Abstract
Numerous secreted virulence factors have been proposed to account for the fulminating and destructive nature of Vibrio vulnificus infections. A mutant of V. vulnificus that exhibited less cytotoxicity to INT-407 human intestinal epithelial cells was screened from a library of mutants constructed by random transposon mutagenesis. A transposon-tagging method was used to identify and clone an open reading frame encoding an RTX toxin secretion ATP binding protein, RtxE, from V. vulnificus. The deduced amino acid sequence of RtxE from V. vulnificus was 91% identical to that reported from Vibrio cholerae. Functions of the rtxE gene in virulence were assessed by constructing an isogenic mutant whose rtxE gene was inactivated by allelic exchanges and by evaluating the differences between its virulence phenotype and that of the wild type in vitro and in mice. The disruption of rtxE blocked secretion of RtxA to the cell exterior and resulted in a significant reduction in cytotoxic activity against epithelial cells in vitro. Also, the intraperitoneal 50% lethal dose of the rtxE mutant was 104 to 105 times higher than that of the parental wild type, indicating that RtxE is essential for the virulence of V. vulnificus. Furthermore, the present study demonstrated that the rtxBDE genes are transcribed as one transcriptional unit under the control of a single promoter, PrtxBDE. The activity of V. vulnificus PrtxBDE is induced by exposure to INT-407 cells, and the induction requires direct contact of the bacteria with the host cells.
The pathogenic marine bacterium Vibrio vulnificus is a causative agent of food-borne diseases such as life-threatening septicemia and possibly gastroenteritis in individuals with underlying predisposed conditions (for recent reviews, see references 18, 27, and 40). Disease caused by infection with V. vulnificus is remarkable for the invasive nature of the infection, the severity of the tissue damage that ensues, and its rapidly fulminating course, indicating that the pathogenicity of the bacteria is a multifactorial and complex phenomenon that involves the products of many genes. The characterization of somatic as well as secreted products of V. vulnificus has yielded a large list of putative virulence factors, including a carbohydrate capsule, a lipopolysaccharide, a cytolysin/hemolysin, an elastolytic metalloprotease, iron-sequestering systems, a lipase, and pili (18, 27, 41). Of these putative virulence factors, however, only a few, such as the capsule, iron acquisition systems, and type IV pilin have been confirmed to be essential for virulence by the use of molecular Koch's postulates (12, 28, 35, 45). Therefore, extensive screening and characterization of more virulence factors are still required for development of improved treatment and prevention as well as for understanding the molecular pathogenesis of the multifaceted host-pathogen interaction of V. vulnificus.
Bacterial infection is primarily a consequence of contributions of many virulence factors, which are often displayed on the bacterial cell surface, secreted into the extracellular environment, or directly injected into the cytosol of a host cell (11). This indicates that bacterial pathogenicity depends greatly on the secretion systems by which bacteria transport virulence factors to the cell exterior (13). Gram-negative bacteria contain several different types of secretion systems to export virulence factors across the outer membranes (42). Among the secretion systems, the type I secretion system (TISS) is composed of three membrane components that span the cell envelop; one is a specific outer membrane protein (OMP), and the other two, an ATP-binding cassette (ABC) and the membrane fusion (adaptor) protein (MFP), are cytoplasmic membrane proteins (8, 9). This widespread TISS mediates sec gene-independent transport of diversely sized proteins lacking N-terminal signal sequences across cytoplasmic and outer cell membranes. The transport is driven by ATP hydrolysis and, therefore, is also designated ABC transport. There is increasing evidence that the TISSs play direct and/or indirect roles, such as the export of exotoxins, in bacterial virulence (9, 16).
The Escherichia coli Hly transporter is a model for the TISS; it is composed of HlyB (ABC), HlyD (MFP), and TolC (OMP) and exports HlyA, a pore-forming cytotoxin belonging to the RTX (repeats in toxin) protein family (9). Recently, it has been reported that the V. cholerae RtxA toxin, which is a key virulence factor, is secreted by a TISS composed of RtxB (ABC), RtxD (MFP), RtxE (ABC), and TolC (OMP). In contrast to the E. coli Hly exporter, the V. cholerae Rtx exporter atypically carries two ABCs that may function as a heterodimer (1). In an effort to screen virulence factors of V. vulnificus, an open reading frame (ORF) encoding a V. cholerae RtxE homologue was identified by a transposon-tagging method. A V. vulnificus null mutant, in which the rtxE gene was inactivated, was constructed by allelic exchanges, and the possible roles of the RtxE protein in the virulence of V. vulnificus were explored. As a result, it was found that V. vulnificus RtxE is essential for virulence in vitro and in mice. Moreover, here we extended our understanding of the regulatory mechanisms of rtxE expression at a molecular level by demonstrating that the rtxBDE genes are organized as a single transcriptional unit and transcribed from a single promoter, PrtxBDE, and that the activity of PrtxBDE is dependent on exposure to host cells.
MATERIALS AND METHODS
Strains, plasmids, culture media, and general genetic methods.
The strains and plasmids used in this study are listed in Table 1. Unless noted otherwise, the V. vulnificus strains were grown in Luria-Bertani media supplemented with 2.0% (wt/vol) NaCl (LBS). All the components of the media were purchased from Difco (Detroit, MI), and the chemicals were purchased from Sigma (St. Louis, MO). The procedures for the isolation of plasmid DNA and genomic DNA and transformation were carried out as described by Sambrook and Russell (39).
