Abstract
This work aimed to establish the role of gne (encoding UDP-GalNAc 4-epimerase activity) and galE (encoding UDP-Gal-4-epimerase activity) in the biosynthesis of surface polysaccharides, as well as in the virulence for eels and humans of the zoonotic serovar of Vibrio vulnificus biotype 2, serovar E. DNA sequence data revealed that gne and galE are quite homologous within this species (≥90% homology). Mutation in gne of strain CECT4999 increased the surface hydrophobicity, produced deep alterations in the outer membrane architecture, and resulted in noticeable increases in the sensitivity to microcidal peptides (MP), to eel and human sera, and to phagocytosis/opsonophagocytosis. Furthermore, significant attenuation of virulence for eels and mice was observed. By contrast, mutation in galE did not alter the cellular surface, did not increase the sensitivity to MP, serum, or phagocytosis, and did not affect the virulence for fish and mice. The change in the attenuated-virulence phenotype produced by a mutation in gne was correlated with the loss of the O-antigen lipopolysaccharide (LPS), while the capsule was maintained. Complementation of a gne-deficient mutant restored the LPS structure together with the whole virulence phenotype. In conclusion, gne, but not galE, is essential for LPS biosynthesis and virulence in the zoonotic serovar of V. vulnificus biotype 2.
Vibrio vulnificus is a pathogenic bacterial species that occurs in warm aquatic environments with intermediate levels of salinity. It is commonly isolated in temperate, subtropical, and tropical areas, where it is a risk to public health. V. vulnificus is able to infect fish and humans and causes a disease called vibriosis (41). Human vibriosis occurs after infection of preexisting wounds with seawater, after an individual is injured while fishing or handling fish, or after ingestion of raw seafood (41). Fish vibriosis is produced after gill or intestine colonization, without previous injury (19, 37). Human and fish vibriosis can lead to septicemia and death if innate defenses do not act properly and in time, which makes the mechanisms of resistance of V. vulnificus to innate defenses a key virulence factor in septicemia. Classically, the human virulent strains are classified in biotypes 1 and 3, while the fish virulent strains are classified in biotype 2 (11, 53). Interestingly, some biotype 2 strains, which are serologically homogeneous, are able to infect both humans and fish (3). These strains are designated serovar E strains (VSE strains) and are distributed worldwide. Recently, the genomes of two strains of V. vulnificus biotype 1 isolated from human blood in Asia have been described and compared (15, 29). Both of these strains are serologically different from VSE strains (unpublished results).
A number of virulence factors that confer resistance to human innate immunity have been described for biotype 1 strains. Two such factors are the capsule that protects bacteria from phagocytosis (58) and an iron acquisition system that depends on vulnibactin, which is used for sequestering iron from host transferrin (28). In VSE strains, the capsule seems not to be essential for resistance to eel serum since spontaneous translucent variants survive in fresh eel serum (10), while the lipopolysaccharide (LPS) could contribute to this resistance since spontaneous O-antigen mutants that develop rugose colonies are sensitive to eel serum and are avirulent (2).
No information concerning either the chemical structure of cell surface polysaccharides or the genes involved in their biosynthesis in V. vulnificus VSE strain cells is available, although several genes, such as wbpO, wbpP, and wza, have recently been described in V. vulnificus biotype 1 (46, 47, 59). wbpO encodes a putative UDP-N-acetyl-d-galactosamine dehydrogenase essential for LPS biosynthesis, wbpP encodes a putative UDP-N-acetylglucosamine 4-epimerase essential for capsule biosynthesis, and wza encodes a sugar transferase also needed for capsule biosynthesis. In other bacterial species, additional genes, such as gne (encoding UDP-N-acetylgalactosamine [UDP-GalNAc] 4-epimerase activity) and galE (encoding UDP-galactose [UDP-Gal] 4-epimerase activity), have been described as genes that are involved in LPS or capsule biosynthesis and virulence (12, 16).
The objective of the present work was to establish the roles played by gne and galE in the biosynthesis of surface polysaccharides, as well as in the virulence for fish and humans, in the zoonotic serovar of V. vulnificus biotype 2. To this end, we looked for both of these genes in the previously published genome of biotype 1 strain YJ016, performed PCR to determine whether they were present in representative VSE strains, and obtained two mutants from a selected VSE strain by allelic exchange. The selected strain was CECT4999, originally isolated from a diseased eel, which is highly virulent for both eels and mice (an animal model used to test human virulence) and is resistant to both eel and human sera. We demonstrated that mutations in gne alter the O antigen of the LPS but not the capsule. The gne-deficient mutant showed significant attenuation of virulence for mice and eels, but the galE-deficient mutant did not show significant attenuation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are shown in Table 1. V. vulnificus strains were grown in tryptic soy broth containing 1% NaCl (TSB-1), on tryptic soy agar containing 1% NaCl (TSA-1), in marine seawater yeast extract (MSWYE), on MSWYE agar (42), and in M9 minimal medium (50) supplemented with 10 mM MgSO4, 10 mM CaCl2, and 20% Casamino Acids (Difco) at 28°C for 24 h. For enzymatic determinations, the strains were cultured in Davis minimal medium (13) with 0.2% glucose (DMM-glu) or galactose (DMM-gal) as the only carbon source. Escherichia coli and Salmonella enterica serotype Typhimurium strains were grown in Luria-Bertani broth and on Luria-Bertani agar at 37°C for 24 h. When required, ampicillin (Amp) (100 μg ml−1), rifampin (Rif) (100 μg ml−1), or chloramphenicol (Cm) (20 μg ml−1) was added to the different media. All the strains were stored in TSB-1 with or without an antibiotic plus dimethyl sulfoxide at −80°C. All bacterial counts were obtained by using the drop plate methodology (26).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Origin, genotype, and/or phenotype | Reference(s) or source |
|---|---|---|
| Vibrio vulnificus strains | ||
| Biotype 1 | ||
| YJO16 | Isolated from human blood in Taiwan, avirulent for eels, virulent for mice, genome sequenced, opaque morphotype | 15, 32 |
| Biotype 2 serovar A | ||
| CECT5768 | Isolated from diseased eel in Spain, virulent for eels, avirulent for mice, opaque morphotype | 21 |
| CECT7030 | Isolated from diseased eel in Denmark, virulent for eels, virulence for mice not tested, opaque morphotype | 21 |
| Biotype 2 serovar E | ||
| CECT4999 | Isolated from kidney of diseased eel in Spain, virulent for mice and eels, opaque morphotype | 51 |
| CECT4864 | Isolated from kidney of diseased eel in Spain, virulent for mice and eels, opaque morphotype | 51 |
| CCUG38521 | Isolated from human wound in Sweden, virulent for mice and eels, opaque morphotype | 51 |
| CECT4866 (= ATCC 33817) | Isolated from human wound in United States, virulent for mice and eels, opaque morphotype | 3 |
| CECT5763 | Isolated from seawater in Spain, virulent for mice and eels, OM | 51 |
| 4999-0 | CECT4999, spontaneous Rifr, opaque morphotype | This study |
| 4999-1a | CECT4999, spontaneous translucent variant | This study |
| 4999-2 | CECT4999 (pCM100-gne), Rifr Cmr, opaque morphotype, gne-deficient mutant | This study |
| 4999-3 | CECT4999 (pCM100-galE), Rifr Cmr, opaque morphotype, galE-deficient mutant | This study |
| C4999-2 | R99-2 with pGEMT-gne, opaque morphotype, complemented strain | This study |
| C4999-3 | R99-3 with pGEMT-galE, opaque morphotype, complemented strain | This study |
| Biotype 2 serovar Ib | ||
| 95-8-6 | Isolated from diseased eel in Denmark, virulent for eels, virulence for mice not tested, opaque morphotype | 51 |
| 95-8-161 | Isolated from diseased eel in Denmark, virulent for eels, virulence for mice not tested, opaque morphotype | 51 |
| Biotype 3 | ||
| 11028 | Isolated from human wound in Israel, avirulent for eels, virulent for mice, opaque morphotype | 11 |
| Escherichia coli strains | ||
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 cTetr)] | Stratagene |
| DH5α | F−endA hsdR17 (rK− mK+) supE44 thi-1 rexA1 gyrA96 φ80lacZ | 23 |
| MC1061-λpir | thi thr-1 leu-6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 | 49 |
| S. enterica serotype Typhimurium strains | ||
| SL3770 | Expresses smooth LPS | 54 |
| SL1306 | galE503, produces rough LPS of Rc chemotype | 54 |
| Plasmids | ||
| pGEM-T Easy | PCR cloning vector, Ampr | Promega |
| pCM100 | pGP704 suicide plasmid, λpir dependent, Cmr | 61 |
| pCM100-gne | pCM100 with an internal fragment of gne, Cmr | This study |
| pCM100-galE | pCM100 with an internal fragment of galE, Cmr | This study |
| pGEMT-gne | pGEMT with the complete gne gene of CECT4999, Ampr | This study |
| pGEMT-galE | pGEMT with the complete galE gene of CECT4999, Ampr | This study |
General DNA methods, DNA sequencing, and computer analysis of sequence data.
