Abstract
Our previous work suggested that streptococcal pyrogenic exotoxin (SPE) B-induced apoptosis is mediated through a receptor-like mechanism. In this study, we have identified αvβ3 and Fas as the SPE B receptors for this function. The SPE B fragment without the RGD motif and G308S, a SPE B mutant with the RSD motif, induced less apoptosis than did native SPE B, suggesting that the RGD motif is critical for SPE B-induced apoptosis. Fluorescein isothiocyanate-SPE B binding assays and immunoprecipitation analysis showed that SPE B specifically interacted with αvβ3. Anti-αvβ3 antibody partially inhibited SPE B-induced apoptosis but had no effect on G308S-induced apoptosis. In addition, Fas binding to SPE B was verified in an affinity column and an immunoprecipitation analysis. Anti-Fas antibody inhibited SPE B- and G308S-induced apoptosis in a dose-dependent manner, suggesting that Fas-mediated SPE B-induced apoptosis also occurs RGD independently. Both anti-αvβ3 and anti-Fas antibodies synergistically inhibited SPE B-induced apoptosis. The apoptotic cascades were activated by SPE B and G308S, with a little delay by the latter. After SPE B binding, the cell surface level of αvβ3, but not of Fas, was decreased. The decreased αvβ3 level was restored by treatment with the proteasome inhibitor MG132, suggesting a SPE B-mediated endocytosis of integrin αvβ3 via the ubiquitin-proteasome system. Taken together, our results demonstrate that SPE B-induced apoptosis is mediated through αvβ3 integrin and Fas in a synergistic manner.
Streptococcal pyrogenic exotoxin (SPE) B is an important virulence factor produced by group A streptococcus (GAS), a human pathogenic bacterium that causes infections of various severities (30). SPE B is a cysteine protease secreted as a 40-kDa zymogen that is autocatalytically converted to a 28-kDa mature form. Studies of cellular reactions of SPE B have focused on its protease activity (10, 15, 43). Its role as a ligand protein has not been critically evaluated. Our recent study showed that SPE B-induced apoptosis in human lung epithelial A549 cells is mediated through a receptor-like mechanism and a mitochondrion-dependent pathway and that the protease activity of SPE B is required to initiate apoptotic signaling, most likely by exposing the binding site for SPE B (44). However, the SPE B receptors and the molecular mechanism of SPE B interaction with its receptors remain to be explored.
Three main SPE B variants have been identified in 200 GAS isolates of the speB gene (39). One variant contains an arginine-glycine-aspartic acid (RGD) sequence and interacts with human integrins αvβ3 and αIIbβ3. The GAS isolates that contain the RGD motif in SPE B are the most common cause of invasive diseases. Moreover, Kagawa et al. (23) demonstrated that the surface location of an RGD motif is a feature unique to SPE B among cysteine proteases and is linked to the pathogenesis of the most invasive strains of GAS. Therefore, understanding the role of the RGD motif in the interaction of SPE B with host cells is important. The RGD sequence present in various ligands is recognized by specific cellular integrins. The recognition of this tripeptide sequence is complex. The flank residues and three-dimensional presentation of the RGD sequence and the special feature of the integrin binding pocket are factors that determine whether the interaction of ligand and integrin occurs (35). Bacterial and viral proteins with the RGD motif bind to host integrins to gain entry into cells when infecting them (2, 20). Integrins are α/β heterodimeric membrane proteins that mediate cell adhesion to the surrounding extracellular matrix; they elicit a wide variety of intracellular signals that modulate cell growth, migration, and differentiation (14, 35). Compared to other integrin receptors, the αvβ3 integrin has a greater capacity to recognize and bind extracellular matrix proteins (i.e., fibronectin, fibrinogen, vitronectin, and thrombospondin) and other proteins with different biological functions (e.g., fibroblast growth factor 2 and metalloproteinase 2) (35). Integrin αvβ3 is preferentially expressed on endothelial cells and is the most important integrin for angiogenesis. However, its inhibition leads to apoptosis (4, 5). It has been reported that molecules containing the RGD motif act as integrin αvβ3 antagonists and induce epithelial cell apoptosis (3).
Accumulated evidence indicates that apoptosis is important for modulating the pathogeneses of many bacterial and viral diseases and that Fas signaling significantly contributes to the interaction of pathogens and their host cells (12, 29). Fas, a member of the tumor necrosis factor superfamily, is ubiquitously expressed in various tissue types, and its interaction with ligand on the cell surface is critical for inducing apoptosis. Apoptosis is critical in the development of acute lung injury, during which the Fas and Fas ligand pathway is activated (28). Fas is distributed mostly in cytoplasm and minimally on the cell surface in A549 cells in an unstimulated condition (13, 31). In the presence of hydrogen peroxide, Fas was significantly increased on the plasma membrane and decreased in the cytoplasmic fraction (13), indicating that hydrogen peroxide increased the translocation of Fas from the cytosol to the plasma membrane. The expression of the Fas in A549 cells is also regulated by various external stimuli. Fas mRNA and protein levels increased two- to fourfold after a respiratory syncytial virus infection (32). Fas was upregulated in response both to cigarette smoke extracts and to ionizing radiation (21, 47). Inhibitors of epidermal growth factor receptor tyrosine kinase, such as gefitinib and ZD1839, and extracts of the Chinese herbs gossypol and casuarinin all upregulated Fas signaling and apoptosis (7, 8, 25, 36). The stimulation of Fas by an agent other than physiological Fas ligand might also trigger apoptosis. Epigallocatechin gallate, a component of green tea, can bind to Fas and trigger the apoptotic cascade (16).
In this study, we used A549 cells to elucidate the SPE B receptors and the mechanism by which SPE B induces apoptosis after binding to the receptors. We identified αvβ3 and Fas as the receptors that mediate SPE B-induced apoptosis, which is not a usual one-ligand-to-one-receptor reaction; it is, instead, a one-ligand-to-two-receptors synergistic reaction.
MATERIALS AND METHODS
Expression and purification of rSPE B, C192S, and G308S.