TABLE 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| M06-24/O | Clinical isolate; virulent | Laboratory collection |
| MW061 | M06-24/O with rtxE::nptI; Kmr | This study |
| MW064 | M06-24/O with ΔrtxA::nptI; Kmr | 22 |
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal (DE3) | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λ pir; Kmr; host for π-requiring plasmids; conjugal donor | 31 |
| Plasmids | ||
| Mini-Tn5 lacZ1 | R6Kγori; suicide vector; oriT of RP4; Apr | 10 |
| pCOS5 | Cosmid vector; oriT of RK2; Apr Cmr | 6 |
| pDM4 | Suicide vector; oriR6K; Cmr | 32 |
| pET28a | Protein expression vector; Kmr | Novagen |
| pGEM-T easy | PCR product cloning vector; Apr | Promega |
| pJH0311 | 0.3-kb NruI fragment containing multicloning site of pUC19 cloned into pCOS5; Apr Cmr | This study |
| pJH0607 | pET28a with truncated-rtxA; Kmr | This study |
| pMW0502 | pGEM-T Easy with rtxE; Apr | This study |
| pMW0503 | pGEM-T Easy with rtxE::nptI; Apr, Kmr | This study |
| pMW0504 | pCOS5 with rtxBDE; Apr Cmr | This study |
| pMW0611 | pDM4 with rtxE::nptI; Cmr Kmr | This study |
| pMW0612 | pJH0311 with rtxE; Apr Cmr | This study |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant.
Identification of the V. vulnificus rtxE.
A mutant exhibiting decreased cytotoxic activity against INT-407 intestinal epithelial cells was screened from a library of V. vulnificus mutants generated by random transposon mutagenesis using a mini-Tn5 lacZ1 (10, 20). A DNA segment flanking the transposon insertion was amplified by PCR as previously described (20). Since a database search for homology to the amino acid sequence deduced from the resulting PCR product singled out the product of Vibrio cholerae rtxE (Fig. 1A), a part of the rtxE ORF was amplified from the genomic DNA of V. vulnificus by PCR using primers RTXE0501 and RTXE0502 (Table 2). The primers were designed using the genomic sequence of V. vulnificus YJ016 (GenBank accession numbers BA 000037 and BA 000038; www.ncbi.nlm.nih.gov). The amplified 1.2-kb rtxE was ligated into pGEM-T Easy (Promega, Madison, WI), resulting in pMW0502 (Table 1).
FIG. 1.
Physical map of the rtx gene cluster on the V. vulnificus chromosome and plasmids used in this study. (A) The open arrows represent the transcriptional directions and coding regions of the genes. The figure was derived using the nucleotide sequences of the V. vulnificus YJ016 genome in the GenBank databases (NCBI). The gene identifications are shown below each coding region. The size of rtxA is reduced, as indicated by the dashed lines. The primers RTXBD001 and RTXDE002, used for the RT-PCR analysis, are depicted by solid bars. (B) Regions cloned in each of the plasmids. Plasmid pMW0611 was used for the construction of the rtxE::nptI mutant, and pMW0504 was used for transcription analysis of rtxBDE. The insertion position of the nptI cassette is indicated by an open triangle.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a | Locationb | Use |
|---|---|---|---|
| RTXE0501 | CTTAAAAGCCAAGTCACCAC | rtxE | rtxE mutant construction |
| RTXE0502 | CTAAACTCAATGCCACCTTC | rtxE | rtxE mutant construction |
| RTXE0401 | AAACTGCAGCCAAATGGTTGAGCTGAC | Chromosomal DNA | Amplification of rtxE |
| RTXE0402 | GGAATTCCAATAAATAGCTCTGGACGATG | Chromosomal DNA | Amplification of rtxE |
| RTXA-Ab28aF | TAAGAATTCACCAGCAAGCTCAGCCGCGCA | rtxA | Amplification of rtxA |
| RTXA-Ab28aR | TATCTCGAGTATGAACAACGTCTGCCCACGC | rtxA | Amplification of rtxA |
| RTXS0401 | CGACCATTATTCACTCAATTCC | Chromosomal DNA | Amplification of rtxBDE |
| RTXS0402 | GACGATGTCGTTGATAGGCT | Chromosomal DNA | Amplification of rtxBDE |
| RTXBD001 | CTAGACGATGAATCTCAGGC | 3′ end of rtxB | RT-PCR |
| RTXDE002 | GAGCTGGACGATTTCTTATG | 5′ end of rtxE | RT-PCR |
| RTXB0701F | GCTCAACCTGCCAGTAGAACAAC | rtxB | Real-time PCR |
| RTXB0701R | TCACCCGCTCGTATGTCCAATG | rtxB | Real-time PCR |
| RRSH0701F | ACGACACCACCTTCCTCACAAC | rrsH | Real-time PCR |
| RRSH0701R | ACACGGTCCAGACTCCTACGG | rrsH | Real-time PCR |
| RTXB-PE001 | AATTACTTAGCCGATGTTTATTGCAGCG | rtxB | Primer extension |
Regions of oligonucleotides not complementary to the corresponding genes are underlined.
Location to which the nucleotides were hybridized.
Generation of rtxE::nptI mutant.
