine-dependent granulomatous response in M. tuberculosis infection.
Chaperonin 60 (Cpn60), also termed heat shock protein 60 (Hsp60), is a prototypical molecular chaperone. Much of our knowledge about molecular chaperones and their role in protein folding has emerged from the study of this protein and its cochaperone, chaperonin 10 (Cpn10). In Escherichia coli, these oligomeric proteins (the Cpn60 homologue GroEL is a tetradecamer and the Cpn10 homologue GroES is a heptamer) are essential for growth because the GroEL/GroES chaperone system is required for the folding of at least 13 essential proteins (2, 9). Most bacteria have one copy each of the cpn10 and cpn60 genes in a single operon (13). However, analysis of genome sequences shows that about 15% of bacteria possess more than one cpn60 gene. The major human pathogen Mycobacterium tuberculosis is one such organism. It has been known for many years that this bacterium produces a potently immunogenic 60-kDa protein, termed “common antigen” (43), which was subsequently identified as Hsp65 (or Cpn60.2) (10). In 1993, a second homologous gene was discovered and named cpn60.1 (10). The cpn60.1 gene is immediately downstream of the cpn10 gene in M. tuberculosis, which suggested that it might encode the major 60-kDa chaperonin of this organism.
Some uncertainly exists about the chaperone functions of both of the M. tuberculosis Cpn60 proteins because of the findings that both proteins exist as lower-order oligomers and not tetradecamers, possess very weak ATPase activity, and are only weakly active in refolding substrates (27). This biophysical characterization of both Cpn60 proteins is supported by the finding that M. tuberculosis Cpn60.2 crystallized as a dimer (25). This has led to questions being raised about the nature of the biological functions of these mycobacterial Cpn60 proteins (26), such as whether these two proteins are required for intracellular protein folding or have a different role, perhaps associated with cell survival in the various stressful environments inhabited by M. tuberculosis (26).
The Cpn60.2 (Hsp65) and Cpn10 proteins of M. tuberculosis are well established as potent immunomodulatory proteins which have the ability to vaccinate against various rodent models of human disease (39). This may relate to the finding that all of the M. tuberculosis chaperonins have the capacity to act as intercellular signals with human myeloid cells (3, 12, 15, 38) and vascular endothelial cells (40), causing induction of a proinflammatory phenotype. M. tuberculosis Cpn60.1 is a more potent activator of human monocyte cytokine synthesis than is Cpn60.2 (12). Paradoxically, it has also been reported that acute administration of M. tuberculosis Cpn60.1 (31) or Mycobacterium leprae Cpn60.2 (30), but not M. tuberculosis Cpn60.2 (31), can inhibit experimental allergic asthma in the mouse, suggesting that these proteins can also act as anti-inflammatory signals. This raises the question of the role played by any Cpn60 protein released by mycobacteria.
To determine the roles played by these two Cpn60 proteins, we attempted to inactivate each cpn60 gene separately, as well as the cpn10 gene. We demonstrated that it was impossible to produce M. tuberculosis mutants lacking either the cpn60.2 or cpn10 gene, suggesting that these genes are both essential for viability. We successfully generated a cpn60.1 deletion mutant of M. tuberculosis and examined the characteristics of the mutant in (i) an in vitro model of stationary phase (7) and the response to stress, (ii) murine bone marrow-derived macrophages and a macrophage-like cell line, and (iii) an immune resistant model of murine tuberculosis and a guinea pig infection model (19). We demonstrate an unexpected phenotype of the cpn60.1 deletion mutant in that it failed to induce a granulomatous response in the murine experimental model while showing normal behavior in the other systems.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. M. tuberculosis H37Rv was used as the parental strain to construct the mutants. M. tuberculosis H37Rv was grown in 7H9 medium containing 0.05% Tween 80 supplemented with 10% albumin-dextrose complex (Difco Laboratories) without disturbance. Viability (determined by measuring CFU) was estimated at the beginning of culture and then at 4- to 10-day intervals (for up to 100 days) on 7H11 agar medium supplemented with oleic-albumin-dextrose complex (Difco). Escherichia coli XL1 was used as a host strain for cloning and plasmid propagation and was grown on Luria-Bertani (LB) medium. Antibiotics used were as follows: ampicillin (Sigma), 100 μg/ml; kanamycin (Sigma), 20 μg/ml; gentamicin (Sigma), 20 μg/ml; and hygromycin (Life Technologies), 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli XL1 | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lacZΔM15 Tn10 | 32a |
| M. tuberculosis strains | ||
| H37Rv | WT | Trudeau mycobacterial culture collection |
| YHΔcpn60.1 | M. tuberculosis H37Rv cpn60.1 mutant | This work |
| YHcpn60.1Comp | M. tuberculosis H37Rv complemented cpn60.1 mutant | This work |
| YHcpn60.2C | M. tuberculosis H37Rv WT with an additional copy of cpn60.2 | This work |
| YHcpn60.2CD | cpn60.2 mutation in YHcpn60.2C | This work |
| YHcpn10C | M. tuberculosis H37Rv WT with an additional copy of cpn10 | This work |
| YHcpn10CD | cpn10 mutation in YHcpn10C | This work |
| Plasmids | ||
| pGEM3Z | ColE1 replicon; Ampr | Promega |
| p2NIL | Mutation vector | 20 |
| pGOAL19 | hyg PAg85-lacZ Phsp65-sacB pacI cassette vector; Ampr | 20 |
| pUC-GM-int | Integrase of phage L5, attP site, and Genr marker cassette | 11 |
| pYH10 | Insertion of HindIII fragment of pUC-Gm-Int in pGEM3Z | This work |
| pYHcpn60.1 | Insertion of cpn60.1 gene in pGEM3Z | This work |
| pYHcpn60.2 | Insertion of cpn60.2 gene in PGEM3Z | This work |
| pYHcpn10 | Insertion of cpn10 gene in PGEM3Z | This work |
| p2NILcpn60.1 | Insertion of disrupted cpn60.1 in p2NIL | This work |
| p2NILcpn60.2A | Insertion of disrupted cpn60.2 in p2NIL | This work |
| p2NILcpn60.2B | Insertion of NruI-disrupted cpn60.2 in p2NIL | This work |
| p2NILcpn10 | Insertion of disrupted cpn10 in p2NIL | This work |
| p2NIL60.1HS | Insertion of Hygr gene and sacB in p2NILcpn60.1 | This work |
| p2NIL60.2HS | Insertion of Hygr and sacB in p2NILcpn60.2A | This work |
| p2NIL10HS | Insertion of Hygr and sacB in p2NILcpn10 | This work |
| pYH10cpn60.1C | Insertion of cpn60.1 in pYH10 | This work |
| pYH10cpn60.2C | Insertion of cpn60.2 in pYH10 | This work |
| pYH10cpn10 C | Insertion of cpn10 in pYH10 | This work |
Mutant construction and complementation.