General DNA manipulations were performed as previously described (50). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers. Double-stranded DNA sequencing was performed by using the dideoxy chain termination method (50) with an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). Oligonucleotides were purchased from Pharmacia LKB Biotechnology. The DNA sequence was translated in open reading frames with the Frameplot program (27) and was compared with DNA sequences from the nonredundant GenBank (National Center for Biotechnology Information) and EMBL (European Biotechnology Information Service) databases by using the BLAST network service (1). ClustalW was used for multiple-sequence alignment.
DNA amplification and plasmid and mutant construction.
The VV3026 and VV2639 genes of biotype 1 strain YJ016 (accession number BA000037) were selected for primer design to detect putative gne and galE genes in VSE cells due to their homology to gne and galE of Aeromonas hydrophila AH-3 (12) (Fig. 1). The primers used for gne amplification were gne for (5′-TTTCAGTTCGTCAGCCACC-3′) and gne rev (5′-TCAGCAATATCACCGGCAC-3′), which resulted in a 531-bp amplicon; and the primers used for galE amplification were galE for (5′-TGACATTCGTGACGAAGCC-3′) and galE rev (5′-TCCCCACCGATTTCAATGC-3′), which resulted in a 584-bp amplicon. These amplicons were subcloned as incomplete copies (with an internal sequence missing both the 5′ and 3′ ends) of the gne and galE genes in the vector pGEMT, creating pGEMT-gne and pGEMT-galE. After EcoRI digestion of plasmids pGEMT-gne and pGEMT-galE, the gel-purified DNA was treated with E. coli DNA polymerase I (Klenow fragment) to create blunt ends in order to ligate the band to pCM100 (suicide plasmid, λpir dependent) and obtain plasmids pCM100-gne and pCM100-galE. Plasmids pCM100-gne and pCM100-galE were used to obtain gne- and galE-deficient mutants from CECT4999 by a single recombination event using previously described procedures (50). To this end, pCM100-gne and pCM100-galE were isolated, transformed into E. coli MC1061-λpir (12), and transferred by triparental conjugation to strain CECT4999-0 (a spontaneous rifampin-resistant [Rifr] mutant) as previously described (12). The Cmr Rifr transconjugants should have contained the mobilized plasmid integrated into the chromosome by homologous recombination, leading to two incomplete copies of the gne and/or galE gene (defined insertion mutants). Complementation analysis of the different VSE mutants was performed by conjugal transfer of wild-type complete galE and gne genes cloned in pGEMT to the corresponding mutants. Recombinants were selected on TSA-1 containing Cm, Rif, and Amp. Additional transformation experiments with pGEMT-gne and pGEMT-galE were performed with strains DH5α (which naturally lacks UDP-GalNAc 4-epimerase activity) (23) and SL1306 (a mutant that lacks UDP-Gal 4-epimerase activity) (54).
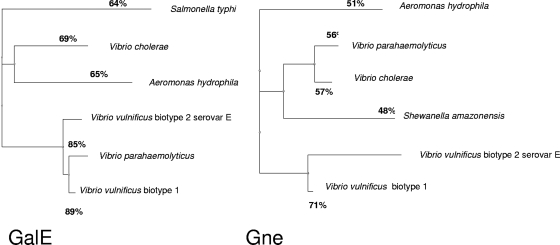
FIG. 1.
Dendrogram showing the amino acid sequence relatedness of V. vulnificus GalE and Gne with UDP-glucose 4-epimerases of other vibrios and related species using the TreeView software (45).
Cell extract production and measurement of enzymatic activity (UDP-Gal 4-epimerase and UDP-GalNAc 4-epimerase assays).
Suspensions of bacteria (25%, wt/vol) that were washed in 25 mM Tris-HCl buffer (pH 7.5) containing 1 mM MgCl2 were disrupted with a Branson model 350 Sonifier at 0°C. The disrupted bacteria were subjected to high-speed centrifugation (180,000 × g for 2 h) at 5°C to obtain cell extracts. Protein concentrations of extracts were determined by using the Bio-Rad Bradford assay as recommended by the manufacturer with bovine serum albumin as the standard.
The assays for UDP-Gal 4-epimerase and UDP-GalNAc 4-epimerase activities were performed as previously described (17, 57). For UDP-Gal 4-epimerase, the initial rates of NADH formation were determined by using the kinetics program installed in the spectrophotometer (Beckman DU640). A molar extinction coefficient (E340) of 6.220 M−1 cm−1 was assumed for all calculations. For UDP-GalNAc 4-epimerase, the conversion of UDP-GalNAc to UDP-N-acetylglucosamine (GlcNAc) was measured after acid hydrolysis by the 3.6-fold-greater absorbance at 585 nm (A585) of free GlcNAc than of GalNAc in the Morgan-Elson reaction. Following hydrolysis and completion of the Morgan-Elson reaction, color development was measured at 585 nm. Control assays with extract and with only substrate were performed simultaneously. All assays were performed in triplicate. Product formation (i.e., formation of GlcNAc by hydrolysis of UDP-GlcNAc) was measured using standard plots prepared by subjecting UDP-GlcNAc, UDP-GalNAc, GlcNAc, and GalNAc (Sigma-Aldrich, St. Louis, MO) to the same procedures.
Electron microscopy.
To stain flagella, cell suspensions were mixed with 2% phosphotungstic acid (pH 7) and air dried. Ultrastructural studies of the bacterial envelopes were performed by using ruthenium red staining for polysaccharides as described by Biosca et al. (10). Briefly, agar pieces with bacteria were placed into sodium cacodylate buffer (0.2 M, pH 7.2) containing 2.5% glutaraldehyde plus 0.075% ruthenium red, washed with sodium cacodylate buffer, and incubated for 2 h at 4°C in 1% osmium tetroxide. Then samples were washed in ultrapure MilliQ water, dehydrated using a graded ethanol series (30 to 100% ethanol, 10 min per step), and transferred sequentially into 3:1, 1:1, and 1:3 ethanol-epoxy resin (Aname) for 1 h per step and finally into 100% resin overnight. Then samples were embedded in flat silicone molds and placed inside a 60°C oven to polymerize for 2 days. Blocks were removed from the molds, and 100-nm ultrathin sections were obtained using a Leica Ultracut R ultramicrotome (Leica Microsystems, Milton Keynes, England). Observations were made with a JEOL 1010 transmission electron microscope.
MICs of microcidal peptides (MP) and human transferrin.
Stationary-phase bacteria were suspended in phosphate-buffered saline (pH 7) containing 1% NaCl (PBS-1) at a concentration of approximately 107 CFU ml−1, and 10 μl was inoculated into MSWYE plates containing polymyxin B sulfate (500 to 105 U ml−1; Sigma), poly-l-lysine (30 to 300 μg ml−; Sigma), or transferrin (2.5 to 50 μM; Sigma) and incubated for 24 h at 28°C. The MIC endpoint was defined as the lowest antibiotic concentration at which there were no visible colonies. Experiments were carried out in triplicate.
Antigen extraction, separation, and visualization.
