Abstract
The Cpx two-component system regulates an extracytoplasmic stress response that functions to rid the envelope of misfolded and mislocalized proteins that may interfere with normal cellular processes. The Cpx pathway is also involved in pathogenesis. This study investigated the role of the Cpx response in enteropathogenic Escherichia coli (EPEC) type III secretion (T3S). It was determined that a functional Cpx pathway is not required for T3S but that pathway activation inhibits secretion by reducing the cellular pools of T3S substrates. The EPEC T3S system structural components, as well as a number of its substrates, are encoded on the locus of enterocyte effacement (LEE) pathogenicity island. Transcriptional fusions to the five major operons of the LEE were constructed and examined under Cpx pathway-activating conditions. Induction of the Cpx response caused a decrease in the transcription of several LEE operons, with the most pronounced effect on LEE4 and LEE5. Collectively, these two operons encode components of the T3S translocation apparatus, the bacterial adhesin intimin, and the translocated bacterial receptor Tir. These data show for the first time that activation of the Cpx envelope stress response in EPEC inhibits T3S of both translocators and effectors, likely through down regulation of LEE transcription. Coupled with recent findings, our results suggest that Cpx-mediated down regulation of virulence is a conserved theme in a number of bacterial pathogens.
The Cpx two-component system regulates an envelope stress response adapted by Escherichia coli to monitor and maintain cell envelope integrity. This is accomplished by the interplay between a membrane-bound histidine kinase, CpxA, and a cytoplasmic response regulator, CpxR. In the absence of stress, CpxA autokinase activity is inhibited by the small periplasmic protein CpxP (20). Upon presentation of an inducing cue, such as alkaline pH or the accumulation of misfolded or mislocalized envelope proteins, CpxP is degraded and CpxA takes on autokinase and kinase activities that lead to the accumulation of CpxR∼P (2, 31, 54). A major subset of genes regulated by CpxR∼P encode folding and degrading factors involved in the biogenesis of periplasmic proteins as well as proteins en route to the outer membrane (8, 32, 51, 58). It is generally accepted that a key physiological role of the Cpx pathway is to rid the envelope of potentially toxic misfolded proteins and to return homeostasis. Another subset of the Cpx regulon includes genes involved in adherence. CpxR∼P has been shown to bind upstream of the promoter regions of genes involved in motility and chemotaxis and those encoding the adhesive appendages curli and P pili and the Yersinia adhesin, invasin (5, 13, 17, 29). These findings, coupled with the discovery that the Cpx pathway is activated upon contact with hydrophobic surfaces through the outer membrane lipoprotein NlpE (50), suggest that the Cpx response is intimately involved in attachment.
In conjunction with its activities in maintaining cell envelope integrity and responding to cellular attachment, the Cpx pathway is implicated in the elaboration and expression of multiple virulence-associated structures. The regulon member DsbA has been shown to catalyze the folding of secreted virulence factors as well as assembly components of type IV pili, P pili, and the type III secretion systems (T3SS) in numerous gram-negative pathogens (15, 25, 34, 44, 59). A second regulon member, DegP, enables intracellular survival, likely by degrading proteins which are unfolded due to exposure to high temperatures and oxidative stress inside the host (33, 41). The Cpx pathway has also been implicated in posttranscriptional regulation of the InvE regulatory protein in Salmonella (45) and of the major subunit of the type IV bundle-forming pilus (BFP) in enteropathogenic E. coli (EPEC) (49).
In addition to impacting virulence determinants at the posttranscriptional and assembly levels, there are multiple examples of the Cpx pathway having a direct effect on transcription of virulence genes and key virulence regulators. In Shigella sonnei, CpxR∼P binds upstream of a master regulator of virulence, VirF (47, 48). Elegant work with uropathogenic E. coli demonstrated that CpxR∼P binds to a region controlling pap phase variation and inhibits P pilus expression in vivo (26, 29). In Salmonella enterica serovar Typhimurium, a cpxA mutant decreases expression of a major regulator of invasion genes, HilA (46), and exhibits a diminished ability to attach to and invade eukaryotic cells (28). Lastly, a CpxR homologue positively regulates transcription of the icm-dot type IV secretion system in Legionella pneumophila (23).
The present study investigated the role of the Cpx pathway in EPEC T3S. EPEC is an attaching and effacing pathogen that initiates infection by adhering to host epithelial cells, using a type IV BFP, and then translocating virulence determinants into the host cells via a T3SS. Both the T3SS structural components and numerous EPEC secreted proteins (Esps) are encoded on a 35-kb pathogenicity island called the locus of enterocyte effacement (LEE). The LEE contains 41 genes and is organized into five main polycistronic operons (LEE1-5) (19). In addition to encoding the T3SS, the LEE also encodes the bacterial adhesin, intimin, its translocated receptor, Tir, and several T3S chaperones and effector proteins (7, 10, 19). The regulation of EPEC virulence gene expression involves a complex network of regulators (42). In the present communication, we demonstrate that activation of the Cpx pathway negatively affects EPEC T3S by down regulating the expression of several key components of the translocation apparatus as well as the translocated bacterial receptor Tir. We argue that while low levels of pathway activity can facilitate pathogenesis (e.g., BFP biogenesis), full activation down regulates nonessential protein traffic in the envelope.
MATERIALS AND METHODS
Medium and growth conditions.
Bacterial strains were maintained in Luria-Bertani (LB) broth with the appropriate antibiotics and grown at 37°C with shaking. Strains E2348/69 cpxA24 and TR10 were maintained at 30°C with shaking. The antibiotics used in this study included ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), amikacin (3 μg/ml), and spectinomycin (30 μg/ml) (Sigma-Aldrich).