Mutant construction was based on plasmids p2NIL and pGOAL19 as described previously (20). To construct the cpn60.1 mutant, a 3,852-bp PCR product containing the cpn60.1 gene and approximately 1 kb of flanking sequences at each end of the gene was amplified using M. tuberculosis genomic DNA as the template and primers cpn60.1F (5′-ATTGAATTCGTTCAGCTTCTCCGGGCTC-3′) and cpn60.1R (5′-ATTGAATTCGGACACCACCAACGGATCC-3′). The PCR product was cloned into the HindIII site of pGEM3Z (Promega) to form pYHcpn60.1. An 873-bp MluI fragment in the cpn60.1 coding region was deleted. The disrupted cpn60.1 gene was cloned into the HindIII site of p2NIL (20) to make p2NILcpn60.1. To construct the cpn60.2 mutant, a 3,667-bp PCR product containing the cpn60.2 gene and approximately 1 kb of flanking sequence was amplified using primers cpn60.2F (5′-ATTAAGCTTTGTTGGAGCCGCCAGGGTGA-3′) and cpn60.2R (5′-ATTAAGCTTGGCTCCCTGAACAGCGGCAT-3′). The PCR product was cloned into the HindIII site of pGEM3Z (Promega) to form pYHcpn60.2. Two attempts were carried out to delete the cpn60.2 gene. Firstly, the cpn60.2 gene was deleted by PCR with primers cpn60.2MF (5′-ATTCCATGGGGGTGGCATGGATTTCTGAC-3′) and cpn60.2MR (5′-ATTCCATGGGCGAAGTGATTCCTCCGGAT-3′), which were designed outwardly starting from the start and stop codons of the cpn60.2 gene, using pYHcpn60.2 as a template. The PCR product, which contained only the flanking sequences of cpn60.2, was cloned into the HindIII site of p2NIL (20) to make p2NILcpn60.2A. Secondly, a 473-bp NruI fragment which included 173 bp of upstream sequence from the cpn60.2 start codon was deleted. The disrupted cpn60.2 gene with flanking sequences was cloned into the HindIII site of p2NIL to make p2NILcpn60.2B. To construct the cpn10 mutant, a PCR product of 2,324 bp, including the cpn10 gene and the flanking sequences, was amplified using primers cpn10F (5′-AATAAGCTTATTGTCGCGGCCACCATCG-3′) and cpn10R (5′-AATAAGCTTAACGCCTTACGGCGGTCAC-3′) and cloned into pGEN3Z to form pYHcpn10. The cpn10 gene was deleted by PCR with primers cpn10MF (5′-AGCGGCCGCGATTGGAGCCCTCCACTA-3′) and cpn10MR (5′-AGCGGCCGCTGGCCGTCGTTTCCAAGT-3′), which were oriented outwardly starting from the start and stop codons of the cpn10 gene, using pYHcpn10 as a template. The PCR product, containing only two flanking sequences, was cloned into p2NIL to form p2NILcpn10. A hyg lacZ-sacB marker cassette from pGOAL19 (20) was cloned into the PacI sites of p2NILcpn60.1, p2NILcpn60.2, and p2NILcpn10 to form the final mutant constructs p2NIL60.1HS, p2NIL60.2HS, and p2NIL10HS, respectively. Electroporation of the final constructs into M. tuberculosis H37Rv cells and the selection of mutants were performed as described previously (21).
To make the complemented strains, a 2,670-bp HindIII fragment from plasmid pUC-Gm-Int (11), which contains a gene encoding the integrase of phage L5, the attP site, and a gentamicin resistance marker cassette, was cloned into pGEM3Z and generated plasmid pYH10. For complementation of cpn60.1, a 2,188-bp DNA fragment containing the cpn60.1 gene and a 360-bp upstream sequence was amplified by PCR, using primers cpn60.1CF (5′-AATGAATTCTTCTCGTGCAGGCCAACGA-3′) and cpn60.1CR (5′-AATGAATTCTGATCGAGGCGTGCCGATC-3′), and then cloned into the EcoRI site of plasmid pYH10 to form pYH10cpn60.1C. The construct was transformed into the cpn60.1 deletion mutant by electroporation, and gentamicin-resistant transformants were selected.
To introduce an additional copy of the cpn60.2 gene into the wild-type (WT) strain, a 2,102-bp PCR product, which contained the cpn60.2 gene and a 304-bp upstream sequence, was amplified using primers cpn60.2CF (5′-ATTGAATTCTTGGCGGTCATGGGCCGAA-3′) and cpn60.2CR (5′-ATTGAATTCGCTGCGGTGTCGTACCCATC-3′) and then cloned into the EcoRI site of plasmid pYH10 to form pYH10cpn60.2C. Similarly, to introduce an additional copy of the cpn10 gene into the WT strain, a PCR product of 706 bp containing the cpn10 gene and 272 bp of upstream sequence was amplified using cpn10CF (5′-AATGAATTCCGGTGACCCGGACATTGCA-3′) and cpn10CR (5′-AATGAATTCGACGCGCGGTTTCGTCGTA-3′) and cloned into pYH10 to produce pYH10cpn10C. After electroporation of the constructs into the WT strain, gentamicin-resistant colonies were selected. All PCR products were sequenced using both primers to confirm that the sequences matched those expected.
Estimation of cell viability under stress conditions.
The M. tuberculosis strains were grown to late log phase for 10 days. A series of 10-ml cultures were used for the determination of CFU counts after exposure to environmental stresses. To induce thermal stress, the cultures were shifted to 55°C for 2 h. To expose cells to toxic macrophage radicals, cells were incubated with hydrogen peroxide or diethylenetriamine/nitric oxide adduct (to generate NO) at a final concentration of 5, 10, or 20 mM. For acidic stress, the culture medium was replaced with acidic 7H9 medium (pH 4). The cultures were then incubated at 37°C. At different times, CFU counts in the control and stressed cultures were determined. Each stress experiment was repeated three times.
Infection of bone marrow-derived macrophages.
Bone marrow-derived macrophages were prepared as described previously (34). Briefly, bone marrow cells were flushed from the femurs of female BALB/c mice (6 to 8 weeks old) and cultured in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 20% L cell conditioned medium, 100 U/ml of penicillin, and 100 μg/ml of streptomycin at 37°C in 5% CO2 for 7 days. Adherent macrophages were harvested and seeded at 106 cells per well in 24-well plates in the above culture medium without L cell conditioned medium, penicillin, and streptomycin. For the macrophage cell line J774A.1, the cells were grown to 80% confluence using the above medium without L cell conditioned medium and seeded as described above. The bone marrow-derived and J774A.1 macrophage cells were activated by the addition of gamma interferon (IFN-γ) (100 U/ml; R&D Systems) for 24 h, and this was followed by the addition of lipopolysaccharide (200 ng/ml; Sigma) for 3 h. The cells were infected with M. tuberculosis H37Rv (WT), the Δcpn60.1 mutant (YHΔcpn60.1), and the complemented strain (YHΔcpn60.1Comp) at a multiplicity of infection of 1:1 for 4 h. On day 0, 1, 2, 4, or 7 after infection, the cells were washed five times with warm culture medium and lysed with 0.1% Triton X-100. Various dilutions of the lysed cell suspension were made, and the numbers of bacteria present were enumerated by counting CFU. These macrophage infection experiments were repeated three times. On days 1, 2, and 3 after infection, tumor necrosis factor alpha (TNF-α) in the supernatants of the resting and activated J774A.1 macrophages was analyzed using enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems) according to the manufacturer's instructions.
Mouse infection model.
BALB/c mice (Harlan UK Ltd.) weighing 18 to 20 g were used for the mouse infection model. M. tuberculosis H37Rv WT, mutant, and complemented strains (2 × 104 CFU) were injected intravenously into mice. After 2 h or 1, 2, 3, 6, 12, or 15 weeks after infection, spleens and lungs from groups of four mice were removed rapidly after sacrifice, and sterile autopsy was performed. The organs were transferred to 2-ml tubes, with each containing 1 ml sterile distilled water and 2-mm-diameter glass beads. Lungs and spleens of the mice were homogenized using a reciprocal shaker (Thermo Hybaid Ltd.) for 40 seconds at speed 6.5. The homogenates were diluted, and CFU counts were performed. A part of each lung was fixed in 10% buffered formalin, paraffin embedded, and sectioned for histological examination, which was carried out commercially by a professional pathologist (Finn Pathologists UK). The mouse infection experiments were performed twice.
Guinea pig infection.
Female Hartley guinea pigs (450 to 500 g) (Harlan UK Ltd.) were used. The guinea pigs were infected with M. tuberculosis H37Rv WT, mutant, and complemented strains by intranasal challenge. The animals were anesthetized with a mixture of halothane and nitrous oxide. Twenty microliters of M. tuberculosis cell suspension, which contained approximately 104 CFU of bacilli, was administered into each nostril. After 12 weeks of infection, two animals in each group were sacrificed. The whole spleen and one of the two lungs from each animal were removed aseptically for bacteriological examination. The other lung was fixed in 10% buffered formalin, paraffin embedded, and sectioned for histological examination.
Cytokine mRNA levels in infected mice.