Outer membrane proteins (OMP) were obtained from cells after sonication and differential solubilization of the inner membranes in a Sarkosyl solution (0.05%) as described by Biosca et al. (9). The extracellular proteins (ECP) were obtained after separation by differential centrifugation from cells grown on cellophane sheets covering TSA-1 plates using the protocol of Liu (34). OMP and ECP concentrations of samples were determined by the method of Lowry et al. (35). Crude fractions of bacterial polysaccharides (LPS and capsule) were obtained from cell lysates after proteinase K digestion (Boehringer Mannheim) as described by Hitchcock and Brown (25). The LPS concentration was determined with ProQ Emerald 300 staining for glycoproteins (Invitrogen) by following the manufacturer's instructions. OMP, ECP, LPS, and capsule antigens were separated and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (30) in discontinuous gels (4% stacking gel, 12% separating gel). Protein bands were visualized by Coomassie brilliant blue staining, and LPS and capsule bands were visualized by immunostaining after transfer from the polyacrylamide gels to nitrocellulose sheets (0.45 μm; Bio-Rad) as described by Towbin et al. (55). Blotting was done at 200 mA for 2 h in Tris-glycine-methanol transfer buffer (25 mM Tris, 192 mM glycine [pH 8.3], 20% [vol/vol] methanol). Immunostaining was performed with previously obtained serovar-specific sera (4) (dilution for immunostaining, 1:1,000) or with the same sera after absorption with one of the mutants. The absorption process involved incubating serum with mutant cells (1 × 109 CFU per ml in PBS-1) with gentle shaking for 30 min at 37°C, followed by filtration through 0.22-μm-pore size membranes (Millipore filters) (43). Absorbed sera were diluted 1:10 for immunostaining (43).
Eel serum, mucus, and phagocyte collection.
Humoral (serum and gill mucus) and cellular (head kidney, spleen, and blood phagocytes) defensive factors were obtained from adult eels (Anguilla anguilla) as described by Esteve-Gassent et al. (18) and Castro et al. (14), respectively. For the cellular factors, the head kidney and spleen were aseptically removed and crushed, and blood was taken from the caudal vein; these factors were mixed (1:3) with Leibovitz's medium (L-15) (Gibco) containing 2% fetal calf serum, heparin (10 IU ml−1), penicillin (100 IU ml−1), and streptomycin (100 μg ml−1), layered onto a 34 to 51% (vol/vol) Percoll gradient, and centrifuged at 400 × g for 30 min at 4°C. The interface cells were collected, washed twice, and suspended at a concentration of 1 × 106 viable cells ml−1 in the same medium. Viability was estimated by the trypan blue exclusion test. Cell suspensions were immediately used in phagocytosis assays.
Colonization assays. (i) Motility.
Motility was measured by transferring bacterial colonies on MSWYE into motility medium (1% tryptone, 0.5% yeast extract, 1% NaCl) supplemented with 0.3% (swimming) and 0.6% (swarming) agar. Halo diameters were measured after 24 h of incubation at 28°C.
(ii) Chemotaxis.
Serum and mucus from eels, as well as human serum (Sigma), were used as substrates. Chemotaxis was evaluated using the capillary assay described by Larsen et al. (31). To equalize differences in random motility and experimental variations, the chemotactic responses were expressed as the ratio of the number of bacteria in attractant capillaries to the number of bacteria in control capillaries filled with chemotaxis buffer.
(iii) Adherence to mucus.
Ninety-six-well microtiter plates covered with eel mucus were inoculated with 100 μl of a bacterial suspension (103 to 104 CFU ml−1) in PBS-1 per well, incubated at 28°C for 24 and 72 h and 1 and 2 weeks, washed three times with distilled water, stained with a 0.1% crystal violet solution, washed twice, and destained with 95% alcohol. The optical density at 540 nm was determined with a spectrometer (Termospectronic Helios λ).
(iv) Cell surface hydrophobicity.
Hydrophobicity was measured by the salt aggregation test (SAT) using ammonium sulfate solutions (0, 0.01, 0.05, 0.1, 0.5, 1, 2, and 4 mol liter−1) (33).
(v) Biofilm formation.
Cells were grown in MSWYE (42). Biofilm formation was evaluated as described by O'Toole and Kolter (44) by determining the ability of cells to adhere to polystyrene tubes (hydrophobic surface) or glass tubes (hydrophilic surface). The tubes were incubated at 28°C for 5 and 10 h. Biofilm formation was quantified by addition of 200 μl of 95% (vol/vol) ethanol to crystal violet-stained samples, and the absorbance at 540 nm was determined with a plate reader (Multiskan Askcent; Labsystems).
Eel complement fixation assay.
Anti-sheep red blood cell (SRBC) (Biomedics) serum (anti-SRBC serum) was obtained by injecting an SRBC suspension (5% in PBS-1) plus Freund's incomplete adjuvant (1:1) into eels using the protocol of Esteve-Gassent et al. (18). After 2 weeks, anti-SRBC serum was collected and inactivated by heating it at 56°C 30 min. The assay was performed on 96-well microtiter plates as follows. One hundred microliters of inactivated anti-SRBC serum was added to 100 μl of a 5% SRBC solution in 0.01 M EDTA (tetrasodium salt)-gelatin-Veronal buffer (EDTA-GVB2+), and the mixture was incubated for 30 min at 28°C to sensitize the SRBC. Then 200-μl aliquots of the bacterial suspensions in PBS-1 were added to 200 μl of nonimmune eel serum, incubated at 28°C for 1 h, and centrifuged (400 × g, 5 min, 4°C). The supernatants serially diluted in EDTA-GVB2+ (1:100 to 1:51,200) were added either to 100 μl of sensitized SRBC (1:1) to calculate 50% hemolysis due to complement activated by the classical pathway (CH50) or to nonsensitized SRBC to calculate 50% hemolysis due to complement activated by the alternative pathway (ACH50) after incubation at 28°C for 2 h and assessment of the relative hemoglobin content by determination of the optical density at 540 nm with a microplate reader (Bio-Rad). Experiments were done in triplicate. A complement fixation index (CFI) was calculated by determining the ACH50 or CH50 ratio before and after bacterial incubation (60).
Resistance to phagocytosis and opsonophagocytosis by eel phagocytes.
To obtain opsonized bacteria, a bacterial suspension containing 108 CFU ml−1 in PBS-1 was incubated with 10% nonimmune serum for 30 min. Then the bactericidal activity of eel phagocytes was determined as previously described (52), with some modifications. Briefly, eel leukocytes (107 cells ml−1 in Hanks balanced salt solution) were dispensed into Eppendorf plates and incubated with opsonized and nonopsonized bacteria (100 μl). After 0, 1, 2, 3, and 4 h of incubation, 50 μl of saponin (0.15% in distilled water; Sigma) was added to each well, and the microplates were incubated for 15 min at 4°C to lyse the cells. Bacterial survival was measured both by determining bacterial culturability on TSA-1 plates and by determining bacterial viability by reduction of 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). For the latter assay, a solution of MTT (2 mg ml−1 in distilled water) was added (50 μl per well), and plates were incubated for 15 min. MTT reduction was measured with a multiscan spectrophotometer (Bio-Rad) at a wavelength of 630 nm. A bactericidal index was determined by dividing the absorbance/count for each sample by the absorbance/count for the control. Experiments were performed in triplicate.
Resistance to serum and mucus.
Fresh gill mucus from eels, fresh and heat-inactivated (56°C for 60 min) human serum, and fresh and heat-inactivated (56°C, 30 min) eel serum were used in resistance assays (2, 36). A suspension containing 103 to 104 CFU ml−1 in PBS-1 was mixed with serum mucus samples, and the mixtures were incubated at 28°C (for eels) or 37°C (for humans) for 4 h. Viability counting was performed on TSA-1 plates at zero time and after 4 h of incubation.
Virulence for fish and mice.