To inactivate rtxE in vitro, 1.2 kb of nptI DNA conferring resistance to kanamycin (33) was inserted into a unique SalI site present within the rtxE ORF. The 2.4-kb rtxE::nptI cartridge was then liberated from the resulting construct (pMW0503) and ligated with SphI-SpeI-digested pDM4 (32) to form pMW0611 (Fig. 1B; Table 1). To generate the rtxE::nptI mutant MW061 by homologous recombination, E. coli SM10λpir tra (containing pMW0611) (31) was used as a conjugal donor to V. vulnificus M06-24/O. Inactivation of rtxE by replacing wild-type rtxE on the chromosome with the rtxE::nptI allele was confirmed by PCR using previously described methods (20, 36). The V. vulnificus rtxE mutant chosen for further analysis was named MW061 (Table 1). To complement the rtxE mutant, pMW0612 was constructed by subcloning rtxE amplified by PCR using the primers RTXE0401 and RTXE0402 (Table 2) into the broad-host-range vector pJH0311 (Table 1) and transferred into M06-24/O cells by conjugation.
Cytotoxicity assays.
Two different assays were performed using INT-407 (ATCC CCL-6) human intestinal epithelial cells. The preparation of the INT-407 cells and infection with the bacterial cultures were performed in a 96-well tissue culture plate (Nunc, Roskilde, Denmark) as previously described (36). The cytotoxicity was then determined by measuring the activity of lactate dehydrogenase (LDH) in the supernatant using a cytotoxicity detection kit (Roche, Mannheim, Germany) and expressed using the total LDH activity of the cells completely lysed by 1% Triton X-100 as 100%. Morphological studies were also carried out using INT-407 cells which were seeded onto glass coverslips placed at the bottom of the tissue culture plate and infected with the V. vulnificus strains as previously described (36).
LD50 determination.
The 50% lethal doses (LD50s) of the wild type and the rtxE mutant were compared using ICR mice (specific pathogen free; Seoul National University), as described elsewhere (23, 36). The infected mice were observed for 24 h, and the LD50s were calculated using the method described by Reed and Muench (38). Mice were injected intraperitoneally with 250 μg of iron dextran for each gram of body weight immediately before injection with bacterial cells. All manipulations of mice were approved by the Institute of Laboratory Animal Resources of the Seoul National University.
Purification of truncated RtxA protein and Western blot analysis of secreted RtxA.
The DNA fragment encoding the C-terminal 640 amino acids of RtxA (truncated RtxA) was amplified by PCR using the primers RTXA-Ab28aF and RTXA-Ab28aR (Table 2). The 1.9-kb PCR product was digested with BamHI and EcoRI and then ligated with six-histidine-tagging expression vector pET28a (Novagen, Darmstadt, Germany) digested with the same enzymes. The resulting plasmid, pJH0607, encodes truncated RtxA with a six-His tag at the amino terminus. His-tagged truncated RtxA protein was expressed in E. coli BL21(DE3), and the protein was purified by affinity chromatography according to the manufacturer's procedure (Qiagen, Valencia, CA). The purified truncated RtxA was used to raise an anti-RtxA polyclonal antibody, and immunoblotting was performed according to the procedure previously described by Lee et al. (22).
For Western blot analyses, proteins of the cell lysates or the concentrated culture supernatants were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (21). Secreted proteins in the culture supernatants of V. vulnificus were precipitated and concentrated with ammonium sulfate using the methods described by Park et al. (37) with slight modifications (22). The protein concentrations were determined by the method of Bradford (3), with bovine serum albumin as the standard.
Cloning of rtxBDE.
Analyses of the rtxBDE transcript in total RNA isolated from the V. vulnificus by either reverse transcription-PCR (RT-PCR) or primer extension assay were unsuccessful in several attempts. The 5.9-kb DNA fragment carrying the rtxBDE genes was amplified by PCR using RTXS0401 and RTXS0402 as a pair of primers (Table 2) and cloned into broad-host-range vector pCOS5 (6), resulting in pMW0504 (Fig. 1B and Table 1). In order to increase the cellular level of the rtxBDE transcript, pMW0504 was transferred into M06-24/O cells by conjugation.
RNA isolation.
Total cellular RNA was isolated from M06-24/O(pMW0504) cells grown to mid-exponential phase with different media, such as LBS, minimum essential medium (MEM; Gibco, Invitrogen), and INT-407-spent MEM using an RNeasy minikit (Qiagen) (24). The supernatant of the INT-407 cell line grown as a monolayer in MEM was filtrated with Steritop (Millipore, Billerica, MA), and the resulting filtrate was used as an INT-407-spent MEM. Also, the RNA was prepared from M06-24/O(pMW0504) exposed to INT-407 cells. For this purpose, M06-24/O(pMW0504) was incubated with INT-407 cells at a multiplicity of infection (MOI) of 10 for 2 h. The mixture of the INT-407 and V. vulnificus cells was centrifuged at 250 × g for 10 min to precipitate INT-407 cells, and the bacterial cells were then harvested from the supernatant by centrifugation at 2,430 × g for 20 min.
Transcript analysis.
The prepared RNA was used for RT-PCR, real-time PCR, and primer extension analyses. For RT-PCR, a series of reactions was performed with SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's procedures to synthesize cDNA. PCR amplification of the cDNA was performed using standard protocols with a pair of primers, RTXBD001 and RTXDE002, which are designed to hybridize to the 3′ end of rtxB and the 5′ end of rtxE (Table 2; Fig. 1).