To determine the presence of proinflammatory cytokine mRNAs in murine tissues, the right lungs of BALB/c mice were removed, immediately fixed with RNAlater RNA stabilization reagent (Qiagen), and kept at −20°C. Thirty milligrams of the fixed lungs was used for RNA extraction, using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNAs were treated with RNase-free DNase Ι (Qiagen) to remove contaminating genomic DNA. The sequences of the oligonucleotides for the cytokines and β-actin were as follows: for interleukin-6 (IL-6), 5′-AGGAGACTTCACAGAGGAT-3′ (sense) and 5′-TCATGTACTCCAGGTAGCT-3′ (antisense); for TNF-α, 5′-CACGCTCTTCTGTCTACTG-3′ (sense) and 5′-TTGAAGAGAACCTGGGAGT-3′ (antisense); for IFN-γ, 5′-GCTACACACTGCATCTTG-3′ (sense) and 5′-CTGTTGCTGAAGAAGGTAG-3′ (antisense); for IL-12p40, 5′-GTAGAGGTGGACTGGACT-3′ (sense) and 5′-TGGTGCTTCACACTTCAG-3′ (antisense); and for β-actin, 5′-ATGGATGACGATATCGCT-3′ (sense) and 5′-ATGAGGTAGTCTGTCAGGT-3′ (antisense). Total RNA (5 μg) was used to synthesize cDNA. The RNA was transcribed in a total volume of 20 μl containing a 0.5 mM concentration (each) of dATP, dCTP, dGTP, and dTTP, 2.5 μM of antisense primer, 5 mM dithiothreitol, 40 units of RNasin (Promega), 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, and 200 units of Superscript II (Life Technologies). The reverse transcription (RT) reaction was carried out at 42°C for 1 h. Ten microliters of diluted cDNA was used for PCR amplification using the gene-specific primers described above. Each RT-PCR was repeated twice. The PCR products were separated in a 1% agarose gel, followed by transfer in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a Hybond-N+ filter (Amersham). The specific probe for each cytokine was amplified using mouse DNA as a template and the gene-specific primers described above and then labeled with [α-32P]dCTP (specific activity, >3,000 Ci mmol−1; ICN) by using the random priming method according to the instructions of the manufacturer (Amersham). The blots were hybridized with the specific probes overnight. After being washed, the blots were scanned using a Storm 840 phosphorimaging instrument at a 50-μm pixel resolution. The image files were analyzed using ImageQuant software (Molecular Dynamics). Quantity analysis of each PCR was standardized by determining the ratio of the intensity of the cytokine PCR band to that of the β-actin band and expressing it as the relative level of mRNA.
Extraction and analysis of free lipids and mycolic acids.
M. tuberculosis H37Rv WT and the YHΔcpn60.1 mutant were grown on 7H11 agar medium or in 7H9 medium (without Tween 80) to mid-log phase (about 10 days). The cells were harvested by centrifugation, washed with phosphate-buffered saline, and freeze-dried. The extraction of lipids was performed according to the method described previously (1). The apolar and polar lipid extracts were resuspended in chloroform-methanol (2:1), and 50 μg of crude lipid was applied to the corners of 6.6- by 6.6-cm pieces of Merck 5554 aluminum-backed thin-layer chromatography (TLC) plates. The plates were developed in a series of solvent systems, designed to cover the whole range of lipid polarities, as detailed previously (1). For apolar lipid extracts, these systems were named systems A to D, and for polar lipid extracts, they were named systems D and E (1). Lipids were visualized using 5% ethanolic molybdophosphoric acid and charring, and glycolipids were visualized by either spraying plates with α-naphthol-sulfuric acid followed by gentle charring of plates or using the Dittmer and Lester reagent, which is specific for phospholipids and glycophospholipids. Mycolic acid methyl esters were redissolved in diethyl ether. After centrifugation, the clear supernatant was again dried and resuspended in dichloromethane (100 μl), and a 50-μg aliquot was subjected to one-dimensional high-performance TLC, using two developments of hexane-ethyl acetate (95:5). Mycolic acid methyl esters were visualized using 5% ethanolic molybdophosphoric acid and charring.
Analysis of chaperonin gene transcription in M. tuberculosis H37Rv.
For the analysis of RNA accumulation under stress conditions, a series of 10-ml cultures of M. tuberculosis H37Rv were used. For heat shock, the cultures were shifted to 45°C for 30 min. For oxidative stress and alcohol shock, H2O2 (10 mM) or ethanol (5%) was added to the cultures for 30 min. For starvation stress, the cultures were thoroughly washed with phosphate-buffered saline, resuspended in 10 ml H2O, and then kept at 37°C for 2 h. To induce osmotic stress, the cultures were exposed to 1.5 M NaCl for 1 h. For acidic and alkaline stress, the culture medium was replaced with 7H9 medium at pH 4 and pH 10, respectively, for 30 min. RNAs were extracted after exposure to various stresses and analyzed by a previously constructed M. tuberculosis gridded DNA array (5) containing 82 M. tuberculosis genes. Each RNA sample was analyzed using three separate arrays. Array images were scanned using a Storm 840 phosphorimaging instrument at a 50-μm pixel resolution. The image files were analyzed using ImageQuant software (Molecular Dynamics). The pixel density of each spot was analyzed and exported to a Microsoft Excel spreadsheet. Background was subtracted by the software, using the local average background subtraction method. The mean intensity value for each pair of gene spots was divided by the total intensity of all gene spots in the miniarray and expressed as the percentage of total genes. The consistency of the percent intensities between duplicate spots on an array was 0.9. Changes in mRNA levels between two different conditions were determined by comparing the ratio of the mean percentage values of the corresponding gene spots on the two blots. A threshold value which was equal to the background level was used to determine the detectable expression of genes in each sample. Increases in gene expression are expressed as x-fold induction. Changes in gene expression, either up-regulation or down-regulation, under different conditions were considered significant if the changes in the mean percentage values among different cDNA probes exhibited at least a twofold difference (5). The hybridization data derived from the triplicate arrays were reproducible (P > 0.05; determined by a paired t test). These experiments were repeated twice with different preparations of RNA samples.
In vivo complementation of chaperonin genes.
The expression vector pTrc99AgroESL carries the lacIq gene and the trc promoter upstream of the E. coli groES-groEL operon. DNA fragments containing the M. tuberculosis cpn10, con60.1, and cpn60.2 genes were amplified by PCR using the following primers: cpn10F, 5′-ATTGAATTCATCGGCTAACCCCTGCGT-3′; cpn10R, 5′-ATTTCTAGACGGAAATCACCCGTGGTG-3′; 60.1F, 5′-AATACTAGTATGAGCAAGCTGATCGAATAC-3′; 60.1R, 5′-AATACTAGTGGTGAGCCCTATGGCGTT-3′; 60.2F, 5′-ACGACTAGTATGGCCAAGACAATTGCGTA-3′; and 60.2R, 5′-AATACTAGTCACAAAGGGACCGGGCTC-3′. The constructs containing different combinations of the M. tuberculosis chaperonin and groE genes were made by replacing groES or groEL in pTrc99AgroESL. The constructs were transformed into E. coli MGM100, in which the groESL operon is expressed under the control of the pBAD promoter (17). The expression of protein by E. coli MGM100 harboring each construct was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Viability of strains was determined at a range of temperatures by CFU counts on LB agar plates in the presence of glucose (0.5%), which leads to tight repression of the pBAD promoter, with or without IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 mM) to induce high expression of the chaperonin genes.
Statistical analysis.
The difference between different experimental groups was determined by a Student t test. P values of <0.05 were considered significant.
RESULTS
Inactivation of chaperonin genes in M. tuberculosis H37Rv.
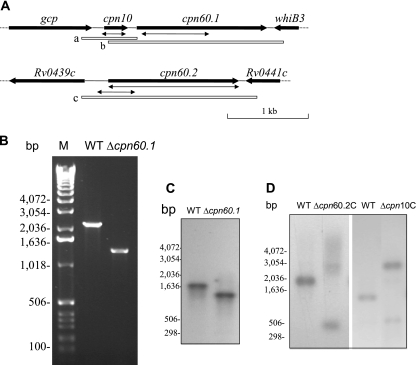
In order to clarify the function of the chaperonin genes in M. tuberculosis, a two-step mutagenesis strategy (20) was employed to disrupt and individually replace the cpn10, cpn60.1, and cpn60.2 genes. In M. tuberculosis, the cpn60.2 gene is not adjacent to the cpn10 gene but is located independently in the genome. In contrast, the cpn10 and cpn60.1 genes are adjacent in a possible operon with a 95-bp intergenic region (Fig. 1A).
FIG. 1.