The expression and purification of recombinant SPE B (rSPE B) and C192S, a protease-negative mutant, have been previously described (10). Briefly, the genomic DNA of Streptococcus pyogenes was extracted from strain A-20 (type M1, T1, opacity factor negative), which was isolated from a culture of blood from a patient with necrotizing fasciitis in National Cheng Kung University Hospital, Tainan, Taiwan. The ProSPE B gene was amplified using PCR with a His6 tag and BamHI recognition sites. The PCR product was purified and cloned into the BamHI site of the pET21a vector. A wild-type construct was used to produce a G308S mutation using overlap extension PCR (1). Recombinant constructs were transformed into Escherichia coli strain BL21(DE3) pLys. Bacteria were cultured in 250 ml of Luria-Bertani (LB) medium containing 100 μg/ml of ampicillin until the optical density at 600 nm (OD600) reached 0.5 to 1.0. An aliquot of 250 μl of IPTG (isopropyl-β-d-thiogalactopyranoside) (100 mg/ml) was added to induce protein expression. For the expression of rSPE B, C192S, and G308S, bacteria were cultured at 35°C, 28°C, and 37°C overnight, respectively. After induction, the bacteria were collected using centrifugation at 5,000 rpm for 15 min. The resuspended pellet was broken using a French press three times at 1,500 kg/cm2 for 30 s. Whole-cell lysates were centrifuged at 12,000 rpm for 10 min at 4°C to collect the supernatant. Proteins in the supernatant were then separated on a Ni2+-chelated column (Amersham Biosciences) and eluted using a 0 to 200 mM imidazol gradient.
Preparation of GST-fused SPE B fragments.
The genes of 639, 519, and 399 nucleotides that encode the SPE B fragments S1 (Gln146 to Gly358), S2 (Gln146 to Gln318), and S3 (Gln146 to Pro278) were amplified using PCR with ProSPE B/pET21a as a template. The sense primer 5′-GAATTCGAATTCCAACCAGTTGTTAAATCT-3′ with EcoRI recognition and the various antisense primers, 5′-GCGGCCGCGCGGCCGCACCCCAGTTAACATGG-3′, 5′-GCGGCCGCGCGGCCGCTTGTGATTCCCAATCT-3′, and 5′-GCGGCCGCGCGGCCGCTGGACCATAATCCATG-3′, for S1, S2, and S3 with a NotI site were used. The PCR product was purified and then cloned into the PGEX 4T-1 vector. The recombinant plasmid was transformed into the E. coli BL21(DE3) pLys strain. Bacteria were grown at 37°C for 5 to 6 h in LB medium containing 100 μg/ml of ampicillin until the OD600 reached 0.6 to 0.8. IPTG was added to the culture, which was then incubated at 20°C for 5 h. Glutathione S-transferase (GST) fusion-fragmented SPE B was in inclusion bodies, and a standard procedure using a denaturing condition was performed to refold the protein (10). The inclusion bodies were solubilized in denaturing solution (4.5 M urea, 20 mM Tris-HCl, and 200 mM NaCl [pH 8.0]), and the solutions were diluted to an OD280 of <0.1. The proteins were renatured by dialysis against 20 mM of Tris-HCl and 200 mM of NaCl (pH 8.0) and purified using a GSTrap FF column (Amersham Biosciences). The collected fractions were concentrated using Amicon ultrafiltration with a 10-kDa cutoff membrane and exchanged with phosphate-buffered saline (PBS). The purity was verified using SDS-PAGE. The purified proteins were passed through Detoxi-Gel (Pierce Biotechnology) to remove possible lipopolysaccharide contaminant. The 28-kDa C192S was prepared from 42-kDa C192S digested by a low dose of rSPE B, and undigested 42-kDa C192S was removed using a G-50 column (Amersham Biosciences).
Antibodies.
The antibodies used in integrin experiments were mouse monoclonal antibodies against α1 (FB12), α2 (P1E6), β1 (P4G1), αvβ3 (LM609), β3 (25E11) (Chemicon International), and αv (Becton Dickinson Transduction) and rabbit anti-αvβ3 polyclonal antibody (Santa Cruz Biotechnology) and mouse immunoglobulin G (IgG) (Chemicon International). Rabbit anti-SPE B polyclonal antibody was prepared and purified as previously described (24). The antibodies used in Fas experiments were mouse anti-Fas monoclonal antibody (Becton Dickinson Transduction, clone 13; Upstate Biotechnology, clone ZB4) and rabbit anti-Fas polyclonal antibody (Santa Cruz Biotechnology). The antibodies used in apoptotic-signaling experiments were mouse anti-caspase 8 monoclonal antibody, rabbit anti-caspase 9 polyclonal antibody (Cell Signaling Technology), rabbit anti-Bid polyclonal antibody (Biosource International), mouse anti-cytochrome c monoclonal antibody (Pharmingen), mouse anti-Bax and anti-caspase 3 monoclonal antibodies (Oncogene Research Products), and mouse anti-β-actin monoclonal antibody (Sigma-Aldrich). The secondary antibodies used in this study were fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) and peroxidase-conjugated goat anti-mouse and goat anti-rabbit antibodies (Calbiochem).
Cell culture.
A549 cells were routinely maintained in our laboratory in complete medium (Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and 50 μg/ml gentamicin). For experiments, cells at a density of 8 × 104/well were seeded in 24-well plates. After 24 h, the cells were washed with sterile PBS and then incubated in the complete medium containing the indicated agents for the time periods specified.
Fluorescence-activated cell sorter analysis of integrin and Fas levels.
Cells were trypsinized, washed with binding buffer (Dulbecco's modified Eagle's medium containing 2% fetal calf serum and 0.01% sodium azide [pH 7.4]), and then resuspended at 2 × 105 cells/100 μl in the same buffer. The cell suspension was incubated with primary antibodies at a 1:100 dilution for 60 min at 4°C. The cells were then washed twice with binding buffer and incubated with a 1:100 dilution of FITC-conjugated goat anti-mouse IgG for 60 min at 4°C. After two washes, the cells were resuspended in 1 ml of binding buffer and then analyzed on a FACScan using CELLQuest software (Becton Dickinson).
FITC-conjugated rSPE B binding assay.
FITC (freshly dissolved in dimethyl sulfoxide) was added to rSPE B in bicarbonate buffer (1.7 g Na2CO3 and 2.8 g NaHCO3 in 100 ml H2O [pH 9.4]) at 27 μg/mg of protein. The mixture was incubated overnight in the dark at 4°C with continuous rocking, and NH4Cl (50 μM) was then added to stop the reaction. Free FITC was removed from the mixture using a salt exchange on a PD-10 column (Amersham Biosciences). Cells at a density of 1 × 105/well were seeded onto a 12-well plate. After 24 h, the cells were washed with PBS and pretreated with a low dose of rSPE B (2 μg/ml) for 20 min to ensure better FITC-rSPE B binding. After two washes, the cells were incubated with various concentrations of FITC-rSPE B or FITC-rSPE B combined with 100-fold rSPE B for 2 h at 4°C. After incubation, the cells were washed twice and then lysed in 0.1 N NaOH. The fluorescence level on the cells was detected using a fluorescence enzyme-linked immunosorbent assay reader (Labsystems Fluoroskan; Global Medical Instrumentation). For the antibody-blocking assay, the cells were pretreated with the indicated antibodies (5 μg/ml) for 30 min at 4°C and then incubated with FITC-rSPE B for 2 h at 4°C.