Quantitative real-time PCRs were performed in a final volume 20 μl of 2XiQ Sybr green Supermix (Bio-Rad Laboratories) containing cDNA synthesized as described above and the specific primer pair. Real-time PCRs were performed in triplicate using the iCycler iQ real-time detection system (Bio-Rad Laboratories). Relative expression levels of rtxBDE were calculated by using a standard curve obtained from PCR on serially diluted genomic DNA (as templates), with the 16S rRNA expression level as the internal reference for normalization, as described previously (5). Primers RTXB0701F and RTXB0701R for amplification of rtxBDE cDNA and RRSH0701F and RRSH0701R for V. vulnificus 16S rRNA cDNA are listed in Table 2.
For the primer extension, end-labeled 28-base primer RTXB-PE001, complementary to the coding region of rtxB, was added to the RNA and then extended with SuperScript II RNase H reverse transcriptase (Invitrogen) as previously described (4, 19). The cDNA products were purified and resolved on a sequencing gel alongside sequencing ladders generated from pMW0504 (Table 1) with the same primer used for the primer extension. The primer extension products were visualized and quantified using a phosphorimage analyzer (BAS1500; Fuji Photo Film Co. Ltd., Tokyo, Japan) and the Image Gauge (version 3.12) program.
RESULTS
Identification of rtxE gene of V. vulnificus.
The amino acid sequence deduced from the rtxE nucleotide sequence revealed a protein, RtxE, composed of 722 amino acids with a theoretical molecular mass of 79,845 Da and a pI of 6.67. The amino acid sequence of the V. vulnificus RtxE was 91% identical to that of the RtxE of V. cholerae (data not shown; http://www.ebi.ac.uk/clustalw). The V. cholerae RtxE has been characterized as a transport ATPase, a component of TISS (1). The predicted profile of the hydrophobicity (http://www.expasy.org) of the V. vulnificus RtxE was significantly similar to that of the RtxE of V. cholerae and is consistent with the fact that the RtxE protein is a membrane-associated protein (data not shown).
Effects of mutation in the rtxE gene on the secretion of RtxA.
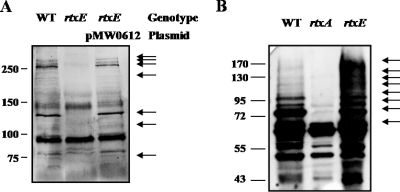
It has been proposed that RtxA is a key cytotoxic virulence factor and secreted by the TISS composed of the RtxE ATPase in V. cholerae (1, 14, 40). In order to determine whether the TISS with RtxE is required for the secretion of the V. vulnificus RtxA, supernatant fluids and cell lysates were collected from cultures of the wild type and MW061 and assayed for the presence of RtxA. A Western blot analysis revealed that the RtxA proteins, which are fragmented, were primarily found in the supernatant of the wild type (Fig. 2A), which was consistent with the prediction that the RtxA protein is a secreted protein. RtxA toxin breakdown products were not detected in the supernatant of the rtxE mutant, and the lack of RtxA secretion in the rtxE mutant was restored by the reintroduction of pMW0612 (Table 1) carrying a recombinant rtxE. Since RtxA toxin breakdown products were not detected in the supernatant of the rtxE mutant, it is reasonable to assume that RtxA of V. vulnificus is also secreted by TISS composed of RtxE as previously noted for V. cholerae (1). Consistent with this assumption, more RtxA toxin was detected in cell lysates of the rtxE mutant than in cell lysates of the wild type (Fig. 2B), indicating that the rtxE mutation inhibits secretion of RtxA toxin.
FIG. 2.
Effects of mutation in the rtxE gene on the secretion of RtxA. (A) For each lane, 30 μg of the proteins of concentrated supernatants from M06-24/O (WT), mutant MW061 (rtxE), or a complemented strain incubated with INT-407 (MOI of 100; 2 h) was loaded. (B) The bacterial cells were harvested, washed, and broken by sonication (ultrasonic processor; Sonics & Materials, Inc., CT). After centrifugation, 10 μg of the proteins of the clarified lysates was loaded for each lane. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and RtxA was detected by a Western blot analysis using rabbit anti-V. vulnificus RtxA antibody. The fragmented RtxA proteins are indicated by arrows.
RtxE is required for cytotoxicity to epithelial cells in vitro.
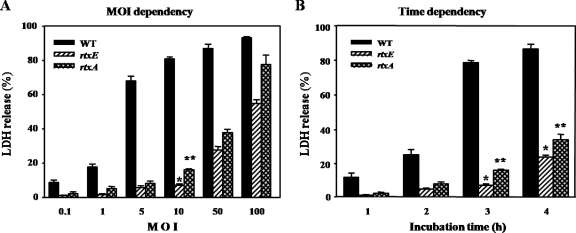
To examine the effects of the rtxE mutation on the ability of V. vulnificus to damage epithelial cells, the LDH activities from monolayers of INT-407 cells infected with 20 μl of a suspension of the wild-type and MW061 strains at different MOIs and incubated for 3 h were determined (Fig. 3A). The rtxE mutant MW061 exhibited significantly less cytotoxic activity when the MOI was up to 100. When the MOI was 10, the level of LDH activity from the INT-407 cells infected with MW061 was almost 10-fold less than that from the cells infected with the wild type. The INT-407 cells were also infected at an MOI of 10, and the LDH activities from the cells at different incubation times were compared (Fig. 3A). The cells infected with MW061 exhibited lower levels of LDH activity than the cells infected with the wild type when the cells were incubated with the bacterial suspension for as long as 4 h.