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. All primers used in strain and plasmid construction are listed in Table 2. The construction of an E2348/96 cpxA24 mutant and luxCDABE reporter fusions is described below. The CpxR overexpression vector pUC19-cpxR was constructed by amplifying the cpxR gene and a portion of the cpxP gene from the chromosome of MC4100, using primers finpho and cpxR3′Eco. The PCR product was purified and cloned into the BamHI and EcoRI sites on pUC19 by use of standard molecular techniques.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| EPEC E2348/69 | Prototype O127:H6 EPEC strain | 40 |
| CFM-14-21 | E2348/69 sepB::TnphoA | 16 |
| E2348/69 Δler | E2346/69 with in-frame deletion in ler | 3 |
| ALN88 | E2348/69 cpxR::Kanr | 49 |
| ALN195 | E2348/69 cpxA24 | This study |
| ALN234 | E2348/69 cpxR::Camr | This study |
| MC4100 | F araD139Δ(argF-lac)U169 rpsL150 StrrrelA1 ref-4 flbB5301 deoC1 ptsF25 rbsR | 6 |
| TR10 | MC4100 cpxA24 | 54 |
| TR51 | MC4100 cpxR::Spcr | 54 |
| Plasmids | ||
| pUC19 | High-copy-number vector; Ampr | Invitrogen |
| pRE112 | Positive selection suicide vector | 18 |
| pACYC184 | Low-copy-number plasmid; Tetr Camr | NEB |
| pCVD450 | pACYC184 containing the per regulator | 24 |
| pCA24N | High-copy-number cloning vector; Camr | 37 |
| pCA-nlpE | nlpE cloned downstream of the IPTG-inducible promoter on pCA24N; Camr | 37 |
| pUC-cpxR | cpxR in pUC19 | This study |
| pNLP10 | pSC26 derivative with expanded multiple cloning site | 1; Price and Raivio, unpublished data |
| pJW15 | pNLP10 with p15 ori | This study |
| pJW17 | LEE1 promoter in pJW15 | This study |
| pJW18 | LEE2 promoter in pJW15 | This study |
| pJW19 | LEE3 promoter in pJW15 | This study |
| pJW20 | LEE4 promoter in pJW15 | This study |
| ptir::lux | LEE5 promoter in pJW15 | This study |
| pJW25 | cpxP promoter in pJW15 | This study |
| pNLP56 | dsbA promoter in pJW15 | Price and Raivio, unpublished data |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Target | Nucleotide sequence (5′-3′)a |
|---|---|---|
| p15ori-F | p15 ori | GCTTAATTAACCTCCTGTTCAGCTATGACG |
| p15ori-R | GCTTAATTAACGATGATAAGCTGTCAAACATGA | |
| BH1 LEE2/3-F | LEE2 promoter | ATGGATCCTATGCGATGCGATGATTAGG |
| ER1 LEE2/3-R | CGGAATTCATCCTGCGAACTCGTTCAAT | |
| ER1 LEE2/3-F | LEE3 promoter | CGGAATTCTATGCGATGCGATGATTAGG |
| BH1 LEE2/3-R | ATGGATCCATCCTGCGAACTCGTTCAAT | |
| proLEE4B-F | LEE4 promoter | GGGAATTCTCACGCTAGCCAGGATAAGA |
| proLEE4B-R | ATGGATCCAAACAGATGCGGTGTTTTGA | |
| ER1 protir-F | LEE5 promoter | CGGAATTCTGAACAGAAATTTGGTGGTTTG |
| BH1 protir-R | ATGGATCCGGTGCAGGTGGAATTAAAGC | |
| ER1 cpx UP | cpxP promoter | CGGAATTCCGGCAGCGGTAACTATGCGC |
| BH1 cpx DN | CGGGATCCCTGGCTCTCGACTGAAGGG | |
| finpho | cpxR | ACATTAACAGGAGGCTGTTCGTGC |
| cpxR3′Eco | CGGAATTCCGGTTAAGCTGCCTATCATG | |
| pNLP10-F | Multiple cloning site | GCTTCCCAACCTTACCAGAG |
| pNLP10-R | CACCAAAATTAATGGATTGCAC | |
| cpxA24F2 | cpxA24 | GGGGTACCGCCGTTCGCACTGGAG |
| cpxA24R2 | GCGAGCTCGCGCGTTTTTTTGCTGATTAC |
Underlined sequences indicate restriction sites introduced at the ends of the primers.
Construction of E2348/69 cpxA24 mutant (ALN195).
The cpxA24 locus of TR10 was PCR amplified using the primers cpxA24R2 and cpxA24F2 and cloned into the TOPO cloning vector according to the manufacturer's instructions (Invitrogen). The resulting clone was sequenced to confirm the presence of the cpxA24 mutation and no others, digested with SacI and KpnI, and cloned into the same sites of the gene replacement vector pRE112 (18). This clone was again sequenced to confirm the presence of the cpxA24 mutation and the absence of others, and the resulting construct was used to integrate the cpxA24 mutation into the chromosome of E2348/69, using published techniques (14).
Construction of luxCDABE reporters.
The pSC101 origin was removed from the promoterless luxCDABE reporter plasmid pNLP10 (1; N. L. Price and T. L. Raivio, unpublished data) by digestion with PacI. The p15 origin of pACYC184 was amplified using primers p15ori-F and p15ori-R. The PCR product was purified and cloned into the PacI sites of pNLP10 by use of standard molecular techniques to generate pJW15. All lux reporter plasmids utilized in this study are derivatives of pJW15. The pJW17 vector was constructed by digesting pJLM164 (43) with the EcoRI and BamHI restriction enzymes and cloning the subsequent fragment into the EcoRI and BamHI sites of pJW15. The pJW18, pJW19, pJW20, and ptir-lux reporter fusions were all constructed by amplifying the promoter regions of the genes of interest from the chromosome of E2348/69, using BamHI- and EcoRI-tagged primers (Table 2). The PCR products were purified and cloned into the BamHI and EcoRI sites of pJW15, using standard techniques. Inserts were confirmed by sequencing the multiple cloning site of pJW15 with the pNLP10-F and pNLP10-R primers and comparing the sequence to the partial E2348/69 sequence available on the NCBI website (http://www.ncbi.nlm.nih.gov). The pJW25 vector was constructed similarly, with the cpxP promoter amplified from the chromosome of MC4100, using the ER1 cpx UP and BH1 cpx DN primers (Table 2).
Bioluminescence assay.
Single colonies of strains of interest were inoculated into 5 ml of LB plus antibiotics in triplicate and grown at 37°C or 30°C with aeration overnight. The next day, a 1:100 dilution was made in 2 ml of LB or Dulbecco's modified Eagle's medium (DMEM)/F-12 supplemented with 0.1 M Tris, pH 7.5, and antibiotics and grown at 37°C with shaking. After 2 hours of growth, 200 μl of culture was transferred to a 96-well white-sided plate (Gibco), and the absorbance (A600) and level of bioluminescence (counts per second [cps]) were measured using a Wallac 420 multilabel plate reader (Perkin-Elmer). The 96-well plate was returned to 37°C, and the A600 and bioluminescence were read every 2 hours for 8 to 10 hours. The data presented here represent values collected after 6 hours of growth or at an A600 of 0.4 to 0.6. In the case of strains harboring the pCA-nlpE vector, strains were grown for 2 hours, induced with 60 μM IPTG (isopropyl-β-d-thiogalactopyranoside), and returned to 37°C. The A600 and the level of bioluminescence were read every hour for up to 5 hours postinduction, as described above. For each time point, the cps and A600 values were corrected by subtracting the values obtained from those of a blank medium control. The final bioluminescence (cps/A600) values were calculated by dividing the corrected cps by the corrected A600. Data presented represent the means and standard deviations for three replicates. Each experiment was repeated a minimum of three times.
Secretion assay.