Apoptosis analysis.
Cells were fixed with 70% ethanol overnight at −20°C. After two washes with PBS, the cells were stained with propidium iodide working solution (40 μg/ml propidium iodide and 100 μg/ml RNase in PBS) for 30 min at room temperature in the dark. Apoptotic cells were quantified on a FACScan using CELLQuest software (Becton Dickinson) and presented as the percentage of hypodiploid cells (43).
Western blotting.
Cells at a density of 2 × 105/well were seeded onto a six-well plate. After 24 h, the cells were washed with cold PBS and treated with the indicated agents in complete medium for the time periods specified for each experiment. After incubation, the cells were washed twice with cold PBS, lysed in Laemmli sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.001% bromophenol blue), and then boiled for 5 min. After SDS-PAGE, proteins were transferred to a PVDF membrane, blocked with PBS containing 5% skim milk, and probed with different primary antibodies. After three washes with PBS containing 0.05% Tween 20, the membranes were incubated with the secondary antibody. The membranes were washed three times with PBS containing 0.05% Tween 20, and the protein bands were visualized using enhanced chemiluminescence (ECL) (Amersham Biosciences).
Immunoprecipitation.
Cells at a density of 2 × 105/well were seeded onto a six-well plate for 24 h. After treatment with rSPE B or G308S for the times indicated in each experiment, the cells were lysed in modified RIPA lysis buffer (50 mM Tris [pH 7.4], 0.1% SDS, 1% Triton X-100, 5 mM EDTA, 150 mM NaCl, 0.5 mM sodium deoxycholate, and protease inhibitor cocktail). Total cell lysates were incubated with 1 μg of primary antibodies with continuous rocking for 2 h at 4°C and then mixed with protein A agarose beads and left overnight. After the mixtures had been washed to remove unbound material, the immune complexes were eluted using Laemmli sample buffer and Western blotted with the indicated antibodies.
Isolation of cytosolic, mitochondrial, and membrane fractions.
After the cells had been treated with rSPE B or G308S for the times indicated for each experiment, they were harvested by scraping them on ice, washed with ice-cold PBS, and then resuspended in 500 μl of buffer A (20 mM HEPES [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 40 μg/ml leupeptin, and 1 mM PMSF). After the cells had been on ice for 1 h, the lysed cells were passed through a 27-gauge needle 20 to 30 times and centrifuged at 750 × g for 10 min at 4°C. The supernatants were recentrifuged at 10,000 × g for 15 min at 4°C. The supernatants were recentrifuged at 100,000 × g for 30 min to collect cytosolic fractions. The mitochondrial pellets were washed once in buffer A, lysed in Laemmli sample buffer, and then saved. To isolate membrane proteins, A549 cells were scraped in buffer A and incubated on ice for 1 h. The cell suspension was homogenized (25 strokes) manually using a Dounce homogenizer on ice and centrifuged at 750 × g for 10 min, and then the supernatant was centrifuged at 100,000 × g for 30 min to collect the cytosolic fraction. The pellet was resuspended in buffer A containing 1% Triton X-100 and incubated on ice for 1 h to solubilize the membrane proteins. After the protein had been centrifuged at 100,000 × g for 30 min, the soluble membrane fraction was collected.
Identifying membrane binding proteins for SPE B.
The coupling efficiency of rSPE B to CNBr-activated Sepharose 4B (Pharmacia Biotech) was influenced by its protease activity. The functional activity of 28-kDa C192S was better than that of 42-kDa C192S; thus, 28-kDa C192S-immobilized Sepharose 4B was used to screen the binding membrane proteins of SPE B. The column was equilibrated with 50 mM of Tris (pH 7.0) containing 1% Triton X-100. A549 cell membrane proteins were loaded on the affinity column, which was sequentially washed with the same buffer to remove unbound proteins. The bound materials were eluted with 0.1 M of glycine (pH 3.0) containing 1% Triton X-100, and differential fractions were collected and analyzed using SDS-PAGE with silver staining and Western blotting.
Statistical analysis.
Two-way analysis of variance and Student's t test were used for statistical analysis. Differences were considered significant at a P value of <0.05.
RESULTS
αvβ3 partially mediates SPE B-induced apoptosis.
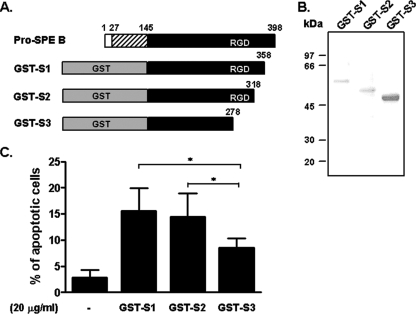
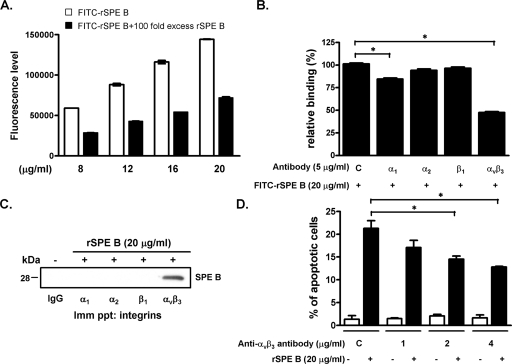
The RGD sequence is critical to many integrins for ligand recognition. SPE B contains an RGD motif and binds to human integrins αvβ3 and αIIbβ3, which suggests that integrins are candidate receptors for SPE B (39). To evaluate the role of the RGD motif in SPE B-induced apoptosis, we generated fragments of GST-conjugated SPE B using series deletions from the carboxyl terminus and tested the role of each fragment in apoptosis. Three fragments of SPE B—S1 (Gln145 to Gly358), S2 (Gln145 to Gln318), and S3 (Gln145 to Pro278)—were fused with the GST tag in their amino termini (Fig. 1A). The size of each fragment displayed on an SDS gel corresponded to the expected molecular masses of 55, 51, and 46 kDa, respectively (Fig. 1B). The SPE B fragment without the RGD motif (GST-S3) induced significantly less apoptosis than did the GST-S1 and GST-S2 SPE B fragments (Fig. 1C), which suggested that the RGD motif is important for SPE B-induced apoptosis. To determine the role of integrins in SPE B-induced apoptosis in A549 cells, we first assessed the interaction of SPE B with A549 cells using a FITC-rSPE B binding assay. FITC-rSPE B dose-dependently bound to the surfaces of A549 cells, and excess non-FITC-conjugated rSPE B effectively reduced the binding, which indicated a specific binding characteristic (Fig. 2A). When monoclonal antibodies against integrin subunits α1, α2, αvβ3, and β1 were added to the binding assay, we found a reduction in FITC-rSPE B binding to A549 cells by anti-α1 (15.5%) and anti-αvβ3 (52.4%) antibodies (Fig. 2B). In addition, the immunoprecipitation experiment showed that rSPE B was coimmunoprecipitated with αvβ3, but not with other integrins (Fig. 2C). These results indicated that SPE B preferentially bound to integrin αvβ3 on A549 cells. We also found that pretreating cells with anti-αvβ3 antibody dose-dependently inhibited SPE B-induced apoptosis compared to the control group pretreated with rSPE B alone (Fig. 2D).