Virulence was determined by calculating the 50% lethal dose (LD50) by the method of Reed and Muench (48), using elvers (average weight, 10 g) and BALB/c mice (average weight, 20 g). The elvers were kept in a 60-liter fiberglass tank containing freshwater at 26°C with a filtration and recirculation system. Groups of six fish and six mice were inoculated intraperitoneally with 0.1 and 0.2 ml, respectively, of serial 10-fold dilutions of bacteria in PBS-1 as previously described (5, 6), and mortality was recorded daily for a 7-day period. Mortality was considered to be caused by V. vulnificus only if the inoculated bacterium was recovered in pure culture from internal organs (5, 6).
Statistical assays.
One-way analysis of variance using the Tukey test was used to evaluate the significance of differences in assays and was performed with the GraphPad Prism program (version IV). Values were considered significantly different if the P value was <0.05. Experiments were performed in triplicate.
Nucleotide sequence accession numbers.
The nucleotide sequence accession number for the VSE gne region described in this paper is EU041623, and the nucleotide sequence accession number for the galE region is EU041622.
RESULTS
Cloning, sequence analysis, and isolation of gne and galE mutants.
Two genes (annotated VV2639 and VV3026) with high levels of homology to the A. hydrophila epimerase genes galE and gne (12) were found in the genome of human-pathogenic V. vulnificus biotype 1 strain YJ016. All DNA samples from the V. vulnificus strains shown Table 1 were positive for the gne and galE genes in a PCR assay. The coding region of galE of CECT4999 was 1,020 nucleotides long and showed 92% homology to VV2639, while the coding region of gne was 849 nucleotides long and showed 90% homology to VV3026. The deduced amino acid sequence revealed that galE codes for a protein containing 339 amino acids with 89% homology to the VV2639 protein and 65% homology to GalE of A. hydrophila, while gne codes for a protein containing 282 amino acids with 71% homology to the VV3026 protein and 51% homology to Gne of A. hydrophila (Fig. 1). galE and gne did not show significant homology to wpbO, wpbP, or wza of V. vulnificus biotype 1.
In order to isolate deficient mutants of strain CECT4999, two plasmids that carried 584- and 531-bp DNA fragments corresponding to incomplete copies of galE and gne, respectively, were constructed. Rif- and Cm-resistant isolates were confirmed by Southern blot hybridization with appropriate DNA probes and were used to obtain insertion mutants.
Enzymatic activities and growth curves.
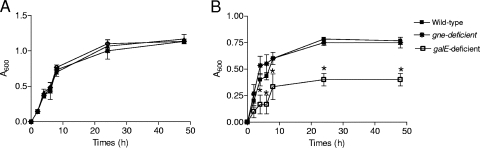
Figure 2 and Table 2 show growth curves in rich and minimal media and epimerase activities for the wild-type strain and its derivatives. The curves for the wild-type and mutant strains were statistically similar in TSB-1 and DMM-glu and different in DMM-gal (Fig. 2). In DMM-gal, the generation time of the galE-deficient mutant was twice the generation time of the wild-type strain and the gne-deficient mutant and the maximum population size of the galE-deficient mutant was threefold less than the maximum population size of the wild-type strain and the gne-deficient mutant, which suggests that mutations in galE significantly reduce but do not eliminate the ability of the bacteria to grow with galactose as the sole carbon source (Fig. 2). In fact, the wild-type strain and the galE-deficient mutant showed both epimerase activities, although the UDP-Gal 4-epimerase activity was lower in the mutant (significant difference, P < 0.05) (Table 2). In contrast, the gne-deficient mutant completely lacked UDP-GalNAc 4-epimerase activity and had UDP-Gal 4-epimerase activity in DMM-glu or DMM-gal that was not significantly different from that of the wild-type strain (Table 2). E. coli strain DH5α, which lacks UDP-GalNAc 4-epimerase activity, had UDP-GalNAc 4-epimerase activity after complementation with gne but not after complementation with galE (Table 2). Moreover, S. enterica serotype Typhimurium SL1306, which is not able to grow on DMM-gal and shows no detectable UDP-Gal 4-epimerase activity (54) (Table 2), grew on this medium and displayed this epimerase activity after complementation with pGEMT-gne or pGEMT-galE (Table 2). Finally, complementation of the V. vulnificus gne-deficient mutant with pGEMT-gne and complementation of the galE-deficient mutant with pGEMT-galE restored the wild-type phenotype.
FIG. 2.
Growth curves in TSB-1 (A) and DMM-gal (B) of strain CECT4999 and its derivatives, the gne-deficient mutant (4999-2) and the galE-deficient mutant (4999-3), incubated at 28°C. An asterisk indicates that a value is significantly different from the value for the wild-type strain (CECT4999) (P < 0.05).
TABLE 2.
UDP-Gal 4-epimerase and UDP-GalNAc 4-epimerase activities in V. vulnificus wild-type and mutant cell extracts
| Strain | Carbon sourcea | UDP-Gal 4-epimerase activity (nmol min−1 mg−1)b | UDP-GalNAc 4-epimerase activity (nmol min−1 mg−1)c |
|---|---|---|---|
| CECT4999 (wild type) | Glucose | 78.3 ± 1.1 | 20.6 ± 0.8 |
| Galactose | 32.6 ± 1.0 | 11.9 ± 1.2 | |
| 4999-2 (gne mutant) | Glucose | 75.8 ± 1.5 | <0.08d,e |
| Galactose | 31.5 ± 1.3 | <0.08d,e | |
| 4999-3 (galE mutant) | Glucose | 16.7 ± 2.1f | 20.1 ± 0.5 |
| Galactose | 12.8 ± 1.4f | 11.1 ± 1.6 | |
| C4999-2 (pGEMT-gne) | Glucose | 76.0 ± 1.8 | 21.6 ± 1.2 |
| Galactose | 30.4 ± 1.9 | 10.7 ± 2.3 | |
| C4999-3 (pGEMT-galE) | Glucose | 77.3 ± 1.7 | 20.8 ± 1.4 |
| Galactose | 30.9 ± 2.2 | 11.7 ± 0.8 | |
| E. coli DH5α | Glucose | NTg | <0.08d,e |
| E. coli DH5α (pGEMT-gne) | Glucose | NT | 8.4 ± 0.9 |
| E. coli DH5α (pGEMT-galE) | Glucose | NT | <0.08d,e |
| SL3770 (wild type) | Glucose | 63.5 ± 2.8 | <0.09d,e |
| SL1306 (galE mutant) | Glucose | <0.05d,e | <0.09d,e |
| SL1306 (pGEMT-gne) | Glucose | 12.8 ± 0.6 | 7.9 ± 1.2 |
| SL1306 (pGEMT-galE) | Glucose | 56.1 ± 1.4 | <0.09d,e |
Each strain was grown in DMM-glu (containing 0.2% glucose) or DMM-gal (containing 0.2% galactose).
Nanomoles of UDP-Gal converted to UDP-Glc per minute per milligram of protein.
Nanomoles of UDP-GalNAc converted to UDP-GlcNAc per minute per milligram of protein.
No detectable activity.
Significantly different from the value for the wild-type strain (P < 0.001).
Significantly different from the value for the wild-type strain (P < 0.05).
NT, not tested.
Surface antigens.