Secretion assays were performed as previously described (36). Briefly, an overnight culture of the strain of interest was subcultured 1:100 in DMEM/F-12, with 0.1 M Tris, pH 7.5, supplemented with the appropriate antibiotics and grown at 37°C with shaking. Strains harboring pCA24N-based vectors were induced with 60 μM IPTG after 2 hours of growth. All cultures were grown until they reached an A600 of 0.5 to 0.7. At that time, 1 ml of culture was collected, cells were pelleted, and the supernatant was transferred to a fresh tube. The pelleted cells were resuspended in 100 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye (56). The secreted proteins were precipitated from the supernatant by adding 10% trichloroacetic acid (TCA) and incubating the samples at 4°C overnight. Precipitated proteins were pelleted by centrifugation at 14,000 rpm at 4°C for 15 min and resuspended in 15 μl of SDS-PAGE loading dye. Ten microliters of precipitated protein was run in a 12% SDS-polyacrylamide gel and stained with Coomassie blue stain (10% Coomassie blue dye, 10% methanol-H2O [1:1], 10% acetic acid). Additionally, all secretion assays were repeated by subculturing an overnight culture 1:100 in DMEM/F-12 with the appropriate antibiotics and incubating it in a 5% CO2 incubator at 37°C statically. Cultures were grown to an A600 of 0.5 to 0.7, and the same protocol as that outlined above was followed. All secretion assays were repeated a minimum of three times, and data for one representative experiment are shown.
Western blot analysis.
Western analysis was performed as previously described (49). The blots were incubated with a 1:50,000 dilution of rabbit polyclonal antiserum raised against a maltose binding protein-CpxR fusion protein, a 1:50,000 dilution of bacterial alkaline phosphatase (BAP) polyclonal antiserum (Research Diagnostics, Inc.), or a 1:300 dilution of mouse monoclonal antiserum raised against EPEC Tir (a gift from R. DeVinney).
RESULTS
Activation of the Cpx pathway reduces secretion of EPEC effectors and translocators.
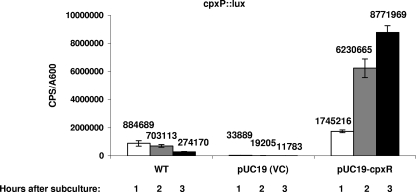
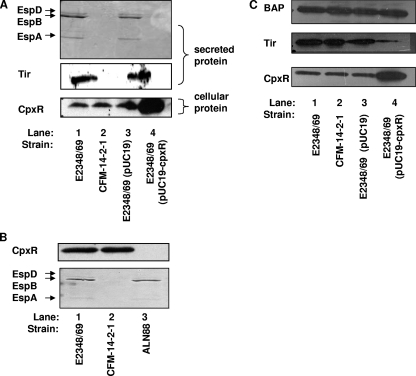
To determine if activation or inactivation of the Cpx pathway had an effect on the assembly and/or function of the EPEC T3S apparatus, we examined the secretion profiles of EPEC strains overexpressing the response regulator CpxR from a lac promoter on pUC19 or with cpxR interrupted by a kanamycin resistance cassette (ALN88). Cpx pathway activation or removal in these strains was confirmed by observing CpxR levels via Western analysis, probing with anti-CpxR antiserum, and by looking at the promoter activity of the known Cpx regulon member cpxP (Fig. 1). Cells overexpressing CpxR grew slower than the vector control but were still viable and capable of producing bioluminescence after 10 to 12 hours of growth. We observed that overexpression of CpxR from pUC19 led to 51-, 324-, and 744-fold increases in cpxP::lux reporter activity compared to that of the empty vector after 1, 2, and 3 h, respectively, of growth in DMEM/F-12 buffered with Tris (Fig. 1). Thus, overexpression of CpxR mimics activation of the Cpx response, as previously seen in E. coli K-12 (54). Western analysis of an E2348/69 cpxR null strain (ALN88) and the E2348/69 strain overexpressing CpxR revealed that there was no CpxR present in ALN88 (Fig. 2B, lane 3), while greatly elevated levels were present in the CpxR overexpression strain (Fig. 2A, bottom panel, lane 4). Examination of the levels of the prominent T3S substrates EspA, EspB, EspD, and Tir revealed that activation of the Cpx pathway by overexpression of CpxR drastically reduced the secretion of these proteins (Fig. 2A, compare lanes 3 and 4). In contrast, knocking out cpxR had no discernible effect on T3S (Fig. 2B, compare lanes 1 and 3). These data indicate that while exaggerated levels of pathway activation adversely affect T3S, the Cpx pathway is not essential for this stage in EPEC pathogenesis.
FIG. 1.
Overexpressing cpxR from pUC19 constitutively activates the Cpx pathway. Expression from the cpxP promoter was measured by monitoring levels of bioluminescence from a cpxP::lux fusion in pJW25 in E2348/69 carrying the vector control pUC19 or pUC19-cpxR. Strains were grown in DMEM/F-12 buffered with Tris at 37°C with aeration, and the A600 and bioluminescence (cps) were measured every hour for 3 hours. Bars represent the means for three replicates; error bars represent the standard deviations.
FIG. 2.
Overexpression of CpxR diminishes T3S of EspA, EspB, EspD, and Tir and leads to decreased intracellular levels of Tir. Secretion assays were performed by growing EPEC strains in DMEM/F-12 in 5% CO2 at 37°C. Bacterial cells were pelleted, and the secreted proteins were precipitated from the supernatant with 10% TCA. (A) Secretion profiles for E2348/69 (lane 1), the T3S mutant CFM-14-2-1 (lane 2), the vector control E2348/69(pUC19) (lane 3), and E2348/69(pUC19-cpxR) (lane 4). Levels of the T3S substrates EspA, EspB, and EspD secreted from the cell were determined by SDS-PAGE and Coomassie blue staining (top). The secreted proteins were also subjected to Western analysis with anti-Tir antiserum (middle). Western analysis was performed on the bacterial cell pellet, probing with anti-CpxR antiserum (bottom). This experiment was repeated at least three times, and all the blots in this figure come from one single experiment. (B) Secretion profiles of E2348/69 (lane 1), CFM-14-2-1 (lane 2), and the cpxR::kan mutant ALN88 (lane 3). CpxR levels were determined by performing Western analysis on bacterial cells, probing with anti-CpxR antiserum (top). The levels of EspA, EspB, and EspD secreted from the cells were visualized by SDS-PAGE followed by staining with Coomassie blue (bottom). (C) Western analysis of E2348/69 (lane 1), CFM-14-2-1 (lane 2), E2348/69(pUC19) (lane 3), and E2348/69(pUC19-cpxR) (lane 4) cells probed with anti-Tir (middle), anti-CpxR (bottom), and anti-BAP (top).