FIG. 1.
The RGD motif is involved in SPE B-induced A549 cell apoptosis. (A) Three truncated SPE B fragments (S1, S2, and S3) conjugated with a GST tag on the N terminus were constructed as described in Materials and Methods. (B) Purified GST-S1, GST-S2, and GST-S3 were resolved using 12% SDS-PAGE. (C) A549 cells were treated with 20 μg/ml of GST-SPE B fragments for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis and are presented as percentages of hypodiploid cells. The data are the means plus standard deviations of triplicate cultures. *, P < 0.05.
FIG. 2.
Role of integrin αvβ3 in SPE B-induced apoptosis. (A) A549 cells were incubated with various concentrations of FITC-rSPE B with or without a 100-fold excess of rSPE B at 4°C for 2 h. FITC-rSPE B bound to A549 cells was determined using a fluorescence enzyme-linked immunosorbent assay reader. (B) A549 cells were pretreated with the indicated anti-integrin antibodies (C, control IgG; α1, FB12; α2, P1E6; β1, P4G1; and αvβ3, LM609) at 4°C for 30 min and then incubated with FITC-rSPE B for another 2 h. Relative binding was defined as the percentage of the fluorescence level in the presence of FITC-rSPE B and anti-integrin antibody compared to the average fluorescence level in the presence of FITC-rSPE B alone. The data are the means ± standard deviations (SD) of triplicate cultures. *, P < 0.05. (C) Immunoprecipitation was used to confirm the interaction of SPE B with integrin αvβ3. A549 cells were treated with rSPE B for 20 min, and the cell lysates were immunoprecipitated (Imm ppt) with the indicated anti-integrin antibodies and hybridized with anti-SPE B antibody. The immunoreactive proteins were visualized using ECL. (D) A549 cells were pretreated with the indicated concentrations of anti-αvβ3 antibody and then incubated with rSPE B for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis. The data are expressed as the means ± SD of triplicate cultures. *, P < 0.05.
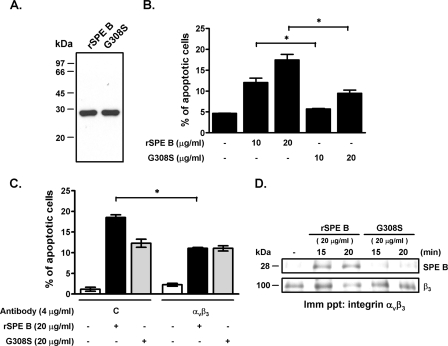
To clarify the role of the RGD motif in SPE B-induced A549 cell apoptosis, we used site mutagenesis to generate an SPE B mutant, G308S (glycine at residue 308 was changed to serine), and then studied its effect on apoptosis. The molecule size of G308S was the same as that of rSPE B (Fig. 3A). G308S induced approximately 50% less apoptosis than did SPE B at each equivalent concentration (Fig. 3B). Pretreatment of A549 cells with anti-αvβ3 antibody partially inhibited SPE B-induced apoptosis, but it had no effect on G308S-induced apoptosis (Fig. 3C), which suggested that G308S-induced apoptosis is integrin αvβ3 independent. Immunoprecipitation was used to distinguish the binding of rSPE B and G308S to integrin αvβ3. We found that integrin αvβ3 interacted with rSPE B, but not with G308S (Fig. 3D), which suggested that SPE B can interact with integrin αvβ3 through its RGD motif and induce cell apoptosis.
FIG. 3.
The RGD motif mutant G308S induces apoptosis. (A) The RGD motif mutant of SPE B, G308S, was purified as described in Materials and Methods. Purified rSPE B and G308S were separated using 12% SDS-PAGE, which resulted in single 28-kDa bands for both rSPE B and G308S. (B) A549 cells were treated with the indicated concentrations of rSPE B or G308S at 37°C for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis. The data are the means ± standard deviations of triplicate cultures. *, P < 0.05. (C) A549 cells were treated with rSPE B or G308S with or without anti-αvβ3 antibody for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis. *, P < 0.05. (D) The interactions of SPE B and G308S with integrin αvβ3 were evaluated using immunoprecipitation. A549 cells were treated with rSPE B or G308S for the times indicated. Cell lysates were immunoprecipitated (Imm ppt) with anti-αvβ3 antibody and hybridized with anti-SPE B antibody. The immunoreactive proteins were visualized using ECL, and the membrane was reprobed with anti-β3 antibody.
Fas mediates SPE B-induced apoptosis.
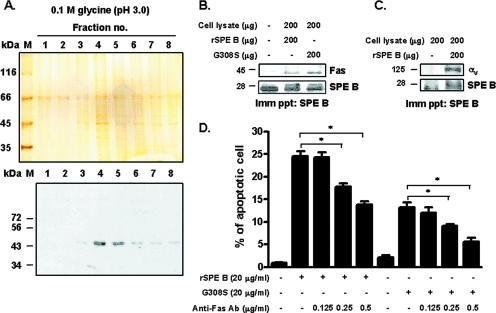
To look for another possible receptor for SPE B-induced apoptosis, we used an affinity column of 28-kDa C192S-linked Sepharose. The bound membrane proteins were eluted by altering the pH. Several bands were detected by silver staining. The 45-kDa molecule in the eluted fractions of no. 4 and 5 (Fig. 4A, top) was of special interest. It is equivalent to the size of Fas that is a well known receptor for mediating apoptosis. This band was then verified as Fas by Western blotting (Fig. 4A, bottom). The interaction of Fas with SPE B was further evaluated by using immunoprecipitation. Cell lysates were incubated with either rSPE B or G308S for 2 h and then precipitated with anti-SPE B antibody. Next, the precipitates were probed with anti-Fas antibody and reprobed with anti-SPE B antibody. Fas was clearly detected in both samples (Fig. 4B), which indicated that Fas interacted with both ligands. When cell lysates were precipitated with anti-SPE B antibody and then probed using anti-αv antibody, integrin αv was identified in the precipitate (Fig. 4C). These results suggested that SPE B reacted with Fas and αvβ3, whereas G308S reacted with Fas. The role of Fas in SPE B-induced apoptosis was further clarified. As shown in Fig. 4D, SPE B-induced apoptosis was inhibited by anti-Fas antibody in a dose-dependent manner. Furthermore, anti-Fas antibody also inhibited G308S-induced apoptosis, which suggested that Fas-mediated SPE B-induced apoptosis occurs in an RGD-independent manner.