Construction of M. tuberculosis cpn60.1 mutant. (A) Genomic context of cpn10, cpn60.1, and cpn60.2. The double arrows indicate the deletions made in these genes. The open boxes a, b, and c show the locations of the fragments used in complementation experiments. (B) PCR confirmation of the deletion of the cpn60.1 gene, using the primers cpn60.1CF and cpn60.1CR. (C) Southern blotting analysis confirming the deletion of the M. tuberculosis H37Rv cpn60.1 gene. DNAs from the WT and the YHcpn60.1Δ strain were digested with NcoI and hybridized with probe b, shown in panel A. (D) Southern blotting analysis confirming that deletion of the M. tuberculosis H37Rv cpn60.2 or cpn10 gene was possible only when a WT copy of the gene was present. DNAs from the WT and the YHΔcpn60.2C strain were digested with AclI and Esp3I and hybridized with probe c, shown in panel A. Also, DNAs from the WT and the YHΔcpn10C strain were digested with PvuII and hybridized with probe a, shown in panel A. Since the WT copy of cpn60.2 or cpn10 was integrated into the chromosomal DNA, digestion with the restriction enzymes resulted in a larger fragment containing the WT copy of the gene. M, molecular weight marker (Invitrogen). The experiments were repeated twice, with identical results.
An M. tuberculosis mutant lacking the cpn60.1 gene was successfully obtained and is referred to as YHΔcpn60.1. YHΔcpn60.1 contains a disrupted cpn60.1 gene which has an 873-bp DNA fragment deleted in its coding region (Fig. 1A). The presence of the mutation was confirmed by PCR (Fig. 1B) and Southern blotting analysis (Fig. 1C). Western blotting with a Cpn60.1-specific antibody confirmed that no Cpn60.1 protein is produced in this mutant (data not shown).
Despite two attempts to construct both complete and partial in-frame deletions, neither cpn60.2- nor cpn10-deleted M. tuberculosis strains were obtained. To verify that the cpn60.2 and cpn10 genes are essential in M. tuberculosis, we cloned an additional copy of cpn60.2 or cpn10 into an integrating vector and introduced this plasmid into the WT strain, which allowed the copy of the gene to integrate into the M. tuberculosis chromosome. Deletion of the chromosomal copy of the cpn60.2 or cpn10 gene in the presence of the insertion copy was now able to be achieved (Fig. 1D), and we termed the strains YHΔcpn60.2C and YHΔcpn10C, respectively. This strongly suggests that cpn60.2 and cpn10 are essential genes for M. tuberculosis.
To complement cpn60.1 in YHΔcpn60.1, the plasmid pYH10cpn60.1C, which contains the cpn60.1 coding region and a 360-bp upstream sequence of the gene, was transformed into YHΔcpn60.1, followed by selection for gentamicin resistance. The complemented strain was confirmed by PCR (data not shown) and is referred to as YHcpn60.1Comp.
In vitro growth and survival of YHΔcpn60.1 under stress conditions.
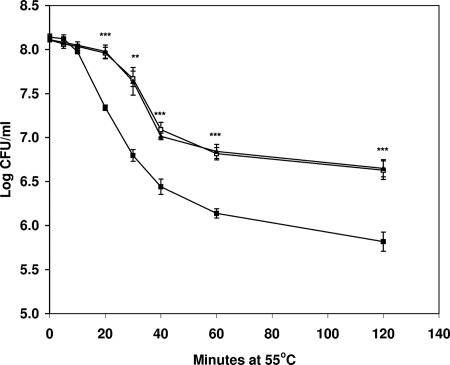
The WT, YHΔcpn60.1, and YHcpn60.1Comp strains were grown in 7H9 broth without disturbance for 100 days. At various times, samples were taken to assess CFU. Similar growth curves were obtained for both the WT and the mutant (data not shown), suggesting that the absence of the cpn60.1 gene had no significant effect on the growth of M. tuberculosis H37Rv in vitro. In order to determine if cpn60.1 plays any role in the response of M. tuberculosis to stress, we examined the survival of the WT, YHΔcpn60.1, and YHcpn60.1Comp strains under various stress conditions. The only major difference between the strains was that exposure to a temperature of 55°C killed YHΔcpn60.1 more rapidly than the WT. The CFU of YHΔcpn60.1 decreased fourfold more than the WT at 20 min, and sixfold more killing of the mutant was seen at 2 h (P < 0.001). The parental level of heat sensitivity was restored in the YHcpn60.1Comp strain (Fig. 2). Thus, the loss of the cpn60.1 gene rendered the strain more sensitive to high temperature. In contrast, no significant differences were observed between the WT and YHΔcpn60.1 strains upon treatment with H2O2 and NO and growth at pH 4 (data not shown), mimicking the stress conditions M. tuberculosis is most likely to encounter during infection.
FIG. 2.
Inactivation of cpn60.1 renders cells more susceptible to elevated temperature. The graph shows a time course of survival for WT cells (□) compared with YHΔcpn60.1 (▪) and YHcpn60.1Comp (▴) cells at 55°C. The data shown are averages for three independent experiments ± standard errors. Statistical significance was determined by Student's t test (n = 3) (**, P < 0.01; ***, P < 0.001).
Cell wall lipid analysis.
A previous study demonstrated that inactivation of cpn60.1 in Mycobacterium smegmatis results in alterations in mycolic acid biosynthesis and changes in biofilm formation (18). To determine if there were changes in cell wall composition in the equivalent M. tuberculosis mutant, we examined the lipids and mycolic acids of the WT and the YHΔcpn60.1 mutant grown on 7H11 agar plates or in 7H9 broth, using TLC. In the previous study, the lipid and mycolic acid analysis of M. smegmatis biofilms used cells which were grown on M63 salts minimal medium (18). We attempted to grow M. tuberculosis H37Rv on the same medium, but the bacilli grew poorly and no biofilms were formed. We then grew the bacterium on 7H11 agar medium. No differences were observed in nonpolar lipid, polar lipid, or mycolic acid composition between the WT strain and the YHΔcpn60.1 mutant grown on 7H11 agar plates (data not shown). We also grew the WT and the mutant in 7H9 medium without Tween 80, in which the bacilli formed clumps and surface pellicles. Again, no differences were found in lipid and mycolic acid composition (data not shown). The lipid profiles of the WT and the mutant were the same under all growth conditions we tested (data not shown).
Growth and survival of YHΔcpn60.1 in macrophages.
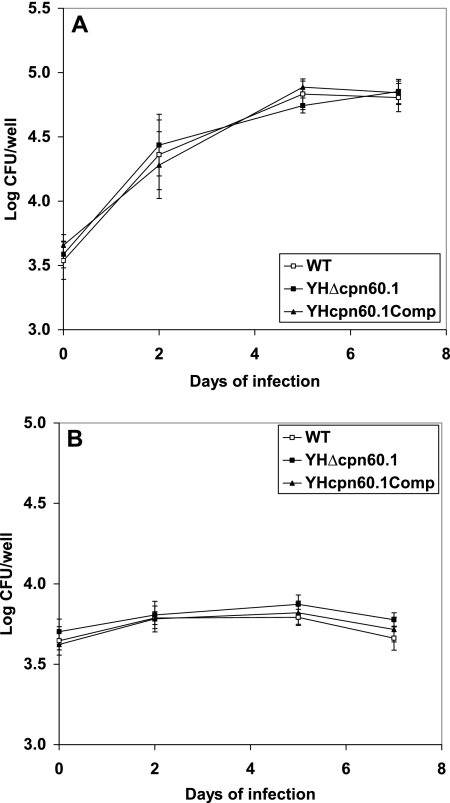
In order to investigate the ability of YHΔcpn60.1 to replicate and persist inside macrophages, resting and activated murine bone marrow-derived macrophages and a macrophage-like cell line, J774A, were infected with log-phase cultures of the WT, YHΔcpn60.1, and YHcpn60.1Comp strains. No significant differences were found between the cell numbers of each strain recovered from either resting or activated bone marrow-derived macrophages (Fig. 3). Similar results were observed with the J774A.1 cell line (data not shown).
FIG. 3.
Growth and survival of M. tuberculosis Δcpn60.1 in resting and IFN-γ-activated macrophages. (A) Infection in resting bone marrow-derived macrophages. (B) Infection in IFN-γ-activated bone marrow-derived macrophages. □, H37Rv WT; ▪, YHΔcpn60.1; ▴, YHcpn60.1 ΔComp. These results are the means and standard deviations derived from three independent experiments carried out in triplicate.
Growth and persistence of YHΔcpn60.1 in immunocompetent mice.