In an effort to establish whether the Cpx pathway was exerting its effect on the T3S apparatus or the effectors themselves, we compared the cellular level of Tir in E2348/69(pUC19-cpxR) to that of the T3S mutant CFM-14-2-1 (16). The T3S mutant had comparable intracellular levels of Tir to the wild-type level (Fig. 2C, compare lanes 1 and 2), while EPEC cells overexpressing CpxR had a decreased intracellular level of Tir (Fig. 2C, compare lanes 3 and 4). The variation in intracellular Tir levels was not due to loading differences, since control blots with antiserum to BAP revealed no differences among the strains (Fig. 2C, top panel). Also, when the same strains were grown under low-Ca2+conditions shown to promote effector secretion (11), there were increased cellular levels of Tir in the T3S mutant CFM-14-2-1 and reduced cellular levels of Tir in E2348/69(pUC19-cpxR) compared to the wild-type level (data not shown). This suggests that Cpx pathway activation results in diminished intracellular levels of T3S substrates.
The Cpx pathway negatively regulates LEE transcription independently of Ler.
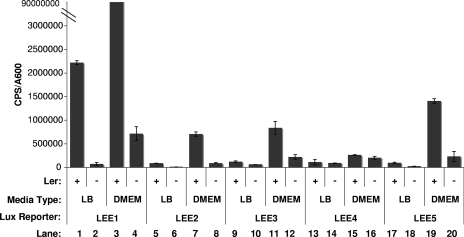
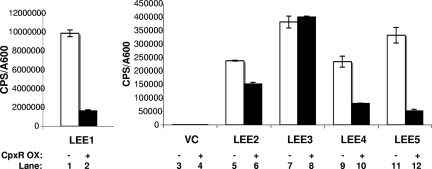
After observing that activation of the Cpx pathway leads to a decrease in cellular levels of Tir, we were interested in ascertaining whether the Cpx pathway was exerting its effect on T3S at the transcriptional level. We generated reporter constructs harboring the promoter regions of each of the five LEE operons cloned in front of a promoterless luxCDABE cassette in pJW15. To confirm that the reporters were functioning properly, bioluminescence levels were assayed in a wild-type EPEC background and in an EPEC strain lacking the master regulator of the LEE, Ler (E2348/69 Δler) (3), grown in either LB or DMEM/F-12. As expected, all of the LEE reporters exhibited some decrease in expression in the absence of Ler (Fig. 3). Furthermore, all LEE::lux fusions were expressed at elevated levels when they were grown in DMEM, a condition thought to mimic infection and known to induce virulence gene expression (36) (Fig. 3). The LEE4::lux fusion was the least sensitive to changes in medium type, which is in agreement with published observations (39). Levels of luminescence produced by the LEE reporter constructs were then assayed in the presence and absence of CpxR overexpression. It was found that Cpx response activation reduced the expression of LEE1 and LEE5 between 3- and 6-fold and that of LEE4 between 3- and 10-fold and had little (<2-fold) to no effect on LEE2 and LEE3 (Fig. 4). The ranges represent the differences obtained from three separate experiments, each of which contained three replicates per strain. These findings coincide with our observations of diminished EspA, EspB, EspD, and Tir secretion (Fig. 2), as LEE4 and LEE5 carry espADB and tir, respectively. This experiment suggested that the decrease in T3S observed for EPEC cells overexpressing CpxR was at least partly due to a decrease in LEE4 and LEE5 transcription. Since LEE1 encodes the master regulator of the LEE, Ler, we were surprised that the LEE2 and LEE3 reporters revealed little to no change in expression when LEE1 expression was down regulated. This observation may be attributed to Ler-independent modes of activation or to the stability of the Ler protein. In support of this, reverse transcription-PCR analysis of LEE1-5 during an in vitro infection found that expression of LEE3-5 increased, while LEE1 transcription decreased (39). These findings, coupled with the results described below, suggest that it is unlikely that Cpx pathway activation influences LEE transcription via Ler.
FIG. 3.
LEE::lux reporters are regulated by Ler and environmental cues. Expression from the LEE::lux fusions was examined in the wild-type E2348/69 and E2348/69 ler strains grown in LB and DMEM/F-12 buffered with Tris at 37°C. The A600 and bioluminescence (cps) were measured every 2 hours for 8 hours. Data represented here are from the 6-hour time point (A600 = 0.4 to 0.6). Bars represent the means for three replicates; error bars represent the standard deviations. The experiment was repeated three times, and data for one representative experiment are shown.
FIG. 4.
Overexpressing CpxR inhibits transcription of LEE4 and LEE5. Expression from the LEE promoters was measured by monitoring levels of bioluminescence from LEE::lux fusions in pJW15 in strains of EPEC carrying the vector control (VC) pUC19 (white bars) or pUC19-cpxR (black bars). Strains were grown in DMEM/F-12 buffered with Tris at 37°C with shaking, and the A600 and bioluminescence (cps) were measured every 2 hours for 8 hours. Data represented here are from the 6-hour time point (A600 = 0.4 to 0.6). Bars represent the means for three replicates; error bars represent the standard deviations. The experiment was repeated three times, and data for one representative experiment are shown.
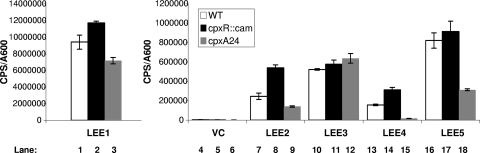
To further support our results, we examined LEE expression in the absence of a functional Cpx pathway in an E2348/69 cpxR::cam strain and in the presence of a constitutively activated Cpx response in an E2348/69 cpxA24 background. The E2348/69 cpxA24 strain was generated by deleting specific residues within the periplasmic sensing domain of CpxA that are important for signal detection (54). This type of mutation maintains CpxA autophosphorylation and kinase activities while eliminating its phosphatase activity, resulting in elevated levels of CpxR∼P (54). In line with the results found for overexpression of CpxR, LEE4 and LEE5 demonstrated a marked decrease in transcription in the cpxA24 background (Fig. 5, compare lanes 13 and 15 and lanes 16 and 18). Interestingly, the cpxA24 allele had a minor affect on LEE1 expression (Fig. 5, lanes 1 and 3). The cpxR null strain caused only minor increases in LEE1, LEE2, LEE4, and LEE5 expression (Fig. 5, compare lanes 1 and 2, 7 and 8, 13 and 14, and 16 and 17).
FIG. 5.
LEE4 and LEE5 expression is down regulated in E2348/69 cpxA24. Expression from the LEE promoters was measured by monitoring the bioluminescence from LEE::lux reporters in wild-type E2348/69 (white bars), E2348/69 cpxR::cam (black bars), and E2348/69 cpxA24 (gray bars) backgrounds. The strains were grown in DMEM/F-12 buffered with Tris at 37°C with shaking, and the A600 and bioluminescence (cps) were measured every 2 hours. The data represented here are from the 6-hour time point (A600= 0.4 to 0.6). Bars represent the means for three replicates; error bars represent the standard deviations.