FIG. 4.
Interaction of SPE B with Fas and integrin αvβ3. (A) A 28-kDa C192S affinity column was used to identify membrane proteins that bound to SPE B as described in Materials and Methods. The eluted fractions were collected and separated using 12% SDS-PAGE, and the resultant gel was analyzed using silver staining (top). The same fractions were immunoblotted using anti-Fas antibody (bottom). (B) A549 cell lysates were incubated with rSPE B or G308S at 4°C for 2 h. The mixtures were precipitated (Imm ppt) using anti-SPE B antibody and blotted with anti-Fas antibody, and then the membrane was reprobed with anti-SPE B antibody. (C) A549 cell lysates were incubated with rSPE B at 4°C for 2 h. The mixtures were precipitated using anti-SPE B antibody and blotted with anti-αv antibody, and then the membrane was reprobed using anti-SPE B antibody. (D) A549 cells were pretreated with the indicated concentrations of anti-Fas antibody and then incubated with rSPE B or G308S at 37°C for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis. The data are shown as the means plus standard deviations of triplicate cultures. *, P < 0.05.
The synergistic effects of αvβ3 and Fas on SPE B-induced apoptosis.
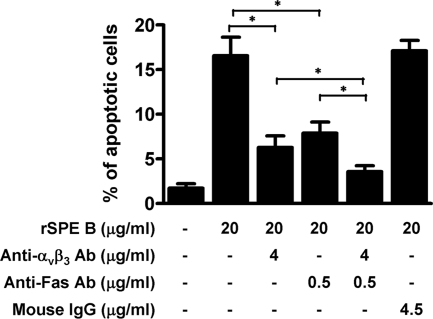
Since SPE B can bind to both αvβ3 and Fas and initiate apoptotic pathways, we next investigated whether there is a synergistic effect of αvβ3- and Fas-mediated pathways in SPE B-induced apoptosis. We found that anti-αvβ3 and anti-Fas antibodies partially inhibited SPE B-induced apoptosis when tested separately. Incubating anti-αvβ3, anti-Fas, and rSPE B with A549 cells simultaneously further reduced apoptosis (Fig. 5). Two-way analysis of variance showed that interaction between αvβ3 and Fas was significant (F = 13.02; P = 0.0069), indicating a synergistic effect between these two pathways.
FIG. 5.
Synergistic effects of anti-αvβ3 and anti-Fas antibodies in SPE B-induced apoptosis. A549 cells were pretreated with anti-αvβ3 or anti-Fas antibodies, or both, or with IgG as a control for 30 min and then incubated with rSPE B for 24 h. After propidium iodide staining, the apoptotic cells were quantified using flow cytometric analysis. The data are shown as the means plus standard deviations of triplicate cultures. *, P < 0.05.
Both αvβ3 and Fas mediated the activation of apoptotic cascades.
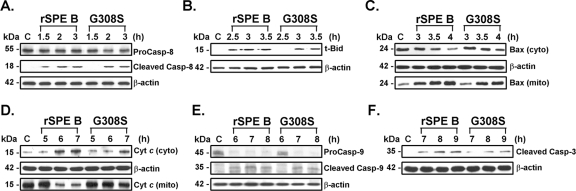
We previously showed sequential events in SPE B-induced apoptosis involving caspase 8 activation, t-Bid activation, Bax translocation, cytochrome c release, caspase 9 activation, and caspase 3 activation (44). We then sought to determine which molecule(s) in the apoptotic cascade is activated by αvβ3 or Fas. Both SPE B and G308S activated all molecules in the apoptotic cascade. However, G308S-induced activation took 30 to 60 min longer than SPE B-induced activation (Fig. 6). These results suggested that the activation processes mediated by different receptors may be regulated differently.
FIG. 6.
Apoptotic cascades activated by G308S and SPE B during apoptosis. A549 cells were treated with rSPE B or G308S for different periods of time, and the activations of caspase (Casp) 8 (A), t-Bid (B), Bax translocation (C), cytochrome (Cyt) c release (D), caspase 9 (E), and caspase 3 (F) were determined using Western blot analysis. cyto, cytosolic fraction; mito, mitochondrial fraction.
Receptor downregulation and recycling.
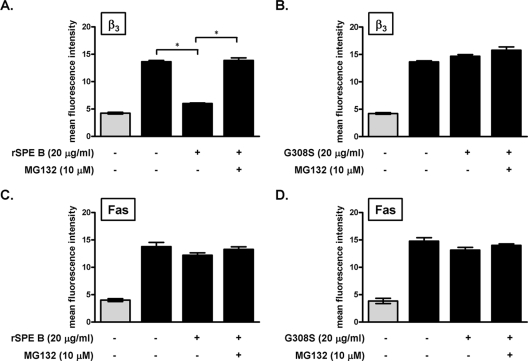
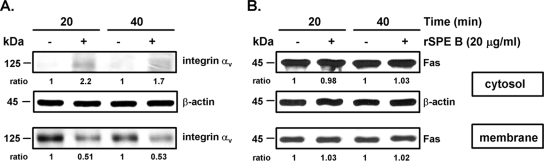
Receptors on the cell surface are usually tightly regulated by ligands or other cellular regulators. To study how the levels of αvβ3 and Fas on the cell surface are regulated by SPE B, we incubated A549 cells with SPE B for 40 min and then added antibody against αvβ3 or Fas to detect the surface expression of αvβ3 or Fas. Treatment with SPE B for 40 min decreased the level of αvβ3 on the cell surface, while treatment with G308S did not change the level of αvβ3 on the cell surface (Fig. 7A and B). However, pretreatment with the proteasome inhibitor MG132 restored cell surface expression of αvβ3 to the original level (Fig. 7A). Since the ubiqutin-proteasome pathway is involved in an endosomal sorting step of membrane proteins to lysosome (45), these results indicated that when the endosomal sorting step is inhibited, αvβ3 is retranslocated to the cell surface. When G308S was used as a ligand, the level of αvβ3 on the cell surface was not changed (Fig. 7B). Further experiments showed that neither SPE B nor G308S caused any effect on cell surface Fas expression (Fig. 7C and D). These results suggest that after SPE B binding, αvβ3 is regulated by the ubiquitin-proteasome system and is recycled, whereas Fas expression is not regulated in a similar way. To further detect receptor translocation from the membrane to the cytosol, we used centrifugation to separate the membrane and cytosol fractions and then determined the distribution of αvβ3 and Fas in each fraction. In the absence of SPE B, αvβ3 was located exclusively on the membrane. After A549 cells had been incubated with 20 μg/ml of rSPE B for 20 or 40 min, approximately 50% of the αvβ3 translocated from the membrane to the cytosol (Fig. 8A). Fas, however, was located primarily in the cytosol; SPE B treatment did not change its distribution regardless of the incubation time (Fig. 8B).