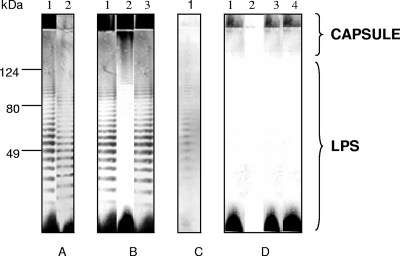
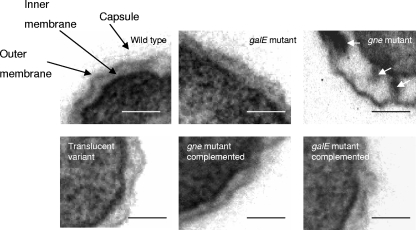
Colonies of the wild-type strain, the gne-deficient mutant, and the galE-deficient mutant on TSA-1 were opaque and had similar degrees of opacity, whereas colonies of strain 4999-1 were translucent. No apparent differences in the OMP and ECP patterns were observed among the strains (data not shown). In contrast, some differences in polysaccharides were detected among the strains (Fig. 3). Thus, after immunostaining of LPS-capsule extracts with anti-serovar E antibodies, the wild-type strain and the galE-deficient mutant showed the same pattern, which corresponded to the pattern for a smooth LPS plus the capsule (Fig. 3A and B). The translucent variant contained the whole LPS and lacked most of the capsular material, especially the part corresponding to the diffuse slowly migrating band that did not enter the separating gel (Fig. 3A), and the gne-deficient mutant lacked most of the LPS bands (Fig. 3B). After immunostaining of the wild-type strain with serum absorbed with the gne-deficient mutant, only the O antigen was observed (Fig. 3C), confirming that gne affected LPS; and after immunostaining with serum absorbed with the translucent variant, only the highest-molecular-weight portion of the capsule was stained (Fig. 3D). As Fig. 3D shows, the gne- and galE-deficient mutants were encapsulated. Electron micrographs of ruthenium red-stained ultrathin sections with capsule staining are shown in Fig. 4. Wild-type and mutant strains were surrounded by an electron-dense layer located outside the outer membrane (OM), which was apparently more dispersed or absent in the translucent variant (Fig. 4). The OM of the wild-type strain, the translucent variant, and the galE-deficient mutant had the same appearance; however, the OM of the gne-deficient mutant was profoundly altered, and there was a concomitant increase in the periplasmic space, which was filled with electron-dense material (Fig. 4). Complementation of the gne-deficient mutant with pGEMT-gne restored the whole structure of LPS (Fig. 3B), as well as the architecture of the OM (Fig. 4), whereas complementation of the galE-deficient mutant with pGEMT-galE did not alter either the OM architecture or the antigenic phenotype, which were identical to those of the wild-type (Fig. 4 and data not shown).
FIG. 3.
Immunostaining of LPS and capsule extracts with (A) rabbit anti-CECT4999 serum diluted 1:1,000 (lane 1, CECT4999; lane 2, 4999-1 [translucent variant]), (B) rabbit anti-CECT4999 serum diluted 1:1,000 (lane 1, 4999-3 [galE-deficient mutant]; lane 2, 4999-2 [gne-deficient mutant]; lane 3, C4999-2 [gne-complemented strain]), (C) rabbit anti-CECT4999 serum after absorption of the antibodies with 4999-2 cells (anti-O-antigen serum) diluted 1:10 (lane 1, CECT4999), and (D) rabbit anti-CECT400 serum after absorption with 4999-1 cells (anticapsule serum) diluted 1:10 (lane 1, CECT4999; lane 2, 4999-1; lane 3, 4999-3; lane 4, 4999-2).
FIG. 4.
Electron micrographs of strains CECT4999, 4999-3 (galE-deficient mutant), 4999-2 (gne-deficient mutant), 4999-1 (translucent variant), C4999-2 (gne-complemented strain), and C4999-3 (galE-complemented strain) after ruthenium red staining of ultrathin sections. The white arrows indicate the electron-dense periplasmic material. Bars = 0.1 μm.
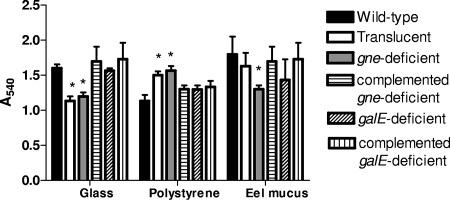
Hydrophobicity and biofilm formation.
There were significant differences between strains in surface hydrophobicity, measured by determining the lowest molarity of ammonium sulfate at which the cells agglutinated (SAT assay) (P < 0.05). The SAT value for the wild-type and galE-deficient strains was 4 mol liter−1, whereas the SAT value for the translucent variant and the gne-deficient mutant was 0.5 mol liter−1. In accordance with the SAT values, the latter strains formed more biofilm on hydrophobic surfaces (polystyrene), whereas the wild-type and galE-deficient strains formed more biofilm on hydrophilic surfaces (glass) (significant differences, P < 0.05) (Fig. 5). When biofilm formation on eel skin mucus was examined, the gne-deficient mutant formed less biofilm than the wild-type strain (significant difference, P < 0.05), and the galE-deficient mutant did not differ significantly from the wild-type strain (Fig. 5). Complementation of the gne-deficient mutant with pGEMT-gne completely restored the hydrophobicity and biofilm formation patterns, whereas complementation of the galE-deficient mutant with pGEMT-galE did not alter the surface and adhesive properties (Fig. 5).
FIG. 5.
Biofilm formation by CECT4999 and its derivatives on glass, polypropylene, and eel mucus measured by determining the absorbance at 540 nm (Multiskan Askcent; Labsystems). The strains used were strains CECT4999 (wild-type strain), 4999-1 (translucent variant), 4999-2 (gne-deficient mutant), 4999-3 (galE-deficient mutant), C4999-2 (gne-complemented strain), and C4999-3 (galE-complemented strain). An asterisk indicates that the value is significantly different from the value for the wild-type strain (P < 0.001).
Chemotaxis and motility.
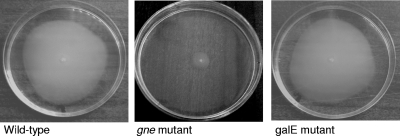
Large amounts of CECT4999 and mutant bacteria accumulated in capillaries containing eel mucus compared with control capillaries containing chemotaxis buffer. The strains also showed positive chemotaxis toward both eel and human sera, and there were no significant difference between strains. Motility assays revealed that the swimming and swarming halo diameters exhibited by the wild-type strain, the translucent variant, and the galE-deficient mutant were not significantly different (diameters for swimming, 10 ± 1 mm; diameters for swarming, 11.0 ± 0.5 mm) and were greater than those of the gne-deficient mutant (diameters for swimming, 5.0 ± 0.3 mm; diameters for swarming, 6.0 ± 0.5 mm). Figure 6 shows the swimming behavior of the strains. Complementation of the gne-deficient mutant with gne increased the swimming and swarming halo diameters to the wild-type strain values. Both the wild-type strain and the mutants, including the gne-deficient mutant, possessed polar flagella with apparently similar lengths and widths.
FIG. 6.
Swimming behavior of strains CECT4999 (wild-type strain), 4999-2 (gne-deficient mutant), and 4999-3 (galE-deficient mutant).
Resistance to eel innate immunity. (i) Resistance to mucus and serum and sensitivity to MP and human serum transferrin.
All the strains grew in gill mucus (Table 3). The wild-type strain and the galE-deficient mutant were also resistant to eel and human serum, whereas the translucent variant was resistant to eel serum and sensitive to human serum and the gne-deficient mutant was sensitive to both sera. The bactericidal effect of human serum on the translucent variant was partially eliminated by heat inactivation; however, the bactericidal effects of human and eel sera on the gne-deficient mutant were maintained even after heat inactivation (Table 3).
TABLE 3.
Survival in serum and mucus, CFI, and MICs of transferrin, lysozyme, and cationic peptides as well as virulence of V. vulnificus strains
| Strain | Survival indexa
|
CFI with live cellsb
|
MICs
|
Virulence (LD50)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eel serum
|
Human serum
|
Eel mucus (gills) | ||||||||||
| NIS | I-NIS | NIS | I-NIS | ACH50 | CH50 | Lysozyme (mg ml−1) | Polymyxin B (IU ml−1) | Polu-l-lysine (μg ml−1) | Eels | Mice | ||
| CECT4999 (wild type) | 50 ± 8 | 47 ± 6 | 57 ± 7 | 51 ± 5 | 49 ± 5 | 6.0 ± 0.5 | 4.8 ± 0.3 | >100 | 5,000 | 150 | 1.7 × 102 | 4.2 × 106 |
| 4999-1 (translucent) | 39 ± 5c | 40 ± 7c | 0d | 20 ± 4c | 43 ± 6 | 6.9 ± 0.5c | 6.8 ± 0.3c | 10d | 500d | 50c | 2 × 104c | 9.7 × 107c |
| 4999-2 (gne deficient) | 0d | 0d | 0d | 0d | 44 ± 3 | 7.0 ± 0.7c | 7.0 ± 0.6c | 10d | 300d | 50c | >107d | 8.5 × 107c |
| 4999-3 (gale deficient) | 46 ± 7 | 43 ± 6 | 53 ± 5 | 54 ± 5 | 48 ± 4 | 6.1 ± 0.4 | 4.6 ± 0.5 | 100 | 1,000 | 100 | 2 × 102 | 7 × 106 |
| C4999-2 (4999-2 complemented) | 48 ± 9 | 45 ± 7 | 53 ± 5 | 48 ± 7 | 50 ± 6 | NTe | NT | 100 | 1,000 | 150 | 3 × 102 | 6.7 × 106 |
| C4999-3 (4999-3 complemented) | 52 ± 9 | 51 ± 5 | 54 ± 5 | 50 ± 6 | 49 ± 4 | NT | NT | 100 | 5,000 | 150 | 2 × 102 | 4.5 × 106 |
The survival index is the ratio of the final bacterial count to the initial bacterial count. NIS, fresh nonimmune serum; I-NIS, inactivated nonimmune serum.