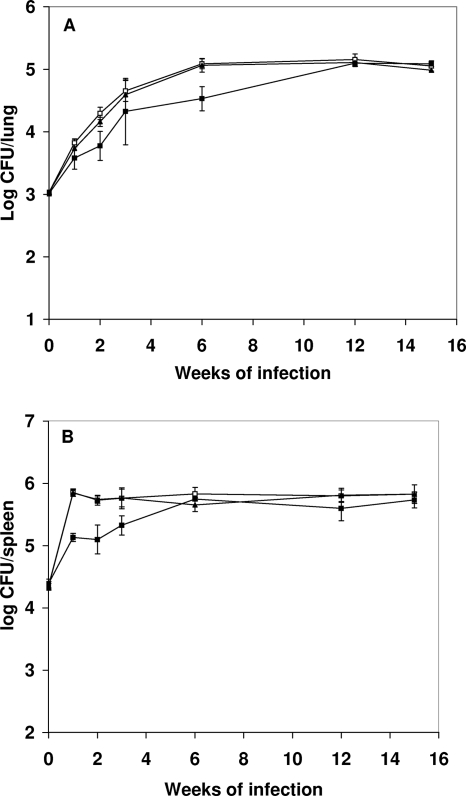
To determine whether the deletion of the cpn60.1 gene affected growth and persistence in vivo, a mouse model in which M. tuberculosis persists in the animals for the duration of the experiment was used (19). In this model, a low dose of M. tuberculosis is used to infect immunocompetent mice. M. tuberculosis exhibits log-phase growth initially. After 2 to 3 weeks of infection, the mice develop an acquired immune response which restricts growth of the bacteria. Using this model, we can distinguish the ability of the WT and the YHΔcpn60.1 mutant to grow and survive in the face of host innate and adaptive immunity. The WT, YHΔcpn60.1, and YHcpn60.1Comp strains were injected intravenously, with each mouse being given 3 × 104 CFU of bacteria. The viability of the injected organisms was determined by taking samples from mice, recovering bacteria, and counting CFU for the 15-week duration of the experiment. In the lungs, there was an initial lower growth rate of YHΔcpn60.1, showing 0.5, 0.33, 0.33, and 0.5 log fewer cells in the lungs at 1, 2, 3, and 6 weeks, respectively, but by 12 weeks there was no significant difference between the mutant and the WT cells (Fig. 4A). In the spleen, the growth of YHΔcpn60.1 was attenuated for the first 3 weeks of infection and then reached the same level as the WT (Fig. 4B). Parental levels of growth and survival were seen in YHcpn60.1Comp.
FIG. 4.
Growth of WT and YHΔcpn60.1 in the lungs and spleens of mice. Mice were infected with 3 × 104 bacteria, and at various times over a 15-week period, groups of mice were sacrificed and the numbers of bacteria in the lung (A) or spleen (B) were measured. The numbers of WT (□), isogenic mutant (▪), and complemented mutant (▴) cells in these two tissues were not significantly different at the last two time points measured. The experiments were repeated twice, with similar CFU counts for lungs and spleens.
Lung pathology in infected mice.
Histopathological examination of the lungs from BALB/c mice infected with the WT and the mutant was performed at 3, 6, and 15 weeks of infection. No significant difference was observed in both WT- and mutant-infected lungs at 3 weeks of infection (Fig. 5A and B), despite the fact that the CFU counts in the WT strain-infected lungs were about 0.33 log higher than those in the mutant-infected lungs. In animals killed after 6 weeks of infection, the lungs infected with the WT strain had moderate to marked diffuse increases in cellularity of the alveolar walls, showing several substantial focal accumulations of mixed inflammatory cells as well as occasional moderate aggregation of alveolar macrophages (Fig. 5D). At 15 weeks of infection, the lungs infected with the WT organism showed large areas of granulomatous inflammation (encompassing approximately 50% of the total lung area), with changes consistent with early granuloma formation (Fig. 5G). In complete contrast, in animals infected with YHΔcpn60.1, the infected lungs showed only slight increases in the numbers of inflammatory cells in the alveolar walls, with normal alveoli and airways evident throughout the lungs (Fig. 5E and H). This indicated that the mutant failed to cause classical tubercular disease. Normal granulomatous inflammation was observed in mouse lungs infected with the complemented strain (Fig. 5F and I). The histopathological examination was performed using three mice per group, and three slides were examined for each lung. The experiment was repeated, with identical results. Representative histology of the mouse lung for each group is shown.
FIG. 5.
Lung histology in mice and guinea pigs infected with the WT, YHΔcpn60.1, and complemented strains. Hematoxylin- and eosin-stained sections (magnification, ×10) show mouse lungs infected with the WT (A), YHΔcpn60.1 (B), and YHcpn60.1Comp (C) strains for 3 weeks; mouse lungs infected with the WT (D), YHΔcpn60.1 (E), and YHcpn60.1Comp (F) strains for 6 weeks; and mouse lungs infected with the WT (G), YHΔcpn60.1 (H), and YHcpn60.1Comp (I) strains for 15 weeks. Histopathological examination was performed using three mice in each group. Three sections from each mouse were examined. The data shown are representative of lung sections from three animals in each experimental group. Guinea pig lungs infected with the WT (J), YHΔcpn60.1 (K), and YHcpn60.1Comp (L) strains for 12 weeks are also shown. Two guinea pigs in each group were used for histopathological examination.
In order to further confirm that the lack of granulomas in mouse lungs was due to the deletion of the cpn60.1 gene, we infected guinea pigs with the WT, YHΔcpn60.1, and YHcpn60.1Comp strains via the nostril route. As shown in Fig. 5J, after 12 weeks of infection, the lungs infected with the WT strain had large numbers of inflammatory cells, including macrophages, lymphocytes, and granulocytes. In contrast, the YHΔcpn60.1-infected lungs (Fig. 5K) were largely normal, with only a slight increase of cellularity of alveolar walls. The same level of granulomatous inflammation as that for the WT strain was seen in the lungs infected by the complemented strain (Fig. 5L). CFU counts in the organs were 3.75 log CFU/spleen and 4.58 log CFU/lung for mice infected with the WT strain, 3.76 log CFU/spleen and 4.51 log CFU/lung for mice infected with the YHΔcpn60.1 strain, and 3.73 log CFU/ spleen and 4.57 log CFU/lung for mice infected with the YHcpn60.1Comp strain.
RT-PCR analysis of mouse cytokine expression in WT- and Δcpn60.1 mutant-infected mouse lungs.
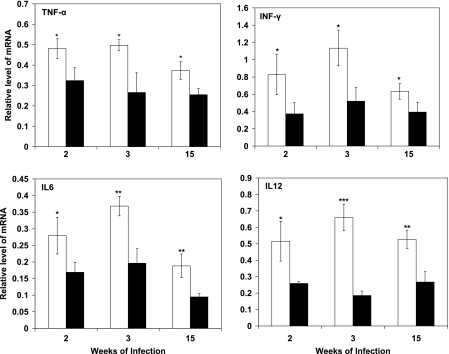
To investigate if the lack of granuloma formation in mice was a result of changes in the host immune response, we monitored the mRNA levels of TNF-α, IFN-γ, IL-6, and IL-12 in lungs infected with the WT or the YHΔcpn60.1 mutant, using semiquantitative RT-PCR. The levels of all cytokine mRNAs were significantly lower in the lungs infected with YHΔcpn60.1. There was a 1.5- to 3.5-fold reduction of these cytokines at 2, 3, and 15 weeks of infection (P < 0.05) (Fig. 6). The mRNA levels in uninfected mouse lungs were indistinguishable from the background and thus were treated as zero. We also examined cytokine expression in a macrophage-like cell line. After infection with the WT or the mutant, similar expression levels of TNF-α, IL-6, IL-12, and IL-10 were observed (data not shown).
FIG. 6.