FIG. 7.
SPE B receptor integrin αvβ3 recycling. A549 cells were pretreated with or without proteasome inhibitor MG132 at 37°C for 30 min and then incubated with rSPE B (A and C) or G308S (B and D) for 40 min. After two washes with PBS, the cells were incubated with anti-β3 antibody (A and B) or anti-Fas antibody (C and D) (black bars), followed by FITC-conjugated anti-mouse antibody at 4°C, and analyzed using flow cytometric analysis. Background fluorescence was measured using FITC-conjugated anti-mouse IgG alone (gray columns). The data are the means ± standard deviations of duplicate cultures. *, P < 0.05.
FIG. 8.
Effect of SPE B on integrin αvβ3 translocation. A549 cells were incubated with or without rSPE B (20 μg/ml) for 20 or 40 min and then fractionated into the cytosol and membrane fractions. The amount of integrin αvβ3 (A) or Fas (B) was determined using Western blot analysis. The density of each band was determined using an imaging densitometer, and the ratio of rSPE B treatment to the untreated group is indicated. β-Actin was an internal control.
DISCUSSION
In the present study, we showed that the interaction of the SPE B RGD motif with integrin αvβ3 is critical for binding of SPE B to cells and mediates SPE B-induced apoptosis. We also showed binding between SPE B and Fas, which is a well-recognized apoptotic receptor, and that Fas-mediated SPE B-induced apoptosis occurs via an RGD-independent pathway. These results suggest that both integrin αvβ3 and Fas are involved in SPE B-induced apoptosis in A549 cells. There may be evolutionary reasons for one ligand with two receptors to ensure that apoptosis occurs. It is not known, however, whether this characteristic is cell specific. In addition, the results from the blocking experiments using anti-αvβ3 and anti-Fas showed that SPE B-induced apoptosis was not completely inhibited, suggesting that other pathways independent of αvβ3 and Fas may also be involved.
The interaction of RGD-containing proteins with integrins has been extensively studied. The preference of any given integrin and its ligands is determined by relative affinity, the specific microenvironment, and the conformational state that controls exposure of the integrin recognition sequence (35). It is generally agreed that integrin recognition specificity can often be reduced to a small peptide sequence. A synthetic heptapeptide corresponding to residues 305 to 311 of SPE B (INRGDFS) inhibited the binding of SPE B to αvβ3-positive 835 cells, while the peptide (INRSDFS) had no effect on binding and INRGDFS caused human umbilical vein endothelial cell detachment whereas INRSDFS did not (39). Two discontinuous regions within β3, Glu65 to Glu220 and Ser211 to Gly222, have been identified for ligand binding in the structure of integrin (9, 38). A point mutation in this region, D119Y, resulted in the loss of ligand binding (26). Interaction between SPE B and αIIbβ3 showed that a mutation at residue 119 or 252 also resulted in the loss of binding (39). Since αvβ3 and αIIbβ3 share the same β3 subunit, the binding of residues 119 and 252 to SPE B can be very important. However, the effect of the α subunit on the binding of αvβ3 to SPE B remains to be clarified.
A variety of external stimuli, such as flavonoid compounds, acacetin, extracts of Chinese herbs, ursolic acid, gossypol, and casuarinin, upregulated Fas and Fas ligand in A549 cells (8, 18, 19, 25). SPE B also increased the expression of Fas, Fas ligand, and p53 in human leukocytes (46) and of soluble Fas ligand in SW480 cells (41). However, in SPE B-treated A549 cells, we detected no soluble Fas ligand in the culture medium after 2 hours of incubation with SPE B, nor did we detect Fas ligand in the immunoprecipitation assay in the SPE B antibody-precipitated whole-cell lysate (data not shown). To identify the binding sites for SPE B and Fas, we aligned SPE B and Fas ligand using BLAST 2 sequences and found no significant similarity (42). However, the study of the interaction between Fas and Fas ligand showed that the tyrosine residue at position 218 of Fas ligand was essential for Fas binding (37). This residue is located in the loop between β-strands D and E of Fas ligand. By comparing the molecular models of SPE B and Fas ligand, we found that the tyrosine residue at position 224 of SPE B is in a similar position between two β-strands. This tyrosine residue may have similar functions in the interaction between SPE B and Fas. Its role in the binding of SPE B and Fas needs to be evaluated using site mutagenesis.
The Fas-mediated death pathway has been well established (17). Binding of Fas with ligand triggers the trimerization of Fas and the formation of death-inducing signaling complex. This complex recruits procaspase 8, which undergoes activation by autocatalytic cleavage. The activated caspase 8 activates caspase 3 directly or indirectly via the mitochondrial pathway, which includes the activation of Bid, cytochrome c release, and activation of caspase 9. However, the integrin-mediated apoptotic pathway is not so well defined. Echistatin, an αvβ3-selective RGD-containing antagonist, induced apoptosis of cultured stellate cells by increasing Bax expression and caspase 3 activation (48). Cansatin, the noncollagenous domain of the collagen type VI α chain, binds to integrins αvβ3 and αvβ5 and activates caspase 9 and caspase 3, whereas angiostatin, the plasminogen fragment, binds to αvβ3 on endothelial cells and triggers caspase 8 activation, which leads to caspase 3 activation (27). In this study, both αvβ3 and Fas mediated caspases 8, 9, and 3 and Bid activation; Bax translocation; and cytochrome c release. However, G308S-induced apoptotic signaling activation was 30 to 60 min later than SPE B-induced activation. The reason for this is not clear. However, we have also observed that Fas and αvβ3 activate p38 and JAK2, respectively, leading activation of STAT1 and expression of Bax and caspase 8 (C. W. Chang, W. H. Tsai, W. J. Chuang, Y. S. Lin, J. J. Wu, C. C. Liu, P. J. Tsai, and M. T. Lin, submitted for publication). The difference in the levels of caspase 8 expressed in two pathways may account for the delay in the G308S-activated cascade.