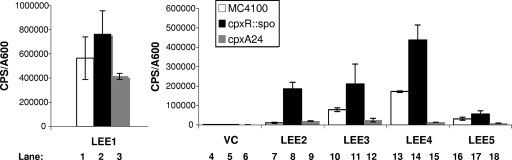
The observation that LEE4 and LEE5 transcription levels were reduced but the LEE1 transcription level was not suggested that the effects on LEE4 and LEE5 might be independent of LEE1-encoded Ler. To test this, the LEE::lux reporter plasmids were moved into a nonpathogenic E. coli K-12 background, and LEE expression was assayed in the presence of mutations that either eliminate (cpxR::spc; TR51) or constitutively activate (cpxA24; TR10) the Cpx pathway. Interestingly, all five LEE operons exhibited some degree of negative regulation by the Cpx pathway in the nonpathogenic E. coli K-12 MC4100 background (Fig. 6). In the absence of a functional Cpx pathway in a cpxR null strain (TR51), all five LEE operons exhibited increases in expression, ranging from 1.3- to 16-fold. Conversely, in a constitutively active cpxA* mutant (TR10), we observed decreases in LEE1 (1.3-fold) (Fig. 6, compare lanes 1 and 3), LEE4 (12-fold) (Fig. 6, compare lanes 13 and 15), and LEE5 (3- to 3.5-fold) (Fig. 6, compare lanes 16 and 18), which mirrored the trends seen in EPEC upon CpxR overexpression (Fig. 4). In addition, the MC4100 cpxA* mutant also led to a 3- to 3.5-fold decrease in LEE3 expression (Fig. 6, compare lanes 10 and 12). It may be that subtle differences in LEE expression caused by activation or inactivation of the Cpx response are more visible in the K-12 strain background. This could be the result of the much lower levels of LEE expression in this strain than in EPEC (compare Fig. 4 and 5 with Fig. 6) due to the absence of several positive regulators not present in the nonpathogenic background. Regardless, we can conclude that the Cpx pathway does not act entirely through Ler or any other EPEC-specific regulator, as there is negative regulation of LEE genes by the Cpx pathway in both pathogenic and nonpathogenic strains of E. coli.
FIG. 6.
Inhibitory effect of the Cpx pathway on LEE transcription is independent of EPEC-specific regulators. Expression from the LEE promoters was measured by monitoring levels of bioluminescence from LEE::lux fusions in pJW15 in MC4100 (white bars), TR51 (black bars), and TR10 (gray bars). Strains were grown in LB at 37°C with shaking, and the A600 and bioluminescence (cps) were measured every 2 hours for 10 hours. Data represented here are from the 8-hour time point (late log phase). Bars represent the means for three replicates; error bars represent the standard deviations. VC, vector control.
The Cpx response inhibits T3S when EPEC-specific positive regulators are overexpressed.
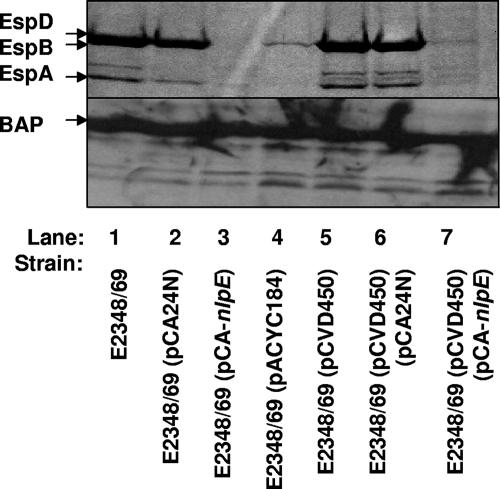
To further examine the relationship between the Cpx response and EPEC-specific positive regulators with regard to T3S, we examined the ability of overexpression of the EPEC-specific PerABC (BfpTVW) positive regulators encoded in plasmid pCVD450 (24) to overcome the effects of Cpx pathway activation on T3S. PerC is responsible for up regulating LEE1/ler expression, which is proposed to result in a cascade of positive regulation leading to increased expression of LEE2-5 (52). The pCVD450 plasmid has been shown to enhance T3S (12, 36). We confirmed this finding, since the presence of pCVD450 leads to a large increase in T3S relative to that in the same strain carrying the pACYC184 vector control (Fig. 7, compare lanes 4 and 5). For unknown reasons, the pACYC184 plasmid caused a decrease in E2348/69 T3S (Fig. 7, compare lanes 1 and 4). In contrast, E2348/69 carrying both pCVD450 and the NlpE overexpression vector pCA-nlpE secreted barely detectable levels of protein (Fig. 7, compare lanes 6 and 7), similar to the E2348/69 strain carrying pCA-nlpE alone (Fig. 7, compare lanes 3 and 7). The variation in levels was not due to loading differences, since control blots with antiserum to BAP revealed no differences among the strains (Fig. 7, lanes 1 to 7). These results indicate that activation of the Cpx response can still inhibit T3S, even in the presence of elevated expression of the LEE transcriptional activators PerABC and Ler. Interestingly, more EspB was detected in the EPEC strain overexpressing PerABC and NlpE than in the strain expressing only NlpE (Fig. 7, compare lanes 3 and 7). This might suggest that the presence of excess PerABC/Ler can partially counter the negative effects of the Cpx response.
FIG. 7.
Activation of the Cpx response decreases T3S when EPEC-specific positive regulators are overexpressed. Secretion profiles for E2348/69 (lane 1), E2348/69(pCA24N) (lane 2), E2348/69(pCA-nlpE) (lane 3), E2348/69(pACYC184) (lane 4), E2348/68(pCVD450) (lane 5), E2348/69(pCA24N)(pCVD450) (lane 6), and E2348/69(pCA-nlpE)(pCVD450) (lane 7) are shown. EPEC strains were grown in DMEM/F-12 at 37°C in 5% CO2 for 2 h, induced with 60 μM IPTG, and allowed to grow to an A600 of ∼0.5 to 0.7. Bacterial cells were pelleted, and secreted proteins were precipitated from the supernatant with 10% TCA. EspA, EspB, and EspD were detected by SDS-PAGE with Coomassie blue staining (top). The bacterial cells were subjected to Western analysis with anti-BAP antiserum (bottom).
Activation of the wild-type Cpx pathway confirms its inhibitory role in EPEC T3S.