Cytokine mRNA expression in the lungs of mice infected with either M. tuberculosis WT or YHΔcpn60.1. RNAs were extracted from the lungs of mice infected with the WT or YHΔcpn60.1 for 2, 3, and 15 weeks and subject to semiquantitative RT-PCR. The intensities of the PCR bands were normalized with those of β-actin. The mRNA level at each time point was examined using groups of three mice. Open bars, WT-infected lungs; filled bars, YHΔcpn60.1 mutant-infected lungs. The values are the means for three RT-PCR analyses of RNAs extracted from three mice, and the results were confirmed in one independent repeat experiment. Statistical significance was determined by Student's t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
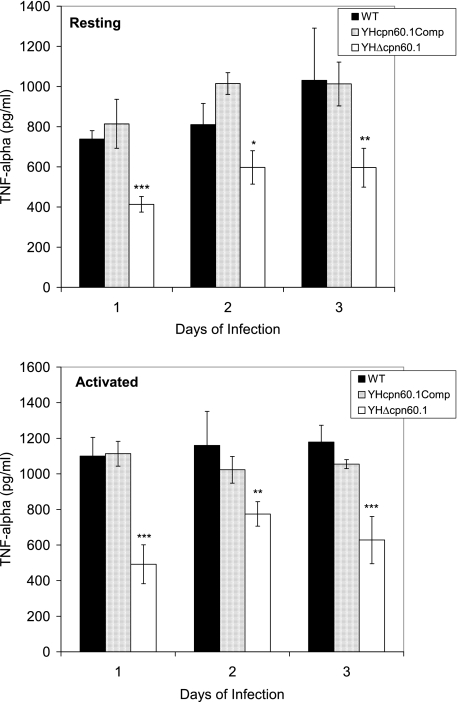
ELISA analysis of TNF-α levels in WT- and mutant-infected macrophages.
To further confirm that the mutant lacking the cpn60.1 gene fails to induce the same levels of cytokines in the host as the WT, we measured the TNF-α levels in infected J774A.1 macrophages. Similar to the cytokine mRNA levels observed in the infected mice, the TNF-α levels of the Δcpn60.1 mutant-infected macrophages were significantly reduced on days 1, 2, and 3 of infection in both resting and activated macrophages (Fig. 7). The TNF-α level was recovered in the macrophages infected with the complemented strain. The CFU counts of the mutant, the WT, and the complemented strain at those time points when the cytokines were measured were the same, indicating that the cpn60.1 mutant retained the ability to invade and survive in macrophages but stimulated significantly lower levels of TNF-α.
FIG. 7.
TNF-α levels in J774A.1 macrophages infected with the M. tuberculosis WT, YHΔcpn60.1, and complemented strains, determined by ELISA. TNF-α levels were measured in both resting macrophages and IFN-γ-activated macrophages. The data shown are the averages for two independent experiments ± standard errors. Statistical significance was determined by Student's t test (n = 2) (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
In vivo complementation of E. coli groEL and groES by M. tuberculosis genes.
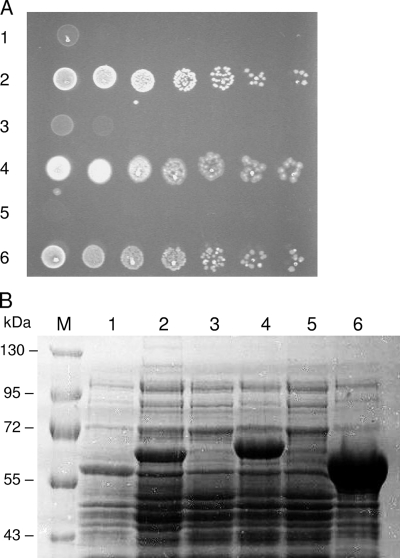
In order to investigate whether M. tuberculosis cpn gene products can function as molecular chaperones, an E. coli in vivo complementation system was used. In this model, the chromosomal groE promoter has been replaced by the pBAD promoter from the arabinose operon, which means that synthesis of native GroEL and GroES is completely repressed by the presence of glucose but can be induced in the presence of arabinose. This allows the ability of heterologous or mutant chaperonins to chaperone the folding of GroEL-dependent proteins in vivo to be determined (36). Constructs expressing the M. tuberculosis chaperonin proteins from the strong trc promoter were transformed into E. coli MGM100 to examine if they supported cell growth and therefore functioned as molecular chaperones. As shown in Fig. 8A, Cpn60.2 coexpressed with either Cpn10 or GroES was able to support normal E. coli growth at 37°C, although cpn60.2 plus cpn10 complemented growth only in the present of IPTG. cpn60.2 plus groES always showed good complementation with and without IPTG. This may be due to a combination of leaky expression and differences in the efficiency of translation of Cpn60.2 depending on the precise sequence of the DNA immediately upstream of it. Interestingly, the molecular mass of Cpn60.2 measured by SDS-PAGE differed slightly depending on whether it was coexpressed with Cpn10 or with GroES (Fig. 8B). The reason for this difference is not clear, but N-terminal sequence and mass spectrometry analysis showed that it was not due to N-terminal cleavage (data not shown). Cpn60.1 was expressed at reasonable levels with GroES but failed to complement E. coli MGM100 at any temperature tested. When it was expressed with Cpn10, expression was extremely poor and it could only be seen on Western blots (data not shown); complementation also did not occur in this case. These results clearly show that Cpn60.2 and Cpn10 can function as molecular chaperones in E. coli but that Cpn60.1 cannot, at least under the conditions of this assay.
FIG. 8.
(A) Cultures of MGM100 expressing different combinations of chaperonin proteins were grown overnight in arabinose, serially diluted in LB, and spotted onto plates containing 0.2% glucose and 0.1 mM IPTG. The strains contained the following plasmids: 1, ptrc-groES-cpn60.1; 2, ptrc-groES-cpn60.2; 3, ptrc-cpn10-cpn60.1; 4, ptrc-cpn10-cpn60.2; 5, ptrc99A (vector only); and 6, ptrc-groES-groEL. (B) 8% SDS-PAGE gel with proteins expressed by the strains in panel A, loaded in the same order. Lane M contains molecular weight markers. These experiments were repeated twice, with similar results.
Effect of stress on chaperonin mRNA levels.
In order to determine how cpn60.1, cpn60.2, and cpn10 were expressed under stress conditions, we examined the mRNA levels of these genes in the WT strain by using a previously described mini DNA array containing 82 M. tuberculosis genes (5). The M. tuberculosis H37Rv cultures were exposed to a variety of stress conditions, including elevated temperature, increased osmolarity, low and high pH, starvation, and hydrogen peroxide. Among the 82 genes, mRNA levels were significantly increased for about 30 genes, including those which were previously described to be induced under stress conditions, for example, katG (16), which was increased about 10-fold after exposure to H2O2; and sigB (6) and sigE (14), which were increased in response to heat shock, H2O2, ethanol, starvation, and hyperosmolarity (data not shown). As shown in Fig. 9, cpn10 showed marked increases in intracellular mRNA levels. mRNA levels of cpn60.2 increased significantly apart from the case under acidic conditions. In contrast, the expression of cpn60.1 showed significant increases only under stress conditions such as elevated temperature and hyperosmolarity (Fig. 9). The cpn10 gene rarely had a similar pattern of expression to cpn60.1, but its expression in response to heat shock, pH 10, and starvation was consistent with that of cpn60.2 (P > 0.1, P > 0.5, and P > 0.05, respectively). The primers which were used to synthesize cpn60.1 and cpn60.2 cDNAs (radioactivity-labeled probes) were designed within the conserved sequence of each gene in order to avoid cross-hybridization between these two genes.
FIG. 9.
Cellular levels of mRNAs for M. tuberculosis Cpn60.1, Cpn60.2, and Cpn10 proteins under stress conditions. M. tuberculosis in culture was exposed to a variety of physical, chemical, and biological stressors, and the mRNA levels of cpn10, cpn60.1, and cpn60.2 were determined. The results are the means for three independent experiments ± standard errors. In each experiment, RNAs from one single extraction were examined by triplicate array analysis.
DISCUSSION
We have taken several approaches to clarify the biological functions of the M. tuberculosis Cpn60 chaperonins. We attempted to inactivate the individual cpn60.1, cpn60.2, and cpn10 genes. It proved impossible to inactivate cpn60.2 or cpn10 unless additional copies of these genes were present on the chromosome. In contrast, it was possible to inactivate the cpn60.1 gene. Thus, Cpn60.2 has an essential role in M. tuberculosis for which Cpn60.1 cannot substitute, at least at its normal levels of expression. This confirms the previous finding that M. smegmatis cpn60.2 is an essential gene whose product functions as a housekeeping molecular chaperone (18). Our results also confirm and extend an earlier study using global transposon mutagenesis which suggested that both the cpn60.2 and cpn10 genes were required by M. tuberculosis for optimal growth and that cpn60.1 was a nonessential gene (33). It is interesting that although cpn10 and cpn60.1 are adjacent in the chromosome, their expression profiles under some stress conditions are distinctively different, which suggests that these two genes are differentially regulated. Previous work demonstrated that cpn60.1 might not be cotranscribed with cpn10 (10), as these two genes contain independent transcriptional start sites. However, in spite of their separate locations in the genome, cpn10 and cpn60.2 share similar patterns of expression and are both required for M. tuberculosis to grow and survive. This indicates that these two genes are under similar mechanisms of regulation and is consistent with the two proteins acting together, as would be predicted from the E. coli chaperonin system.