FIG. 3.
Effects of rtxE mutation on the cytotoxicity of V. vulnificus to INT-407 cells. (A) INT-407 cells were infected with the wild type, the rtxE mutant, or a complemented strain of V. vulnificus at various MOIs for 3 h (left) or at an MOI of 10 for various incubation times (right). The cell cytotoxicity was determined by an LDH release assay. The data represent the means ± standard errors of the means from three independent experiments. *, P < 0.01; **, P < 0.05 (relative to the groups infected with the wild type of V. vulnificus at each MOI or each incubation time). (B) Microscopic observation of INT-407 cells infected with the V. vulnificus strains at an MOI of 10 for 3 h. Shown, from the left, are uninfected (control) cells and cells infected with the wild type (WT), MW061 (rtxE), or the complemented strain.
It was examined whether the reintroduction of pMW0612 (Table 1) carrying a recombinant rtxE could complement the decrease of cytotoxic activity of MW061 cells. The lower LDH activities were restored to levels comparable to the levels obtained from the cells infected with the wild type when the cells were incubated with MW061(pMW0612) (Fig. 3A). Therefore, the decreased cytotoxic activity of MW061 was confirmed to result from the inactivation of functional rtxE rather than from any polar effects on genes downstream of rtxE.
Morphological studies were also carried out using INT-407 cells, which were seeded onto glass coverslips placed at the bottom of the tissue culture plate and infected with the V. vulnificus strains at an MOI of 10 for 3 h (Fig. 3B). The stained cells were assessed for size, regularity of the cell margin, and the morphological characteristics of the nuclei. As shown in Fig. 3B, many Giemsa-stained INT-407 cells exhibited marked cellular damage after infection with the wild type and MW061(pMW0612). Cytoplasmic loss and nuclear material condensation, typical phenotypes of cell death, were observed in the intestinal cells infected with the wild type and MW061(pMW0612). In contrast, fewer dead cells were observed after incubation with MW061. The cells infected with MW061 exhibited a less-damaged surface and less cytoplasmic loss. These results suggest that RtxE plays an important part in the ability of V. vulnificus to infect and injure host cells.
Comparison of cytotoxicity levels for rtxE and rtxA mutants.
Previously a ΔrtxA mutant, MW064, in which two-thirds of the rtxA ORF was deleted, was constructed by replacing rtxA on the chromosome with ΔrtxA (22). We examined the rtxE mutant MW061 and rtxA mutant MW064 for different effects on INT-407 intestinal epithelial cells. LDH activities from INT-407 cells infected with MW061 and MW064 strains at different MOIs and incubated for 3 h were determined (Fig. 4A). MW061 was consistently and significantly less cytotoxic than MW064 at all MOI points studied (Fig. 4A). Similarly, INT-407 cells were infected at an MOI of 10, and LDH activities from the cells at different incubation times were compared (Fig. 4B). The cells infected with MW061 exhibited lower levels of LDH activity than the cells infected with MW064 when the cells were incubated with the bacterial suspension for not less than 2 h. These results indicated that the rtxE mutant is significantly more attenuated in its ability to impair INT-407 cells and suggested that secretions of toxins, in addition to RtxA, involved in cytotoxic activity are inhibited by mutation of rtxE.
FIG. 4.
Cytotoxicities of V. vulnificus rtxE and rtxA mutants to INT-407 cells. INT-407 cells were infected with the rtxE or rtxA mutant of V. vulnificus at various MOIs for 3 h (A) or at an MOI of 10 for various incubation times (B). The cell cytotoxicities were determined by LDH release assays and are presented as described in the legend to Fig. 3. WT, wild type.
Virulence in mice is dependent on rtxE.
The role of the V. vulnificus rtxE gene in virulence was further examined using a mouse model. The LD50s for the iron-overloaded mice after intraperitoneal infection with MW061 and the wild type were 1.78 × 105 and 4.33 × 10°, respectively. Therefore, in the mouse model of intraperitoneal infection, in which the rtxE mutant exhibited more than a 4-log increase in the LD50 over the wild type, the rtxE mutant appeared to be less virulent than its parental wild type. Taking these results together, it is reasonable to conclude that rtxE is essential for the virulence of V. vulnificus in mice as well as in tissue cultures. Our recent work revealed that the rtxA mutant in the mouse model of intraperitoneal infection exhibited about a 2-log increase in the LD50 over the wild type (22). This result suggests again that a TISS composed of the RtxE ATPase is involved in secretion of multiple virulence factors.
rtxBDE is transcribed as a single operon.
To analyze the expression pattern of the rtxBDE genes at the transcriptional level, the presence of transcripts of the intergenic regions of rtxB, rtxD, and rtxE was examined using RT-PCR methods. When cDNA was synthesized from the RNA of M06-24/O(pMW0504) cells grown with LBS and PCR amplification was followed by using the primers RTXBD001 and RTXDE002 (Table 2; Fig. 1), a single, approximately 1.7-kb DNA was detected (Fig. 5). Based on the DNA sequence of rtxBDE, the 1.7-kb PCR product corresponded to the expected size of the amplification product between the RTXBD001 and RTXDE002 primers. This shows that rtxB, rtxD, and rtxE were transcribed as a single transcriptional unit. A positive control reaction with V. vulnificus genomic DNA as a template produced the same-sized 1.7-kb PCR product, and no product was observed when total RNA was directly used as a PCR template (a negative control). Therefore, it appeared that the rtxBDE genes were transcribed as a transcriptional operon rather than as three independent genes.