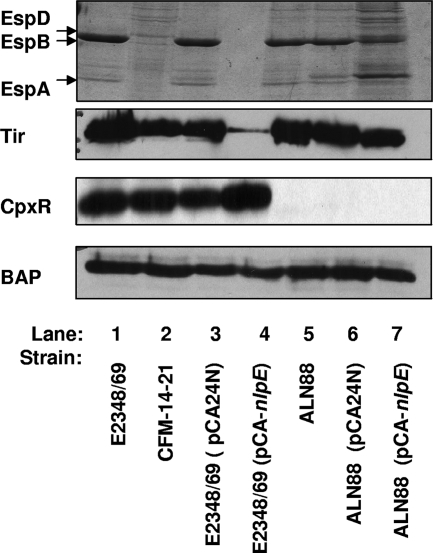
Activation of the Cpx response by CpxR overexpression is artificial, since no inducing cue is present that would lead to phosphorylation of CpxR by CpxA. In addition, activation of the pathway by this means leads to slow-growing EPEC strains (data not shown). To confirm that the effects of CpxR overexpression on T3S were characteristic of the wild-type Cpx pathway and not aberrant or due to growth effects, we repeated the secretion experiments, using an additional and proven mode of Cpx response activation. The outer membrane lipoprotein NlpE was overexpressed from an IPTG-inducible PT5-lac promoter on the pCA24N derivative pCA-nlpE (37). Overexpression of NlpE had only mild effects on growth, in contrast to CpxR overexpression or constitutive activation of the Cpx response by mutation (data not shown). We found that NlpE overexpression negatively affected secretion of EPEC effectors in a manner that was dependent on a functional Cpx pathway. This was established by comparing the secretion profiles of E2348/69 and E2348/69 cpxR::kan (ALN88) carrying the pCA-nlpE vector. In the presence of a functional Cpx pathway in E2348/69, induction of the pCA-nlpE plasmid effectively diminished secretion of T3S effectors (Fig. 8, compare lanes 3 and 4), while the same level of induction in a cpxR::kan mutant resulted in cells that, while slow growing, were still capable of T3S (Fig. 8, compare lanes 5 to 7). The decreases in EspB and Tir levels observed were not due to unequal loading, since Western analysis with antiserum directed against BAP indicated that there were no differences in overall protein amounts between strains (Fig. 8, bottom panel).
FIG. 8.
NlpE overexpression decreases T3S in a Cpx-dependent manner. Secretion profiles for E2348/69 (lane 1), CFM-14-2-1 (lane 2), E2348/69(pCA24N) (lane 3), E2348/69(pCA-nlpE) (lane 4), ALN88 (lane 5), ALN88(pCA24N) (lane 6), and ALN88(pCA-nlpE) (lane 7) are shown. EPEC strains were grown in DMEM/F-12 in 5% CO2 at 37°C for 2 hours, at which time the cultures were induced with 60 μM IPTG and allowed to grow for an additional 3 hours (A600 of ∼0.5 to 0.7). Bacterial cells were pelleted, and secreted proteins were precipitated from the supernatant with 10% TCA. EspA, EspB, and EspD proteins were detected by SDS-PAGE with Coomassie blue staining (top). The bacterial cells were subjected to Western analysis with anti-Tir (second panel), anti-CpxR (third panel), and anti-BAP (bottom).
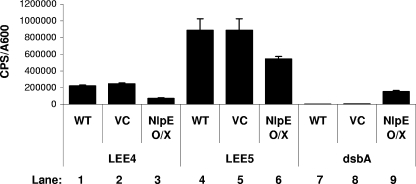
As with activation of the Cpx response by overproduction of the response regulator CpxR, NlpE overexpression impacted T3S by decreasing the transcription of LEE genes (Fig. 9) and, in turn, reducing the cellular pools of T3S substrates (Fig. 8, compare Tir levels in lanes 3 and 4). EPEC strains harboring pCA24N or pCA-nlpE and either the LEE1-lux, LEE4-lux, LEE5-lux, or dsbA-lux reporter plasmid were assayed for promoter activity every hour for 5 hours postinduction. High levels of expression from the dsbA promoter confirmed that the native EPEC Cpx pathway is activated by NlpE overexpression (Fig. 9, compare lanes 7 and 9). Activation of the Cpx response by this method reduced LEE4 and LEE5 transcription (Fig. 9, compare lanes 1 and 3 and lanes 4 and 6), albeit to a lesser extent than that with CpxR overexpression, and had a modest effect on LEE1 (data not shown). This finding suggests that activation of the wild-type Cpx pathway influences LEE transcription in the same fashion as overexpressing CpxR in trans does, although to a lesser degree.
FIG. 9.
NlpE overexpression inhibits transcription of LEE4 and LEE5. Bioluminescence assays were performed with E2348/69 (lanes 1, 4, and 7), E2348/69(pCA24N) (lanes 2, 5, and 8), and E2348/69(pCA-nlpE) (lanes 3, 6, and 9) harboring either pJW20 (lanes 1 to 3), ptir-lux (lanes 4 to 6), or pNLP56 (lanes 7 to 9). Strains were grown in DMEM/F-12 buffered with Tris at 37°C with shaking for 2 hours, at which time cultures were induced with 60 μM IPTG. Bioluminescence and the A600 were read every hour postinduction. The graph represents data from 3 hours postinduction. Bars represent the mean cps/A600 for three replicates, and error bars represent the standard deviations.
DISCUSSION
In EPEC, a functional Cpx pathway is required for the efficient elaboration of the type IV BFP involved in the initial attachment to the host epithelium (49). The present study investigated whether the Cpx response was also integral to the assembly and/or function of the EPEC T3SS. Surprisingly, while the EPEC secretion profile remained unchanged in a cpxR::kan mutant, activation of the Cpx response resulted in a significant decrease in the secretion of both translocator and effector proteins, namely, EspA, EspB, EspD, and Tir (Fig. 2 and 8). EspA monomers polymerize at or around the tip of the T3S needle, forming a long needle extension that is thought to facilitate attachment and formation of the translocation pore (38). EspB is transported through the T3SS and the translocon comprised of EspA and inserted into the host membrane, where it and an additional T3S substrate, EspD, form the pore in the host cell membrane (9, 30). It is through this pore that the bacterial receptor Tir and the other EPEC effectors enter the host (38). It seems likely that cells lacking the ability to efficiently secrete these important virulence factors would have a diminished ability to carry out a successful infection. One future objective of this work will be to determine if activation of the Cpx response reduces virulence of the attaching and effacing class of pathogens in vivo.
Comparison of the intracellular levels of Tir in EPEC cells overexpressing CpxR and in a T3S mutant revealed that activation of the Cpx response inhibits T3S by reducing cellular levels of T3S substrates (Fig. 2). An examination of the transcriptional profiles of the five major operons of the LEE under conditions shown to activate the Cpx response revealed that the pathway inhibits LEE4 and LEE5 expression, has little to no effect on LEE2 and LEE3, and has a variable effect on LEE1 (Fig. 4, 5, and 6). Collectively, LEE4 and LEE5 encode the translocator proteins (EspA, EspD, and EspB), the needle complex protein (EscF), the bacterial adhesin intimin, and its translocated receptor Tir (10). These findings suggest that Cpx pathway activation inhibits EPEC T3S, at least in part, by down regulating the expression of its substrates (Esps and Tir) and several structural components (EscF, EspA, EspB, and EspD) incorporated into the apparatus at the later stages of its assembly. It is also plausible that certain T3S structural components may not be assembled properly in the bacterial envelope when the Cpx pathway is activated. Future experiments will investigate the latter possibility.