Inactivation of cpn60.1 had few consequences other than increased susceptibility to high temperature (55°C), which suggests that cpn60.1 has a role in thermotolerance in M. tuberculosis. It is interesting that the loss of thermotolerance in the cpn60.1 mutant was seen during the first 20 to 40 min of heat shock, when killing of the mutant was about 1 log greater than that of the WT strain, but thereafter the two survival curves became more or less parallel. This suggests that an alternative chaperoning activity may be induced by heat shock, compensating for the loss of Cpn60.1 at later time points. The YHΔcpn60.1 mutant was equivalent to the WT when exposed to pathophysiological stresses, such as H2O2, NO, and low pH, which would be encountered when bacteria come into contact with host leukocytes. The loss of the cpn60.1 gene did not inhibit the ability of M. tuberculosis to survive and grow within resting or activated macrophages, supporting the previous findings.
We demonstrated that YHΔcpn60.1 grew slightly slower than the WT strain or YHcpn60.1Comp in the mouse lung and spleen in early infection, but by 12 weeks the numbers of organisms in these tissues were equivalent. It is not known why the mutant shows slower growth in the first few weeks of infection yet continues to grow after the onset of the acquired immune response and then reaches a plateau 12 weeks after infection. This continued growth could be due to a reduced immune response, and indeed, this was seen during the course of infection in mutant-infected mice (Fig. 6). At the early stage of infection (1 to 3 weeks), the difference in CFU counts per organ (0.3- to 0.5-log difference) led to no significant difference in the inflammation of the lungs infected by both the WT and the mutant, with both showing moderate, diffuse increases in cellularity of the alveolar walls containing inflammatory cells. It was surprising, therefore, to find that the lungs of mice infected with the isogenic mutant showed much less evidence of inflammation than did those of mice infected with the WT at 15 weeks of infection, 3 weeks after the CFU counts reached the same level as that of the WT strain. The mutant lacked the ability to stimulate granulomatous inflammation. Furthermore, the spleen size in mice infected with YHΔcpn60.1 was smaller than that in mice infected with the WT (data not shown), again showing that the mutant is less inflammatory. This lack of granulomatous formation was confirmed in guinea pigs, using an aerosol infection model via nostril administration. As an independent measure of inflammation, the mRNA levels of various key proinflammatory cytokines (IFN-γ, TNF-α, IL-6, and IL-12) in the lungs of mice were measured and shown to be lower in the animals infected with YHΔcpn60. The reduced cytokine production caused by the cpn60.1 mutant was also confirmed in macrophages analyzed by ELISA. These results indicated that the reduced pathology caused by this mutant may be due to failure of the strain to stimulate T-helper 1-type immunity, which is the major contributor to controlling M. tuberculosis disease progress (28). It is not uncommon for a loss of one protein in M. tuberculosis to result in reduced tuberculous disease. Work in other laboratories demonstrated that an M. tuberculosis strain lacking whiB3 failed to cause granulomatous inflammation in mice, despite the CFU counts of the whiB3 mutant and the WT being similar in animal organs during the course of infection (35). WhiB3 may play a role as a transcriptional regulator which controls genes involved in stimulation of the host immune response. Another example is an M. tuberculosis mutant containing a disrupted sigH gene, which grows and survives in mice but produces a reduced inflammatory pathology in mice (8). As an M. tuberculosis sigma factor, sigH regulates the transcription of many genes, any of which could be responsible for the reduction of immunopathology (8). The mechanisms by which the cpn60.1 gene operates to regulate the virulence of the bacterium are unknown. Hatfull et al. recently reported that inactivation of the cpn60.1 gene in M. smegmatis results in an alteration in biofilm formation and suggested that this might be relevant to the virulence of M. tuberculosis (18). The proposed mechanism is that Cpn60.1 associates with the 3-oxoacyl (acyl carrier protein) synthase proteins, KasA and KasB, involved in mycolic acid synthesis (18). These are not among the proteins that interact with GroEL in E. coli. We looked for alterations in cell wall lipid composition in our WT and YHΔcpn60.1 mutant. We grew the bacilli on 7H11 agar or in 7H9 medium without Tween but failed to see any differences between the WT and mutant strains. This discrepancy may due to the fact that the growth of the bacilli which we prepared on agar plates or in 7H9 medium was not the same as that described in the M. smegmatis study. We attempted to grow M. tuberculosis H37Rv strains in M. smegmatis biofilm medium (18), but the bacilli failed to grow and form biofilms, indicating that either M. tuberculosis does not form biofilms or the conditions that promote changes in lipids similar to those in M. smegmatis have yet to be identified. Of course, M. smegmatis is a very distinct organism from M. tuberculosis (41). For example, M. smegmatis is not a pathogen. Also, it contains three cpn60 genes. Analysis of a transposon mutant library of Mycobacterium avium revealed a number of genes (not including cpn60.1) that impair biofilm formation (42). The virtually identical Cpn60.2 proteins from M. tuberculosis and M. leprae have very different anti-inflammatory actions in mice (30, 31), and in general, different Cpn60 proteins exhibit a bewildering variety of activities (4, 32). Given these findings, it is not surprising that M. tuberculosis Cpn60.1 does not behave identically to the M. smegmatis homologue. It will be intriguing to discover whether Cpn60.1 from M. tuberculosis can complement the cpn60.1 defect in M. smegmatis, and such experiments are currently in progress.
We have shown here that M. tuberculosis Cpn60.2 can, in concert with Cpn10 or GroES, restore viability of the MGM100 strain when it is grown in glucose to switch off the endogenous groES/groEL operon. Cpn60.2 combined with Cpn10 or GroES can therefore fold at least some of what have been termed class III proteins in E. coli (9). These are 84 low-abundance proteins in E. coli that are thought to have the common characteristic of the persistence of exposed hydrophobic regions during the folding process and which require the GroEL and GroES proteins in order to fold. They include 13 essential E. coli proteins, thus accounting for the indispensable nature of the chaperonin system (9). This result shows that despite the lower stability of the Cpn60.2 complex compared to that of GroEL (27), Cpn60.2 is highly likely to function as a tetradecamer in vivo, since it can interact with the heptameric E. coli GroES complex. Cpn60.1, however, was unable to complement for the loss of GroEL, even when it was expressed to levels where Cpn60.2 did complement, implying that this protein cannot function as a molecular chaperone in E. coli. Cpn60.1 may bind a different substrate or substrates from those of GroEL and Cpn60.2 or may have some other function altogether in the cell. Current experiments are testing these possibilities.
The chronic granulomatous pathology of tuberculosis is driven, in an as yet undiscovered manner, by M. tuberculosis, and the overproduction of host proinflammatory cytokines, such as TNF-α, plays a key role. There are now a number of reports that bacterial Cpn60 proteins, including those from M. tuberculosis, can stimulate human monocytes to secrete proinflammatory cytokines (3, 12, 22-24, 29, 37). It is interesting that in spite of 70% sequence identity, the Cpn60.1 protein of M. tuberculosis is both more potent and more efficacious than the Cpn60.2 protein and appears to activate myeloid cells through a different receptor (12). The simplest explanation is that it is the absence of the proinflammatory activity of the Cpn60.1 protein that accounts for the failure of the cpn60.1 mutant to induce granulomatous inflammation.
In conclusion, our results show that Cpn60.1 is a nonessential molecular chaperone with minimal cell stress protein activity. It plays an important role in stimulating proinflammatory cytokine production, which is essential for tuberculosis disease manifestation.
Acknowledgments
A European Commission project grant (QLRT-2001-01682) supported this work. The financial support of the Burton Medical Trust is gratefully acknowledged. P.T. was supported by ARC Programme grant HO60. G.S.B. acknowledges support from James Bardrick in the form of a personal research chair, as a former Lister Institute-Jenner Research Fellow, and from the Medical Research Council (United Kingdom). H.L. and P.A.L. acknowledge the generous support of the Darwin Trust of Edinburgh.
We thank Ana Cehovin for helping with the cytokine ELISA analysis.