FIG. 5.
Analysis of the rtxBDE transcript by RT-PCR. Shown is an RT-PCR analysis of RNA isolated from a mid-exponential-phase culture of M06-24/O(pMW0504). The RNA was treated with DNase I (Sigma, St. Louis, MO) and used for the synthesis of cDNA by reverse transcription (SuperScript first-strand synthesis system for RT-PCR; Invitrogen, Carlsbad, CA). The cDNA (lane 1), DNase I-treated RNA (negative control; lane 2), and wild-type genomic DNA (positive control; lane 3) were used as templates for PCR. Molecular size markers (1 kb plus ladder; Invitrogen) and a PCR product are indicated.
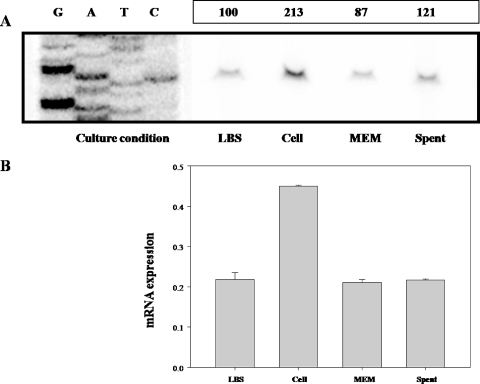
In order to map the promoter of the rtxBDE operon, the transcription start site of rtxBDE was determined by primer extension analysis. A single reverse transcript was produced from primer extension of RNA isolated from log phase cultures (Fig. 6). Using several different sets of primers, we were unable to identify any other transcription start sites by primer extension (data not shown). This result was consistent with the present assumption that rtxBDE is one transcriptional unit. The 5′ end of the rtxBDE transcript is located 102 bp upstream of the translational initiation codon of rtxB and was subsequently designated +1. The putative promoter upstream of the transcription start site was named PrtxBDE.
FIG. 6.
Identification of a transcription start site of the rtxBDE operon and sequence analysis of the rtxBDE upstream region. (A) The transcription start site was determined by primer extension of the RNA derived from M06-24/O(pMW0504) grown to mid-exponential phase in LBS. Lanes G, A, T, and C represent the nucleotide sequencing ladders of pMW0504. The asterisk indicates the site of transcription start. (B) The transcription start site is indicated by a bent arrow, and the putative promoter regions (−10 and −35) are underlined. The TTG translation initiation codon and putative ribosome binding site (GCAA) are in boldface.
rtxBDE is induced by exposure to host cells.
To determine the effect of host cells on the activity of PrtxBDE, the total bacterial RNA was isolated from M06-24/O(pMW0504) cultures exposed to intestinal INT-407 cells and used for primer extension analysis. The primer extension analysis performed with RNA isolated from M06-24/O(pMW0504) exposed to INT-407 for 2 h revealed a band of the reverse transcript, and its intensity was almost twofold greater than that of the reverse transcript obtained with RNA isolated from M06-24/O(pMW0504) grown with LBS (Fig. 7A). Based on the intensity of the bands of the reverse transcripts, it was apparent that the activity of PrtxBDE is induced by exposure of V. vulnificus to host cells. In order to characterize the effect of host cells on the expression of rtxBDE in more detail, primer extension analyses were performed with the RNA isolated from M06-24/O(pMW0504) cultures grown with different media, such as MEM and INT-407-spent MEM. Compared to results for the culture grown with LBS, the intensity of the bands of the reverse transcript was not significantly affected by exposure of the bacterial cells to either MEM to INT-407-spent MEM (Fig. 7A).
FIG. 7.
Activities of the PrtxBDE promoter in V. vulnificus cells grown under different conditions. Results for RNA derived from M06-24/O(pMW0504) exposed to INT-407 cells (cell) or grown with different media such as LBS, MEM, and INT-407-spent MEM (spent) are indicated. (A) The activities of PrtxBDE were determined separately by primer extension of the RNA. Lanes G, A, T, and C represent the nucleotide sequencing ladders of pMW0504. The relative levels of the PrtxBDE activity relative to the level of the PrtxBDE activity of V. vulnificus grown with LBS are presented. (B) For the real-time PCR analysis, the expression level of rtxBDE was normalized to the 16S rRNA expression level. Averages and standard errors of the means were calculated from at least three independent experiments. Details for preparation of total cellular RNA, primer extension analyses, and real-time PCR are given in Materials and Methods.
The induction of rtxBDE expression on the exposure to INT-407 cells was reconfirmed using real-time PCR (Fig. 7B). On exposure to INT-407 cells, the expression level of rtxBDE increased about twofold. However, the expression levels of rtxBDE in cultures grown with MEM and INT-407-spent MEM were not changed significantly. These results indicated that PrtxBDE activity was not induced by any medium components of MEM or by host cell components that are secreted from INT-407 and dissolved into MEM. Thus, taken together, these results make it reasonable to conclude that the activity of V. vulnificus PrtxBDE is induced by host cells and that the induction requires direct contact of the bacteria with the host cells.