Our results agree with and shed light on previous studies of the Cpx response in other pathogens. For example, the interruption of cpxR in Salmonella enterica serovar Typhimurium and Yersinia enterocolitica had no observable effect on virulence in an animal model (27, 28). Alternatively, CpxR∼P has been shown to negatively regulate P pilus expression in uropathogenic E. coli (26). In Salmonella, cpxA mutants caused a decrease in the expression of a key virulence regulator, HilA (46), and experienced a diminished ability to attach and invade in vivo (28). In Shigella, a cpxA mutant affects the posttranscriptional levels of InvE, a regulatory protein involved in relieving H-NS repression of genes involved in T3S (45). The disruption of cpxA in these organisms would eliminate CpxA autokinase, kinase, and phosphatase activities. Since the native CpxR protein is capable of accepting phosphoryl groups from alternate phospho-donors in the cell, it is presumed that in the absence of CpxA phosphatase activity, there would be an accumulation of CpxR∼P, mimicking pathway activation (8, 53, 54). Thus, it may be that some of the negative effects observed on virulence in these cpxA mutants are due to activation of the Cpx response, similar to what we report here. Cumulatively, these findings suggest that the Cpx pathway is not a requisite for gram-negative pathogenesis and that, upon activation, it reduces the pathogenic potential of an organism.
Further support for our findings was published while the manuscript was in preparation. Carlsson et al. (4) investigated the role of the Cpx response in Yersinia pseudotuberculosis T3S and found that a cpxA null mutant secreted lower levels of protein and was defective for translocation. The reduced ability to secrete effectors was hypothesized to be the result of an inability to assemble a functional T3S apparatus, as the cpxA null mutant had decreased cellular levels of needle-associated proteins (including the EspA orthologue LcrV) and reduced expression of several T3S effectors (4). In addition to a decrease in effector expression, we observed a marked decrease in the transcription of translocators that Carlsson et al. did not observe (4). Nonetheless, the similarity of our findings to those of Carlsson et al. (4) suggests that activation of the Cpx pathway may influence T3S at a common point for both EPEC and Yersinia, i.e., late in the assembly of the T3S apparatus. The differences in effector versus translocator regulation by the Cpx response might be due to differences in how each T3SS is regulated (21). In Yersinia, the expression and secretion of effectors are coordinately regulated (22). Extensive work with EPEC has failed to demonstrate a similar mechanism (11, 42).
An interesting observation in this study was that expression from the LEE1 promoter was affected differently by overexpression of the response regulator CpxR than that of a constitutive cpxA* mutant and wild-type activation of the Cpx response by NlpE overexpression (Fig. 4 and 5; data not shown). When CpxR was overexpressed from a high-copy-number plasmid, we observed a considerable decrease in LEE1, LEE4, and LEE5 expression (Fig. 4). When the experiment was repeated with alternate forms of Cpx response induction, LEE4 and LEE5 expression levels were consistently decreased, whereas the LEE1 promoter was not strongly affected (Fig. 5, 6, and 9; data not shown). The cpxA* mutant lacks phosphatase activity but maintains autokinase and kinase activities (54). NlpE overexpression generates a signal that is transduced through the native CpxAR pathway (57). Alternatively, flooding the cell with multiple copies of the response regulator bypasses normal signal transduction entirely. Thus, perhaps a high concentration of unphosphorylated response regulator exerts stronger repressive effects at the LEE1 promoter than do increased levels of CpxR∼P. If so, this effect is not likely relevant in vivo, since the Cpx response is autoregulated (55) and therefore the only time when high levels of CpxR would be present is when the response is activated strongly and CpxR is phosphorylated. This observation poses a strong argument that any results obtained by mimicking Cpx pathway induction through CpxR overexpression should be confirmed with an inducer that acts through the wild-type signal transduction pathway.
Testing the effect of Cpx pathway activation on LEE promoter activity in a nonpathogenic E. coli background revealed that the Cpx pathway is capable of inhibiting LEE expression in the absence of EPEC-specific regulators (Fig. 6). These data suggest that Cpx-mediated inhibition of LEE gene expression and T3S functions independently of Ler. This conclusion is backed up by the weak effects of constitutive activation of the Cpx response and NlpE overexpression on LEE1 expression (Fig. 6; data not shown). Indeed, Cpx pathway activation partially negated the stimulatory effects that overexpressing the positive regulatory locus perABC had on T3S (Fig. 7). Since PerC is known to activate ler gene expression (52), these data again suggest that Cpx pathway activation works independently of Ler to down regulate T3S. At this time, we cannot rule out that the Cpx response might inhibit transcription of the perABC locus from the pCVD450 plasmid used to overexpress these regulators, but we think this unlikely given the weak effects that Cpx pathway activation by mutation or NlpE overexpression had on LEE1 expression (Fig. 6; data not shown). It is also possible that in the presence of excess PerABC/Ler, activation of the Cpx response inhibits T3S partly through posttranscriptional mechanisms. Changes in the status of envelope protein folding/degradation brought on by activation of the Cpx response might disrupt assembly of the T3S machine, as proposed for Yersinia pseudotuberculosis (4).
To elucidate whether CpxR was a direct regulator of LEE expression, the region upstream of the down-regulated operons, LEE4 and LEE5, was screened for the putative CpxR binding site (GTAAN6-7GTAA) (51). The absence of the proposed sequence in the promoter region of LEE4 suggested that the decrease in transcription observed for this operon is an indirect effect of Cpx response activation. Surprisingly, a putative binding site was identified in the region near the transcriptional start site of LEE5. Thus, Cpx regulation of LEE5 may be direct. Alternatively, the Cpx pathway may impose its effect on some unknown regulator found in both pathogenic and nonpathogenic E. coli strains. Among the regulators conserved across both strain backgrounds are H-NS, IHF, Fis, BipA, and QseA (35, 42). All are global regulators that influence a multitude of cellular processes. No one regulator is an obvious target of the Cpx response. The influence of the Cpx pathway on these regulators is currently being investigated. Elucidating the mechanism by which the Cpx pathway negatively regulates LEE4 and LEE5 expression may provide insight into how the Cpx pathway down regulates virulence gene expression in multiple pathogens.