We have no conflicting financial interests.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Besra, G. S. 1998. Preparation of cell-wall fractions from mycobacteria. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 2.Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 1711379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedland, J. S., R. Shattock, D. G. Remick, and G. E. Griffin. 1993. Mycobacterial 65-kD heat shock protein induces release of proinflammatory cytokines from human monocytic cells. Clin. Exp. Immunol. 9158-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson, B., E. Allan, and A. R. Coates. 2006. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 743693-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, Y., and A. R. Coates. 2001. Increased levels of sigJ mRNA in late stationary phase cultures of Mycobacterium tuberculosis detected by DNA array hybridisation. FEMS Microbiol. Lett. 20259-65. [DOI] [PubMed] [Google Scholar]
- 6.Hu, Y., and A. R. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, Y., J. A. Mangan, J. Dhillon, K. M. Sole, D. A. Mitchison, P. D. Butcher, and A. R. Coates. 2000. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J. Bacteriol. 1826358-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 998330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerner, M. J., D. J. Naylor, Y. Ishihama, T. Maier, H. C. Chang, A. P. Stines, C. Georgopoulos, D. Frishman, M. Hayer-Hartl, M. Mann, and F. U. Hartl. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122209-220. [DOI] [PubMed] [Google Scholar]
- 10.Kong, T. H., A. R. Coates, P. D. Butcher, C. J. Hickman, and T. M. Shinnick. 1993. Mycobacterium tuberculosis expresses two chaperonin-60 homologs. Proc. Natl. Acad. Sci. USA 902608-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 883111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewthwaite, J. C., A. R. Coates, P. Tormay, M. Singh, P. Mascagni, S. Poole, M. Roberts, L. Sharp, and B. Henderson. 2001. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect. Immun. 697349-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 4493-140. [DOI] [PubMed] [Google Scholar]
- 14.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41423-437. [DOI] [PubMed] [Google Scholar]
- 15.Meghji, S., P. A. White, S. P. Nair, K. Reddi, K. Heron, B. Henderson, A. Zaliani, G. Fossati, P. Mascagni, J. F. Hunt, M. M. Roberts, and A. R. Coates. 1997. Mycobacterium tuberculosis chaperonin 10 stimulates bone resorption: a potential contributory factor in Pott's disease. J. Exp. Med. 1861241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng, V. H., J. S. Cox, A. O. Sousa, J. D. MacMicking, and J. D. McKinney. 2004. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 521291-1302. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, K. L., N. McLennan, M. Masters, and N. J. Cowan. 1999. A single-ring mitochondrial chaperonin (Hsp60-Hsp10) can substitute for GroEL-GroES in vivo. J. Bacteriol. 1815871-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ojha, A., M. Anand, A. Bhatt, L. Kremer, W. R. Jacobs, Jr., and G. F. Hatfull. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123861-873. [DOI] [PubMed] [Google Scholar]
- 19.Orme, I. M. 1988. A mouse model of the recrudescence of latent tuberculosis in the elderly. Am. Rev. Respir. Dis. 137716-718. [DOI] [PubMed] [Google Scholar]
- 20.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 1461969-1975. [DOI] [PubMed] [Google Scholar]
- 21.Parish, T. S. N. G. 1998. Electroporation of mycobacteria. Humana Press, Totowa, NJ.
- 22.Peetermans, W. E., J. A. Langermans, M. E. van der Hulst, J. D. van Embden, and R. van Furth. 1993. Murine peritoneal macrophages activated by the mycobacterial 65-kilodalton heat shock protein express enhanced microbicidal activity in vitro. Infect. Immun. 61868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peetermans, W. E., C. J. Raats, J. A. Langermans, and R. van Furth. 1994. Mycobacterial heat-shock protein 65 induces proinflammatory cytokines but does not activate human mononuclear phagocytes. Scand. J. Immunol. 39613-617. [DOI] [PubMed] [Google Scholar]
- 24.Peetermans, W. E., C. J. Raats, R. van Furth, and J. A. Langermans. 1995. Mycobacterial 65-kilodalton heat shock protein induces tumor necrosis factor alpha and interleukin 6, reactive nitrogen intermediates, and toxoplasmastatic activity in murine peritoneal macrophages. Infect. Immun. 633454-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qamra, R., and S. C. Mande. 2004. Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J. Bacteriol. 1868105-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qamra, R., S. C. Mande, A. R. Coates, and B. Henderson. 2005. The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 85385-394. [DOI] [PubMed] [Google Scholar]
- 27.Qamra, R., V. Srinivas, and S. C. Mande. 2004. Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. J. Mol. Biol. 342605-617. [DOI] [PubMed] [Google Scholar]
- 28.Raja, A. 2004. Immunology of tuberculosis. Indian J. Med. Res. 120213-232. [PubMed] [Google Scholar]
- 29.Retzlaff, C., Y. Yamamoto, P. S. Hoffman, H. Friedman, and T. W. Klein. 1994. Bacterial heat shock proteins directly induce cytokine mRNA and interleukin-1 secretion in macrophage cultures. Infect. Immun. 625689-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rha, Y. H., C. Taube, A. Haczku, A. Joetham, K. Takeda, C. Duez, M. Siegel, M. K. Aydintug, W. K. Born, A. Dakhama, and E. W. Gelfand. 2002. Effect of microbial heat shock proteins on airway inflammation and hyperresponsiveness. J. Immunol. 1695300-5307. [DOI] [PubMed] [Google Scholar]
- 31.Riffo-Vasquez, Y., D. Spina, C. Page, P. Tormay, M. Singh, B. Henderson, and A. Coates. 2004. Differential effects of Mycobacterium tuberculosis chaperonins on bronchial eosinophilia and hyperresponsiveness in a murine model of allergic inflammation. Clin. Exp. Allergy 34712-719. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Quinones, F., M. Maguire, E. J. Wallington, P. S. Gould, V. Yerko, J. A. Downie, and P. A. Lund. 2005. Two of the three groEL homologues in Rhizobium leguminosarum are dispensable for normal growth. Arch. Microbiol. 183253-265. [DOI] [PubMed] [Google Scholar]
- 32a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 34.Smith, D. A., T. Parish, N. G. Stoker, and G. J. Bancroft. 2001. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect. Immun. 691142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steyn, A. J., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 993147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, Z., D. J. Scott, and P. A. Lund. 2003. Isolation and characterisation of mutants of GroEL that are fully functional as single rings. J. Mol. Biol. 332715-728. [DOI] [PubMed] [Google Scholar]
- 37.Tabona, P., K. Reddi, S. Khan, S. P. Nair, S. J. Crean, S. Meghji, M. Wilson, M. Preuss, A. D. Miller, S. Poole, S. Carne, and B. Henderson. 1998. Homogeneous Escherichia coli chaperonin 60 induces IL-1 beta and IL-6 gene expression in human monocytes by a mechanism independent of protein conformation. J. Immunol. 1611414-1421. [PubMed] [Google Scholar]
- 38.Tormay, P., A. R. Coates, and B. Henderson. 2005. The intercellular signaling activity of the Mycobacterium tuberculosis chaperonin 60.1 protein resides in the equatorial domain. J. Biol. Chem. 28014272-14277. [DOI] [PubMed] [Google Scholar]
- 39.van Eden, W., R. van der Zee, and B. Prakken. 2005. Heat-shock proteins induce T-cell regulation of chronic inflammation. Nat. Rev. Immunol. 5318-330. [DOI] [PubMed] [Google Scholar]
- 40.Verdegaal, M. E., S. T. Zegveld, and R. van Furth. 1996. Heat shock protein 65 induces CD62e, CD106, and CD54 on cultured human endothelial cells and increases their adhesiveness for monocytes and granulocytes. J. Immunol. 157369-376. [PubMed] [Google Scholar]
- 41.Wallace, R. J., Jr., D. R. Nash, M. Tsukamura, Z. M. Blacklock, and V. A. Silcox. 1988. Human disease due to Mycobacterium smegmatis. J. Infect. Dis. 15852-59. [DOI] [PubMed] [Google Scholar]
- 42.Yamazaki, Y., L. Danelishvili, M. Wu, M. Macnab, and L. E. Bermudez. 2006. Mycobacterium avium genes associated with the ability to form a biofilm. Appl. Environ. Microbiol. 72819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young, D. B., J. Ivanyi, J. H. Cox, and J. R. Lamb. 1987. The 65 kDa antigen of mycobacteria—a common bacterial protein? Immunol. Today 8215-219. [DOI] [PubMed] [Google Scholar]