DISCUSSION
The ability to secrete virulence factors that are involved in invasion, colonization, and survival within a host to the cell exterior is important in the pathogenicity of bacteria (11, 13). In particular, gram-negative pathogenic bacteria use several different types of secretion systems to export proteins, and five major pathways (types I, II, III, IV, and V) are known to primarily modulate this protein secretion. Among those, the type III secretion system (TTSS), one the most extensively investigated protein export pathways, plays diverse roles in pathogenesis and has been currently recognized as a major virulence factor in a wide range of gram-negative pathogenic bacteria (7, 15, 17, 43). However, a search of the genome sequences of both V. vulnificus CMCP6 and YJ016 to find TTSS gene homologues with a substantial level of identity with TTSS genes from other gram-negative bacteria at either the nucleotide or amino acid sequence level was not successful (S. H. Choi, unpublished data). Instead, V. vulnificus has been shown to possess a type II secretion system, responsible for extracellular secretion of at least three degradative enzymes including cytolysin (34). While it is unknown if the collective absence of proteins secreted by the type II pathway specifically results in reduced virulence, a mutant unable to express the type IV prepilin leader peptidase (and defective in both type IV pilus biogenesis and type II protein secretion) is significantly less virulent than a mutant defective in the expression of just the PilA type IV pilin subunit (34, 35). Recently, there has been increasing evidence that the widespread TISS is associated with virulence in many pathogenic bacteria (1, 16).
In the present study, the function of the TISS of V. vulnificus, presumably consisting of RtxB, RtxD, and RtxE based on research on V. cholerae (1), was examined by constructing an isogenic rtxE mutant and evaluating the differences between its virulence phenotype and that of the wild type. Compared to the wild type, the rtxE mutant was less toxic to intestinal epithelial cells in vitro and also exhibited significantly diminished virulence in mice. Apparently, this attenuation in the virulence of the rtxE mutant is associated with its ability to block V. vulnificus RtxA (RtxAVv) secretion, and it is reasonable to conclude that RtxAVv is also an important virulence factor in the pathogenesis of V. vulnificus. Since the mutation in rtxE decreased the release of LDH by V. vulnificus (Fig. 3), RtxAVv appeared to disrupt the membrane, as observed in the many pore-forming toxins belonging to the RTX protein family. Consistent with this, it has been demonstrated that the mutations of rtxAVv decreased the cytotoxicity as well as the virulence of V. vulnificus (22, 29). In contrast to RtxAVv, V. cholerae RtxA (RtxAVc) does not disrupt membrane integrity and thus is not likely to be a typical member of pore-forming toxins (14, 26, 40, 44). This different catalytic activity of the two toxins could be related to the extensive sequence divergences found in the two internal regions of RtxA (22, 40). We also hypothesized that this difference may simply account for the observed difference in pathogenesis between the two pathogens, such that V. vulnificus is much more destructive and cytolytic than V. cholerae.
A successful infection of pathogenic bacteria is correlated with their ability to survive and multiply within the harsh environments of the host. Therefore, it has been generally accepted that bacterial genes that are preferentially expressed within the environment of the host are likely important to pathogenesis (25). So far, several experimental approaches, such as in vivo expression technology (30), have been used for the extensive screening of bacterial genes that are specifically induced upon exposure to the host. These screens and subsequent characterization have allowed identification of many bacterial genes encoding potential virulence factors (25). However, until now, there have only been a few studies on the regulatory characteristics of the genes that are highly expressed in the host tissue. In the present study, twofold induction of rtxBDE expression was observed only in V. vulnificus cells exposed to INT-407 cells and not in V. vulnificus cells exposed to MEM or INT-407-spent MEM. Recently, it has been reported that expression of rtxBDE of V. cholerae was dependent on growth phase, decreasing during the stationary phase, and was regulated at the level of transcription (2). However, the growth rates of V. vulnificus with MEM, INT-407-spent MEM, and INT-407 cells were not significantly different (data not shown), and therefore it is reasonable to assume that the induction of V. vulnificus rtxBDE by exposure to INT-407 cells is not due to a different growth phase. In addition, the present study demonstrated that expression of rtxBDE is dependent on promoter PrtxBDE with the same transcription start site regardless of exposure of V. vulnificus to host cells (Fig. 6 and 7A).
Potential promoter sequences consisting of −10 and −35 segments separated by 17 nucleotides have been assigned to PrtxBDE (Fig. 6). The assigned sequences for −35 (TTCACT) and −10 (AACAAT) scored a 67% identity to the E. coli consensus sequences for promoters recognized by RNA polymerase with RpoD (σ70). The similarity of PrtxBDE to the E. coli consensus sequences suggests that PrtxBDE is most probably recognized by the V. vulnificus homolog of the σ70 RNA polymerase holoenzyme. However, it is noteworthy that, even with this substantial level of similarity to consensus sequences, the expression level of rtxBDE from PrtxBDE is extremely low and that detection of the rtxBDE transcript is not possible unless V. vulnificus carries recombinant rtxBDE (data not shown). Although other explanations are possible, we hypothesize that the basal level of PrtxBDE activity is very weak and repressed by a repressor(s), which turns into inactive form in V. vulnificus upon exposure to the host. However, additional work is needed to clarify what repressor(s) is really involved in the differential repression of PrtxBDE and how the signals imposed by the host are delivered to the repressor to inactivate it.