The CFI of eel complement is expressed as the ACH50 or CH50 ratio before and after bacterial incubation.
The value is significantly different from the value for the wild-type strain (P < 0.05).
The value is significantly different from the value for the wild-type strain (P < 0.001).
NT, not tested.
All strains were able to fix the eel complement activated by both pathways, regardless of cell viability (Table 3). There were no significant differences in CFI values between the wild-type strain and the galE-deficient mutant, which fixed more complement when it was activated by the alternative pathway (Table 3). By contrast, the translucent variant and the gne-deficient mutant fixed more complement than the other two strains (significant differences, P < 0.05) and showed similar values for CFI (no significant differences) regardless of the route of complement activation (Table 3).
The translucent variant and the gne-deficient mutant were more sensitive to the selected MP than the wild-type strain (significant differences, P < 0.05), and the most sensitive strain was the gne-deficient mutant (Table 3). The MICs of MP for the galE-deficient mutant were similar to those of the wild-type strain (Table 3). Resistance to MP, as well as to human and eel sera, was reestablished in the gne-deficient mutant by complementation with pGEMT-gne (Table 3). Finally, the MIC of human transferrin was the same for all strains (20 mM).
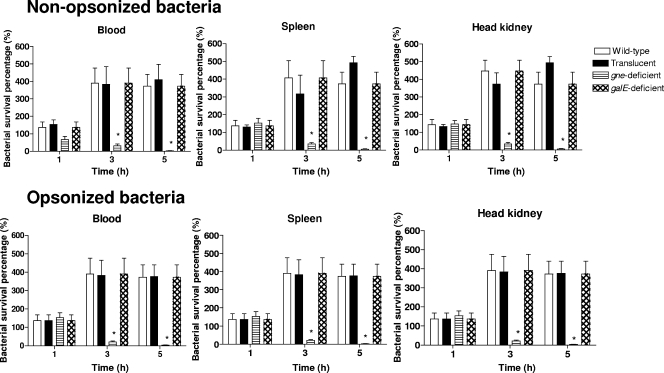
(ii) Resistance to phagocytosis and opsonophagocytosis.
Eel phagocytes were able to ingest V. vulnificus cells regardless of whether they were opsonized by serum complement (Fig. 7), although a significant increase in the average number of ingested bacteria per phagocyte (from two bacteria to five bacteria) was detected when bacteria were opsonized. Bacterial survival after 1, 3, and 5 h of incubation with eel phagocytes was analyzed. The MTT and cultivability data were similar, and only data for cultivable bacteria recovered after phagocytosis are shown in Fig. 7. The wild-type strain and its derivatives, the translucent variant, and the galE-deficient mutant resisted the killing mechanisms of eel phagocytes, and there were no significant differences in bacterial survival (Fig. 7). In contrast, the gne-deficient mutant was sensitive to both phagocytosis and opsonophagocytosis. Thus, the survival percentage compared with the wild-type strain decreased from 1 to 3 h of incubation (significant differences, P < 0.05) for phagocytosis and opsonophagocytosis (Fig. 7).
FIG. 7.
Bactericidal activity of phagocytes from spleen, blood, and head kidney against nonopsonized or opsonized VSE strains. The strains used were strains CECT4999 (wild-type strain), 4999-1 (translucent variant), 4999-2 (gne-deficient mutant), 4999-3 (galE-deficient mutant), C4999-2 (gne-complemented strain), and C4999-3 (galE-complemented strain). An asterisk indicates that the value is significantly different from the value for the wild-type strain (P < 0.001).
Virulence for mice and eels.
The LD50s for mice and eels of the wild-type strain and its derivatives are shown in Table 3. The LD50s of the galE-deficient mutant were similar to the LD50s of the wild-type for both eels and mice (Table 3). However, the gne-deficient mutant was avirulent for eels and showed a significant reduction in virulence for mice, and the translucent variant also showed a significant reduction in virulence for both hosts (Table 3). Complementation with pGEMT-gne completely restored the virulence for fish or mice (the LD50s were similar to those of the wild-type strain) in the gne-deficient mutant, while no changes were observed in the galE-deficient mutant after complementation with and pGEMT-galE (Table 3).
DISCUSSION
This work aimed to establish the role of gne and galE in the biosynthesis of surface polysaccharides, as well as in virulence for eels and humans of the zoonotic serovar of V. vulnificus biotype 2. Putative homologues of these genes were first identified in the genome of the human-pathogenic V. vulnificus biotype 1 isolate YJ016 and were amplified and sequenced in the VSE isolate CECT4999. The sequences of VSE strain genes (galE and gne) were highly similar (90 to 92%) to the sequences of their homologs in biotype 1 strain YJ016. In addition, both genes were found by PCR in all V. vulnificus isolates tested regardless of biotype and serovar.
The enzymatic analyses performed with the mutants and the complemented strains, including the analyses performed with the complemented E. coli and S. enterica serovar Typhimurium strains, confirmed that gne encodes an epimerase with UDP-GalNAc 4-epimerase activity that is responsible for the conversion of UDP-GalNAc to UDP-GlcNAc and that galE encodes a protein with UDP-Gal 4-epimerase activity that is responsible for the conversion of UDP-Gal to UDP-Glc. In addition, the experiments performed with Salmonella suggest that gne encodes an epimerase with both UDP-GalNAc and UDP-Gal activities. In accordance with this observation, the V. vulnificus gne mutant exhibited only a very small reduction in the UDP-Gal 4-epimerase activity, probably because the main enzyme activity is due to GalE. At the same time, the reduced UDP-Gal 4-epimerase activity observed for the V. vulnificus galE mutant is supported by the fact that Gne can have reduced UDP-Gal 4-epimerase activity compared to GalE (Table 2). A similar situation has been described in A. hydrophila (12) and Campylobacter jejuni (8), in which Gne exhibits double activity as a UDP-GalNAc 4-epimerase and a UDP-Gal 4-epimerase and could supply the function of GalE. These results contrast with the results obtained for E. coli and S. enterica serovar Typhimurium, in which the GalE protein is the only UDP-Gal 4-epimerase (54). In these enterobacteria, mutations in galE are correlated with changes in the LPS chemical structure produced by an O-antigen-deficient LPS (54). In addition, gne and galE, as well as the putative proteins that they encode, were highly homologous to Aeromonas gne and galE genes and proteins, respectively, suggesting that the two genes could correspond to gne and galE and could encode UDP-GalNAc 4-epimerase and UDP-Gal 4-epimerase activity, respectively. V. vulnificus VSE gne and galE do not show significant homology with the recently described gene wbpP, which codes for a UDP-GlcNAc epimerase essential for capsule biosynthesis in biotype 1 strains (47).