The distribution of αvβ3 in A549 cells is primarily on the membrane. SPE B treatment, however, translocated αvβ3 from the membrane to the cytosol, which downregulated its level on the surface. In contrast, Fas expression in A549 cells is higher in the cytosol than on the membrane, and treating the cells with SPE B did not change its distribution. Our previous study showed that SPE B was internalized via clathrin-mediated endocytosis and reached the lysosome for degradation (6). In the present study, we showed that SPE B downregulated αvβ3 on the surface, but it had no effect on Fas levels on the surface, which suggested that αvβ3 rather than Fas is responsible for internalizing SPE B. Integrin αvβ3 has been involved in the internalization of various proteins, e.g., it mediates the endocytosis of fibrinogen in A549 cells (34). Foot-and-mouth disease virus entered the cells through the integrin-clathrin-mediated endocytosis pathway via its VP1 protein within the RGD motif, and integrin receptor recycling was regulated by the internalization process (33). The ubiquitin-proteasome system not only removes cellular proteins, but is also important for regulating endocytosis of the ligand-receptor complex (11, 40). The ubiquitin-proteasome pathway is involved in the endosomal sorting of membrane proteins to lysosomes (45). According to this model, MG132 blocks the sorting and degrading process and results in the recycling of the ligand-receptor complex. Our results showing that in the presence of MG132 the αvβ3 recycled back to the cell surface are consistent with this model. However, Fas was not recycled in the presence of MG132. The reason for this difference requires further investigation. The regulation of integrin recycling by the ubiquitin-proteasome system is also not clear. One recent study showed that Cbl-mediated ubiquitination was essential for the α5 integrin degradation induced by FGFR2 activation (22).
Acknowledgments
This work was supported by grant NHRI-EX92-9027SP from the National Health Research Institutes of Taiwan.
We thank Shan-Tair Wang (National Cheng Kung University) for statistical analysis and Bill Franke for editorial assistance.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Aiyar, A., Y. Xiang, and J. Leis. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol 57177-191. [DOI] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108407-419. [DOI] [PubMed] [Google Scholar]
- 3.Brassard, D. L., E. Maxwell, M. Malkowski, T. L. Nagabhushan, C. C. Kumar, and L. Armstrong. 1999. Integrin αvβ3-mediated activation of apoptosis. Exp. Cell Res. 25133-45. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, P. C., R. A. Clark, and D. A. Cheresh. 1994. Requirement of vascular integrin αvβ3 for angiogenesis. Science 264569-571. [DOI] [PubMed] [Google Scholar]
- 5.Brooks, P. C., A. M. Montgomery, M. Rosenfeld, R. A. Reisfeld, T. Hu, G. Klier, and D. A. Cheresh. 1994. Integrin αvβ3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell 791157-1164. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. W., W. H. Tsai, W. J. Chuang, Y. S. Lin, J. J. Wu, C. C. Liu, P. J. Tsai, and M. T. Lin. 2007. The fate of SPE B after internalization and its implication in SPE B-induced apoptosis. J. Biomed. Sci. 14419-427. [DOI] [PubMed] [Google Scholar]
- 7.Chang, G. C., S. L. Hsu, J. R. Tsai, F. P. Liang, S. Y. Lin, G. T. Sheu, and C. Y. Chen. 2004. Molecular mechanisms of ZD1839-induced G1-cell cycle arrest and apoptosis in human lung adenocarcinoma A549 cells. Biochem. Pharmacol. 681453-1464. [DOI] [PubMed] [Google Scholar]
- 8.Chang, J. S., Y. L. Hsu, P. L. Kuo, L. C. Chiang, and C. C. Lin. 2004. Upregulation of Fas/Fas ligand-mediated apoptosis by gossypol in an immortalized human alveolar lung cancer cell line. Clin. Exp. Pharmacol. Physiol. 31716-722. [DOI] [PubMed] [Google Scholar]
- 9.Charo, I. F., L. Nannizzi, D. R. Phillips, M. A. Hsu, and R. M. Scarborough. 1991. Inhibition of fibrinogen binding to GP IIb-IIIa by a GP IIIa peptide. J. Biol. Chem. 2661415-1421. [PubMed] [Google Scholar]
- 10.Chen, C. Y., S. C. Luo, C. F. Kuo, Y. S. Lin, J. J. Wu, M. T. Lin, C. C. Liu, W. Y. Jeng, and W. J. Chuang. 2003. Maturation processing and characterization of streptopain. J. Biol. Chem. 27817336-17343. [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover, A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 679-87. [DOI] [PubMed] [Google Scholar]
- 12.Dockrell, D. H. 2003. The multiple roles of Fas ligand in the pathogenesis of infectious diseases. Clin. Microbiol. Infect. 9766-779. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, T., M. Maruyama, J. Araya, K. Sassa, Y. Kawagishi, R. Hayashi, S. Matsui, T. Kashii, N. Yamashita, E. Sugiyama, and M. Kobayashi. 2002. Hydrogen peroxide induces upregulation of Fas in human airway epithelial cells via the activation of PARP-p53 pathway. Am. J. Respir. Cell Mol. Biol. 27542-552. [DOI] [PubMed] [Google Scholar]
- 14.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 2851028-1032. [DOI] [PubMed] [Google Scholar]
- 15.Gubba, S., D. E. Low, and J. M. Musser. 1998. Expression and characterization of group A Streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect. Immun. 66765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa, S., K. Saeki, M. Sazuka, Y. Suzuki, Y. Shoji, T. Ohta, K. Kaji, A. Yuo, and M. Isemura. 2001. Apoptosis induction by epigallocatechin gallate involves its binding to Fas. Biochem. Biophys. Res. Commun. 2851102-1106. [DOI] [PubMed] [Google Scholar]