Acknowledgments
This study was supported by grants to S.H.C. from the MarineBio 21 Project, MOMAF, and the National Research Laboratory, KOSEF (R0A-2007-000-20039-0), ROK. J.H.L. was supported by a Korea Research Foundation grant funded by the Korean government (MOEHRD) (KRF-2006-352-F00031).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 4 February 2008.
REFERENCES
- 1.Boardman, B. K., and K. J. Satchell. 2004. Vibrio cholerae strains with mutations in an atypical type I secretion system accumulate RTX toxin intracellularly. J. Bacteriol. 1868137-8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boardman, B. K., B. M. Meehan, and K. J. Satchell. 2007. Growth phase regulation of Vibrio cholerae RTX toxin export. J. Bacteriol. 1891827-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Choi, H. K., N. Y. Park, D. I. Kim, H. J. Chung, S. Ryu, and S. H. Choi. 2002. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 27747292-47299. [DOI] [PubMed] [Google Scholar]
- 5.Choi, J., D. Shin, and S. Ryu. 2007. Implication of quorum sensing in Salmonella enterica serovar Typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 754885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, T. D., A. J. Martone, and R. K. Holmes. 1995. A new mobilizable vector for use in Vibrio cholerae and other gram-negative bacteria. Gene 15385-87. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, A., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73241-268. [DOI] [PubMed] [Google Scholar]
- 9.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694149-161. [DOI] [PubMed] [Google Scholar]
- 10.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvaux, M., and M. Hebraud. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30774-805. [DOI] [PubMed] [Google Scholar]
- 12.Falkow, S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10S274-S276. [DOI] [PubMed] [Google Scholar]
- 13.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullner, K. J., and J. J. Mekalanos. 2000. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 195315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galán, J. E., and D. Zhou. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc. Natl. Acad. Sci. USA 978754-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garmory, H. S., and R. W. Titball. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 726757-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goosney, D. L., S. Gruenheid, and B. B. Finlay. 2000. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16173-189. [DOI] [PubMed] [Google Scholar]
- 18.Gulig, P. A., K. L. Bourdage, and A. M. Starks. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43118-131. [PubMed] [Google Scholar]
- 19.Jeong, H. S., K. C. Jeong, H. K. Choi, K. J. Park, K. H. Lee, J. H. Rhee, and S. H. Choi. 2001. Differential expression of Vibrio vulnificus elastase gene in a growth-phase dependent manner by two different types of promoters. J. Biol. Chem. 27613875-13880. [DOI] [PubMed] [Google Scholar]
- 20.Kim, H. J., J. H. Lee, J. E. Rhee, H. S. Jeong, H. K. Choi, H. J. Chung, S., Ryu, and S. H. Choi. 2002. Identification and functional analysis of the putAP genes encoding Vibrio vulnificus proline dehydrogenase and proline permease. J. Microbiol. Biotechnol. 12318-326. [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacterophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. H., M. W. Kim, B. S. Kim, S. M. Kim, B. C. Lee, T. S. Kim, and S. H. Choi. 2007. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45146-152. [PubMed] [Google Scholar]
- 23.Lee, J. H., N. Y. Park, S. J. Park, and S. H. Choi. 2003. Identification and characterization of the Vibrio vulnificus phosphomannomutase gene. J. Microbiol. Biotechnol. 13149-154. [Google Scholar]
- 24.Lee, J. H., and S. H. Choi. 2006. Coactivation of Vibrio vulnificus putAP operon by cAMP receptor protein and PutR through cooperative binding to overlapping sites. Mol. Microbiol. 60513-524. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. H., and A. Camilli. 2000. Novel approaches to monitor bacterial gene expression in infected tissue and host. Curr. Opin. Microbiol. 397-101. [DOI] [PubMed] [Google Scholar]
- 26.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 961071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174207-214. [DOI] [PubMed] [Google Scholar]
- 28.Litwin, C. M., T. W. Rayback, and J. Skinner. 1996. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect. Immun. 642834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, M., A. F. Alice, H. Naka, and J. H. Crosa. 2007. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 753282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259686-688. [DOI] [PubMed] [Google Scholar]
- 31.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1781310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147217-226. [DOI] [PubMed] [Google Scholar]
- 34.Paranjpye, R. N., J. C. Lara, J. C. Pepe, C. M. Pepe, and M. S. Strom. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 665659-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 731411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, N. Y., J. H. Lee, M. W. Kim, H. G. Jeong, B. C. Lee, T. S. Kim, and S. H. Choi. 2006. Identificatioin of Vibrio vulnificus wbpP gene and evaluation of its role in virulence. Infect. Immun. 74721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, S. G., C. W. Kho, S. Cho, D. H. Lee, S. H. Kim, and B. C. Park. 2002. A functional proteomic analysis of secreted fibrinolytic enzymes from Bacillus subtilis 168 using a combined method of two-dimensional gel electrophoresis and zymography. Proteomics 2206-211. [DOI] [PubMed] [Google Scholar]
- 38.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27439-497. [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Sheahan, K. L., C. L. Cordero, and K. J. Satchell. 2004. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. USA 1019798-9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strom, M., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2177-188. [DOI] [PubMed] [Google Scholar]
- 42.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12420-430. [DOI] [PubMed] [Google Scholar]
- 43.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 5969-89. [DOI] [PubMed] [Google Scholar]
- 44.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 25785-111. [DOI] [PubMed] [Google Scholar]
- 45.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 581769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]