The gne-deficient mutant had profound alterations in the cellular envelope that were visible by electron microscopy. Thus, the OM displayed irregular morphology, limiting the enlarged periplasmic space, in which electron-dense material accumulated. The alterations in the OM did not affect the ability of the mutant to grow in complex media, since the growth curves were not statistically different from those of the wild-type strain. The gne-deficient mutant lacked O antigen but contained the capsular polysaccharide. In fact, cells visualized in the micrographs were surrounded by ruthenium red-stained material and developed colonies with an opaque morphotype on agar plates. The gne-deficient mutant produces deficient-LPS, probably because the O-polysaccharide chain contains GlcNAc residues in each of its repeated units, like A. hydrophila serotype 034 (12) or Yersinia enterocolitica serotype 0:8 (7). This sugar was previously found in the capsule of the type strain of the species, which is a biotype 1 strain(22). The capsule of VSE strains does not seem to contain this sugar since the gne-deficient strain is a capsulated strain. In fact, at least 13 different K antigens have been described in the species (24).
Mutation in the gne gene of V. vulnificus VSE strains deeply affected the virulence phenotype of this zoonotic serovar. Eel vibriosis is a septicemic process comprising two steps: gill colonization and blood invasion with subsequent colonization of the internal organs (37, 56). The wild-type strain resisted the bactericidal action of gill mucus, was strongly attracted by it, and formed biofilms on mucus-coated plates, which correlates with its natural route of infection. In contrast, the gne-deficient mutant, although resistant to and chemoattracted by gill mucus, showed a significantly reduced ability to form biofilms on mucus compared with the wild-type strain. The translucent variant, which lacks most of the capsular material and retains the O antigen, formed as much biofilm on gill mucus as the wild-type strain. This result suggests that the O antigen rather than the capsule is involved in gill colonization, probably by facilitating attachment and biofilm formation on gills. Similar functions have been reported for the O antigen in Vibrio cholerae, since O-antigen-deficient V. cholerae strains are also deficient in mucus colonization (39).
The reduced motility observed in the gne-deficient mutant on semisolid surfaces seems not to be due to the loss of the flagellum or to defective flagellar biogenesis, since cells were motile in liquid medium and had a flagellum that was apparently the same length as the flagellum of the wild-type strain. Instead, the reduced swarming and swimming could be explained by the loss of a negatively charged O antigen, which would reduce the surface “wetability” required for colony expansion, as described previously for O-antigen-deficient mutants of S. enterica serovar Typhimurium (54).
It has been hypothesized that resistance to the innate defenses of the eel is the key step for colonization of the internal medium and death by septicemia (56). V. vulnificus VSE strains can survive in eel serum and resist the killing mechanisms of eel phagocytes in vitro, and spontaneous alterations in capsule and LPS that affect serum resistance have been described (2, 5). The gne-deficient mutant was sensitive to fresh eel serum and phagocytosis/opsonophagocytosis. The translucent spontaneous variant, which retains the O antigen and lacks most of the capsule, resisted the bactericidal action of eel serum; however, it grew significantly less than the wild-type strain and also resisted phagocytosis/opsonophagocytosis. Both strains fixed significantly more eel complement than the wild-type strain, especially when complement was activated by the classical pathway. These results suggest that the capsule and the O antigen may interfere with the deposition of natural antibodies and/or lectins on their natural targets (LPS core and other targets). The gne-deficient mutant was still sensitive to serum after inactivation by heating, which suggests that the loss of the O antigen makes the bacterium sensitive to a thermoresistant component naturally present in fresh serum or secreted by eel phagocytes. This extreme sensitivity to both eel serum and killing by phagocytes was correlated with a high level of sensitivity to cationic peptides present in eel serum and secreted by eel phagocytes (40), especially sensitivity to polymyxin B, which was significantly higher than the sensitivity of the translucent variant. The gne-deficient mutant was avirulent for eels, while the translucent variant was virulent for eels but exhibited a significantly higher LD50 than the wild-type strain. Thus, the resistance to the innate defenses of eels was well correlated to the degree of virulence for eels.
To evaluate the role of gne gene in human virulence, additional experiments to examine survival in human serum and virulence for mice were performed. Previous studies suggested that capsule was essential for human serum resistance by VSE cells and for virulence for mice (5). The results obtained in the present work confirm the previous findings since the translucent variant was sensitive to human serum and showed attenuated virulence for mice. As expected, the gne-deficient mutant was sensitive to human complement, even after heating, and showed attenuated virulence for mice, which confirms the role of the O antigen in resistance to both human and fish sera, as well as in virulence for both hosts. All these virulence traits were rescued by complementation of a mutant with the single gene gne.
Currently, eel vibriosis is kept under control on fish farms by vaccination by immersion with Vulnivaccine, a conventional bacterin that provides protection against vibriosis for 6 to 12 months (20). However, this vaccine and vaccination procedure do not eliminate the pathogen, which can survive on the eel surface inside biofilms (56), making the healthy carrier eels a risk to human health. The gne-deficient mutant is avirulent for fish and probably avirulent for humans since it was sensitive to human serum and showed attenuated virulence for mice, and at the same time, it can grow efficiently in complex media. This mutant could be a good candidate for development of a new live vaccine against eel vibriosis, at least against the disease caused by V. vulnificus VSE strains.
In conclusion, gne plays an essential role both in O-antigen biosynthesis of the LPS and in virulence for humans and eels of VSE cells, whereas galE does not play a role in either OM biosynthesis or virulence. Further studies are under way to develop and validate a new live vaccine against fish vibriosis using the gne-deficient mutant, which could also prevent zoonosis caused by V. vulnificus VSE strains.
Acknowledgments
This work was financed by projects MTKD-CT-2004-0145019 (European Union), AGL2005-04688 (Spanish Ministry for Education), PET2005-0053 (Spanish Ministry for Education), ACOMP07/063 (Generalitat Valenciana), and FIS (Spanish Ministry for Education and Science and Ministry of Healthcare and Generalitat de Catalunya). Esmeralda Valiente thanks the Spanish Ministry for Education, Culture and Sport for a Ph.D. fellowship, and Natalia Jimenez thanks the Generalitat de Catalunya for a Ph.D. fellowship.
We thank Maite Polo for her technical assistance and the SCIE of the University of Valencia for technical assistance with microscopy. The English was revised by F. Barraclough and James D. Oliver of the University of North Carolina.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaro, C., B. Fouz, E. G. Biosca, E. Marco-Noales, and R. Collado. 1997. The lipopolysaccharide O side chain of Vibrio vulnificus serogroup E is a virulent determinant for eels. Infect. Immun. 652475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 621454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaro, C., E. G. Biosca, B. Fouz, A. E. Toranzo, and E. Garay. 1992. Electrophoretic analysis of heterogeneous lipopolysaccharides from various strains of Vibrio vulnificus biotypes 1 and 2 using silver staining and inmunoblotting. Curr. Microbiol. 2599-104. [DOI] [PubMed] [Google Scholar]
- 5.Amaro, C., E. G. Biosca, B. Fouz, A. E. Toranzo, and E. Garay. 1994. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice Infect. Immun. 62759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaro, C., E. G. Biosca, B. Fouz, C. Esteve, and E. Alcalde. 1995. Evidence that water transmits Vibrio vulnificus serovar E infections to eels. Appl. Environ. Microbiol. 611133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 1844277-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernatchez, S., C. M. Szymanski, N. Ishiyama, J. Li, H. C. Jarrell, P. C. Lau, A. M. Berghuis, N. M. Young, and W. W. Wakarchuk. 2005. A single bifunctional UDP-GlcNAc/Glc-4 epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J. Biol. Chem. 2804792-4802. [DOI] [PubMed] [Google Scholar]
- 9.Biosca, E. G., E. Garay, A. E. Toranzo, and C. Amaro. 1993. Comparison of the outer membrane protein profiles of Vibrio vulnificus biotypes 1 and 2. FEMS Microbiol. Lett. 107217-222. [DOI] [PubMed] [Google Scholar]
- 10.Biosca, E. G., H. Llorens, E. Garay, and C. Amaro. 1993. Presence of a capsule in Vibrio vulnificus biotype 2 and its relationship to virulence for eels. Infect. Immun. 611611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Israel Vibrio Study Group. Lancet 3541421-1424. [DOI] [PubMed] [Google Scholar]
- 12.Canals, R., N. Jiménez, S. Vilches, M. Regué, S. Merino, and J. M. Tomás. 2007. Role of Gne and GalE in the Virulence of Aeromonas hydrophila serotype O34. J. Bacteriol. 189540-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlton, B. C., and B. J. Brown. 1981. Gene mutation, p. 224-225. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC.