In summary, we have shown that the Cpx envelope stress response inhibits EPEC T3S at the transcriptional level. We propose that high levels of Cpx pathway activation by the presence of misfolded or mislocalized envelope proteins set in motion events leading to the down regulation of unessential protein traffic in the periplasm. In doing so, the Cpx response can prevent the buildup of envelope proteins that could potentially interfere with essential cellular processes while it attempts to rid the cell of the imposing stress. In addition, by down regulating expression of energetically costly higher-level complexes, the cell can conserve energy until it enters a more favorable environment. Clearly, such a response is not likely to be relevant at the site of intestinal infection in the host. Indeed, our data argue that the Cpx response is not important for regulated virulence determinant expression in the intestine, since eliminating CpxR has no effect on T3S or LEE gene expression under conditions thought to mimic this environment (Fig. 2 and 5). However, it may be that shutting down T3S is important en route to the intestine or, alternatively, in the environment during transmission between hosts. Regardless, our observations and those of others suggest that the ability of Cpx envelope stress response activation to inhibit T3S and virulence is conserved in multiple intestinal pathogens and suggest exciting possibilities for the development of novel therapeutic agents.
Acknowledgments
We thank the Alberta Heritage Foundation for Medical Research, The Canadian Institutes of Health Research, and the Natural Sciences and Engineering Research Council of Canada for support.
In addition, we are grateful to M. Donnenberg, B. Finlay, and R. DeVinney for providing strains and reagents.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1843450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buelow, B. R., and T. L. Raivio. 2005. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J. Bacteriol. 1876622-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustamente, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39664-678. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson, K. E., J. Lui, P. J. Edqvist, and M. S. Francis. 2007. Extracytoplasmic stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 83913-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsson, K. E., J. Lui, P. J. Edqvist, and M. S. Francis. 2007. Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect. Immun. 754386-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of lac genes to selected promoters in Escherichia coli using bacteriophages lambda and Mu. J. Mol. Biol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unraveling pathogenesis. FEMS Microbiol. Lett. 2983-98. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., and T. J. Silhavy. 1997. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 111183-1193. [DOI] [PubMed] [Google Scholar]
- 9.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3865-871. [DOI] [PubMed] [Google Scholar]
- 10.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 828-34. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., Y. Li, P. R. Hardwidge, E. A. Frey, R. A. Pfuetzner, A. Lee, S. Gruenheid, N. C. J. Strynadka, J. L. Puente, and B. B. Finlay. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 732135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devinney, R., I. Nisan, S. Ruschkowski, I. Rosenshine, and B. B. Finlay. 2001. Tir tyrosine phosphorylation and pedestal formation are delayed in enteropathogenic Escherichia coli sepZ::TnphoA mutant 30-5-1(3). Infect. Immun. 69559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator of CpxR∼P in Escherichia coli. J. Biol. Chem. 27726652-26661. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 594310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnenberg, M. S., H.-Z. Zhang, and K. D. Stone. 1997. Biogenesis of bundle forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene 19233-38. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg, M. S., S. B. Calderwood, A. Donohue-Rolfe, G. T. Keusch, and J. P. Kaper. 1990. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infect. Immun. 581565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeuene. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178169-175. [DOI] [PubMed] [Google Scholar]
- 18.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207149-157. [DOI] [PubMed] [Google Scholar]
- 19.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 281-4. [DOI] [PubMed] [Google Scholar]
- 20.Fleischer, R., R. K. Heermann, K. Jung, and H. Sabine. 2007. Purification, reconstitution, and characterization of the CpxAR envelope stress system of Escherichia coli. J. Biol. Chem. 2828583-8593. [DOI] [PubMed] [Google Scholar]
- 21.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5166-172. [DOI] [PubMed] [Google Scholar]
- 22.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 421075-1093. [DOI] [PubMed] [Google Scholar]
- 23.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 631767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha, U. H., Y. Wang, and S. Jin. 2003. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 711590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16537-547. [DOI] [PubMed] [Google Scholar]
- 27.Heusipp, G., K. M. Nelson, M. A. Schmidt, and V. L. Miller. 2004. Regulation of htrA expression in Yersinia enterocolitica. FEMS Microbiol. Lett. 16227-235. [DOI] [PubMed] [Google Scholar]
- 28.Humphreys, S., G. Rowley, A. Stevenson, M. F. Anjum, M. J. Woodward, S. Gilbert, J. Kormanec, and M. Roberts. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 724654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 201508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ide, T., S. Laarmann, L. Greune, H. Schilers, H. Oberleithner, and M. A. Schmidt. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3669-679. [DOI] [PubMed] [Google Scholar]
- 31.Isaac, D. D., J. S. Pinkner, S. J. Hultgren, and T. J. Silhavy. 2005. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc. Natl. Acad. Sci. USA 10217775-17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson, M. W., and G. V. Plano. 1999. DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 1815126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Goara, G. Costa, T. Ali, I. Miller, and C. E. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5401-407. [DOI] [PubMed] [Google Scholar]
- 34.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 166394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 36.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 652606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitagawa, M., A. Takeshi, M. Afrifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of orf clones of Escherichia coli ASKA library (a complete set of E. coli H-12 orf archive): unique resources for biological research. DNA Res. 12291-299. [DOI] [PubMed] [Google Scholar]
- 38.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nissan, B. C. Nevesw, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 172166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leverton, L. Q., and J. B. Kaper. 2005. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect. Immun. 731034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152550-559. [DOI] [PubMed] [Google Scholar]
- 41.Li, S. R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 642088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellies, J. L., and A. M. S. Barron. June 2006, posting date. Chapter 8.9.1. Virulence gene regulation in Escherichia coli. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 43.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33296-306. [DOI] [PubMed] [Google Scholar]
- 44.Miki, T., N. Okada, and H. Danbara. 2004. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J. Biol. Chem. 27934631-34642. [DOI] [PubMed] [Google Scholar]
- 45.Mitobe, J., E. Arakawa, and H. Watanabe. 2005. A sensor of the two-component system CpxA affects expression of the type III secretion system through posttranscriptional processing of InvE. J. Bacteriol. 187107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 1492809-2817. [DOI] [PubMed] [Google Scholar]
- 47.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 1775062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 1803522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 992287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pogliano, J. A., S. Lynch, D. Belin, E. C. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 111169-1182. [DOI] [PubMed] [Google Scholar]
- 52.Porter, M. E., P. Mitchell, A. J. Roe, A. Free, D. G. Smith, and D. L. Gally. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 41117-1133. [DOI] [PubMed] [Google Scholar]
- 53.Raivio, T. L. 2005. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 561119-1128. [DOI] [PubMed] [Google Scholar]
- 54.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 1797724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 1815263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 1774216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenson, T. H., and A. A. Weiss. 2002. DsbA and DsbC are required for secretion of Bordetella pertussis. Infect. Immun. 702297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc. Natl. Acad. Sci. USA 924927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]