- 14.Castro, R., N. Couso, A. Obach, and J. Lamas. 1999. Effect of different β-glucans on the respiratory burst of turbot (Psetta maxima) and gilthead seabream (Sparus aurata) phagocytes. Fish Shellfish Immunol. 9529-541. [Google Scholar]
- 15.Chen, C. Y., K. M. Wu, Y. C. Chang, H. C. Tsai, T. L. Liao, Y. M Liu, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 132577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Y., P. Bystricky, J. Adeyeye, P. Panigrahi, A. Ali, J. A Johnson, C. A. Bush, J. G. Morris, Jr., and O. C. Stine. 2007. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creuzenet, C., M. Belanger, W. Wakarchuk, and J. S. Lam. 2000. Expression, purification and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 27519060-19067. [DOI] [PubMed] [Google Scholar]
- 18.Esteve-Gassent, M. D., B. Fouz, R. Barrera, and C. Amaro. 2004. Efficacy of oral immunisation after immersion vaccination against Vibrio vulnificus in farmed European eels. Aquaculture 2319-22. [Google Scholar]
- 19.Fouz, B., and C. Amaro. 2003. Isolation of a new serovar of Vibrio vulnificus pathogenic for eels cultured in freshwater farms. Aquaculture 217677-682. [Google Scholar]
- 20.Fouz, B., M. D. Esteve-Gassent, R. Barrera, J. L. Larsen, M. E. Nielsen, and C. Amaro. 2001. Field testing of a vaccine against eel diseases caused by Vibrio vulnificus. Dis. Aquat. Org. 45183-189. [DOI] [PubMed] [Google Scholar]
- 21.Fouz, B., F. Roig, and C. Amaro. 2007. Phenotypic and genotypic characterization of a new fish-virulent Vibrio vulnificus serovar that lacks potential to infect humans. Microbiology 1531926-1934. [DOI] [PubMed] [Google Scholar]
- 22.Gunawardena, S., G. P. Reddy, Y. Wang, V. S. Kolli, R. Orlando, J. G. Morris, and C. A. Bush. 1998. Structure of a muramic acid containing capsular polysaccharide from the pathogenic strain of Vibrio vulnificus ATCC 27562. Carbohydr. Res. 30965-76. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hayat, U., G. P. Reddy, C. A. Bush, J. A. Johnson, A. C. Wright, and J. G. Morris. 1993. Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J. Infect. Dis. 168758-762. [DOI] [PubMed] [Google Scholar]
- 25.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoben. J., and P. Somasegaran. 1982. Comparison of pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl. Environ. Microbiol. 441246-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawaa, J., and K. Hotta. 1999. Frameplot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174251-253. [DOI] [PubMed] [Google Scholar]
- 28.Kim, C. M., R. Y. Park, J. H. Park, H. Y. Sun, Y. H. Bai, P. Y. Ryu, S. Y. Kim, J. H. Rhee, and S. H. Shin. 2006. Vibrio vulnificus vulnibactin, but not metalloprotease VvpE, is essentially required for iron-uptake from human holotransferrin. Biol. Pharm. Bull. 29911-918. [DOI] [PubMed] [Google Scholar]
- 29.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Chey, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 715461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, M. H., J. L. Larsen, and J. E. Olsen. 2001. Chemotaxis of Vibrio anguillarum to fish mucus: role of the origin of the fish mucus, the fish species and the serogroup of the pathogen. FEMS Microbiol. Ecol. 3877-80. [Google Scholar]
- 32.Lee, C. T., C. Amaro, E. Sanjuan, and L. I. Hor. 2005. Identification of DNA sequences specific for Vibrio vulnificus biotype 2 strains by suppression subtractive hybridization. Appl. Environ. Microbiol. 715593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl, M., A. Faris, T. Wadstrom, and S. Hjerten. 1981. A new test based on “salting out” to measure relative surface hydrophobicity of bacterial cells. Biochim. Biophys. Acta 677471-476. [DOI] [PubMed] [Google Scholar]
- 34.Liu, P. V. 1957. Survey of hemolysin production among species of pseudomonads. J. Bacteriol. 74718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowry, O., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 36.Magre, S., Y. Takeuchi, G. Langford, A. Richards, C. Patience, and R. Weiss. 2004. Reduced sensitivity to human serum inactivation of enveloped viruses produced by pig cells transgenic for human CD55 or deficient for the galactosyl-α(1-3) galactosyl epitope. J. Virol. 785812-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marco-Noales, E., M. Milan, B. Fouz, E. Sanjuán, and C. Amaro. 2001. Transmission to eels, portals of entry, and putative reservoirs of Vibrio vulnificus serovar E (biotype 2). Appl. Environ. Microbiol. 674717-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, S. J., and R. J. Siebeling. 1991. Identification of Vibrio vulnificus O serovars with antilipopolysaccharide monoclonal antibody. J. Clin. Microbiol. 291684-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesper, J., S. Schild, C. M. Lauriano, A. Kraiss, K. E. Klose, and J. Reidl. 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 705990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, M. E., and M. D. Esteve-Gassent. 2006. The eel immune system: present knowledge and the need for research. J. Fish Dis. 2965-78. [DOI] [PubMed] [Google Scholar]
- 41.Oliver, J. D. 2006. Vibrio vulnificus, p. 349-366. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 42.Oliver, J. D., and R. R. Colwell. 1973. Extractable lipids of gram-negative marine bacteria: phospholipid composition. J. Bacteriol. 114897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ørskov, F., and I. Ørskov. 1978. Serotyping of Enterobacteriaceae with special emphasis on K antigen determination. Methods Microbiol. vol. 1137-38. [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 45.Page, R. D. N. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 46.Park, N. Y., J. H. Lee, B. C. Lee, T. S. Kim, and S. H. Choi. 2006. Identification and characterization of the wbpO gene essential for lipopolysaccharide synthesis in Vibrio vulnificus. J. Microbiol. Biotechnol. 16808-816. [Google Scholar]
- 47.Park, N. Y., J. H. Lee, M. W. Kim, H. G. Jeong, B. C. Lee, T. S. Kim, and S. H. Choi. 2006. Identification of the Vibrio vulnificus wbpP gene and evaluation of its role in virulence. Infect. Immun. 74721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed, M. J., and M. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27487-493. [Google Scholar]
- 49.Rubirés, X., F. Saigí, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (024) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 1797581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 51.Sanjuan, E., and C. Amaro. 2007. Multiplex PCR assay for detection of Vibrio vulnificus biotype 2 and simultaneous discrimination of serovar E. Appl. Environ. Microbiol. 732029-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solem, S. T., J. B. Jørgensen, and B. Robertsen. 1995. Stimulation of respiratory burst and phagocytic activity in Atlantic salmon (Salmo salar L.) macrophages by lipopolysaccharide. Fish Shellfish Immunol. 5475-491. [Google Scholar]
- 53.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 1826308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Towbin, H., T. Staehelin, and T. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valiente, E., and C. Amaro. 2006. A method to diagnose the carrier state of Vibrio vulnificus serovar E in eels. Aquaculture 258173-179. [Google Scholar]
- 57.Wang, L., S. Huskic, A. Cisterne, D. Rothemund, and P. R. Reeves. 2002. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J. Bacteriol. 1842620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wright, A. C., J. G. Morris, D. R. Maneval, K. Richardson, and J. B. Kaper. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 581769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright, A. C., J. L. Powell, J. B. Kaper, and J. Glenn Morris. 2001. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect. Immun. 696893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yano, T. 1992. Assays of hemolytic complement activity, p. 131-141. In J. S. Stolen, T. C. Fletcher, D. P. Anderson, S. L. Kaatari, and A. F. Rowley (ed.), Techniques in fish immunology. SOS Publications, New York, NY.
- 61.Yu, H. B., P. S. Srinivasa Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomás, and K. Y. Leung. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 721248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]