- 17.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407770-776. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, Y. L., P. L. Kuo, and C. C. Lin. 2004. Proliferative inhibition, cell-cycle dysregulation, and induction of apoptosis by ursolic acid in human non-small cell lung cancer A549 cells. Life Sci. 752303-2316. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, Y. L., P. L. Kuo, C. F. Liu, and C. C. Lin. 2004. Acacetin-induced cell cycle arrest and apoptosis in human non-small cell lung cancer A549 cells. Cancer Lett. 21253-60. [DOI] [PubMed] [Google Scholar]
- 20.Ishibashi, Y., D. A. Relman, and A. Nishikawa. 2001. Invasion of human respiratory epithelial cells by Bordetella pertussis: possible role for a filamentous hemagglutinin Arg-Gly-Asp sequence and α5β1 integrin. Microb. Pathog. 30279-288. [DOI] [PubMed] [Google Scholar]
- 21.Jiao, Z. X., Q. L. Ao, and M. Xiong. 2006. Cigarette smoke extract inhibits the proliferation of alveolar epithelial cells and induces apoptosis. Sheng Li Xue Bao 58244-254. [PubMed] [Google Scholar]
- 22.Kaabeche, K., H. Guenou, D. Bouvard, N. Didelot, A. Listrat, and P. J. Marie. 2005. Cbl-mediated ubiquitination of α5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 1181223-1232. [DOI] [PubMed] [Google Scholar]
- 23.Kagawa, T. F., J. C. Cooney, H. M. Baker, S. McSweeney, M. Liu, S. Gubba, J. M. Musser, and E. N. Baker. 2000. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc. Natl. Acad. Sci. USA 972235-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, C. F., J. J. Wu, K. Y. Lin, P. J. Tsai, S. C. Lee, Y. T. Jin, H. Y. Lei, and Y. S. Lin. 1998. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect. Immun. 663931-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, P. L., Y. L. Hsu, T. C. Lin, J. K. Chang, and C. C. Lin. 2005. Induction of cell cycle arrest and apoptosis in human non-small cell lung cancer A549 cells by casuarinin from the bark of Terminalia arjuna Linn. Anticancer Drugs 16409-415. [DOI] [PubMed] [Google Scholar]
- 26.Loftus, J. C., T. E. O'Toole, E. F. Plow, A. Glass, A. L. Frelinger III, and M. H. Ginsberg. 1990. A β3 integrin mutation abolishes ligand binding and alters divalent cation-dependent conformation. Science 249915-918. [DOI] [PubMed] [Google Scholar]
- 27.Magnon, C., A. Galaup, B. Mullan, V. Rouffiac, C. Bouquet, J. M. Bidart, F. Griscelli, P. Opolon, and M. Perricaudet. 2005. Canstatin acts on endothelial and tumor cells via mitochondrial damage initiated through interaction with αvβ3 and αvβ5 integrins. Cancer Res. 654353-4361. [DOI] [PubMed] [Google Scholar]
- 28.Martin, T. R., N. Hagimoto, M. Nakamura, and G. Matute-Bello. 2005. Apoptosis and epithelial injury in the lungs. Proc. Am. Thorac. Soc. 2214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menaker, R. J., and N. L. Jones. 2003. Fascination with bacteria-triggered cell death: the significance of Fas-mediated apoptosis during bacterial infection in vivo. Microbes Infect. 51149-1158. [DOI] [PubMed] [Google Scholar]
- 30.Molinari, G., and G. S. Chhatwal. 1999. Streptococcal invasion. Curr. Opin. Microbiol. 256-61. [DOI] [PubMed] [Google Scholar]
- 31.Nambu, Y., S. J. Hughes, A. Rehemtulla, D. Hamstra, M. B. Orringer, and D. G. Beer. 1998. Lack of cell surface Fas/APO-1 expression in pulmonary adenocarcinomas. J. Clin. Investig. 1011102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donnell, D., R. L. Milligan, and J. M. Stark. 1999. Induction of CD95 (Fas) and apoptosis in respiratory epithelial cell cultures following respiratory syncytial virus infection. Virology 257198-207. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell, V., M. LaRocco, H. Duque, and B. Baxt. 2005. Analysis of foot-and-mouth disease virus internalization events in cultured cells. J. Virol. 798506-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odrljin, T. M., C. G. Haidaris, N. B. Lerner, and P. J. Simpson-Haidaris. 2001. Integrin αvβ3-mediated endocytosis of immobilized fibrinogen by A549 lung alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 2412-21. [DOI] [PubMed] [Google Scholar]
- 35.Plow, E. F., T. A. Haas, L. Zhang, J. Loftus, and J. W. Smith. 2000. Ligand binding to integrins. J. Biol. Chem. 27521785-21788. [DOI] [PubMed] [Google Scholar]
- 36.Rho, J. K., Y. J. Choi, B. Y. Ryoo, I. I. Na, S. H. Yang, C. H. Kim, and J. C. Lee. 2007. p53 enhances gefitinib-induced growth inhibition and apoptosis by regulation of Fas in non-small cell lung cancer. Cancer Res. 671163-1169. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, P., J. L. Bodmer, N. Holler, C. Mattmann, P. Scuderi, A. Terskikh, M. C. Peitsch, and J. Tschopp. 1997. Characterization of Fas (Apo-1, CD95)-Fas ligand interaction. J. Biol. Chem. 27218827-18833. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. W., and D. A. Cheresh. 1988. The Arg-Gly-Asp binding domain of the vitronectin receptor. Photoaffinity cross-linking implicates amino acid residues 61-203 of the β subunit. J. Biol. Chem. 26318726-18731. [PubMed] [Google Scholar]
- 39.Stockbauer, K. E., L. Magoun, M. Liu, E. H. Burns, Jr., S. Gubba, S. Renish, X. Pan, S. C. Bodary, E. Baker, J. Coburn, J. M. Leong, and J. M. Musser. 1999. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc. Natl. Acad. Sci. USA 96242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strous, G. J., and J. Gent. 2002. Dimerization, ubiquitylation and endocytosis go together in growth hormone receptor function. FEBS Lett. 529102-109. [DOI] [PubMed] [Google Scholar]
- 41.Tamura, F., R. Nakagawa, T. Akuta, S. Okamoto, S. Hamada, H. Maeda, S. Kawabata, and T. Akaike. 2004. Proapoptotic effect of proteolytic activation of matrix metalloproteinases by Streptococcus pyogenes thiol proteinase (Streptococcus pyrogenic exotoxin B). Infect. Immun. 724836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174247-250. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, P. J., C. F. Kuo, K. Y. Lin, Y. S. Lin, H. Y. Lei, F. F. Chen, J. R. Wang, and J. J. Wu. 1998. Effect of group A streptococcal cysteine protease on invasion of epithelial cells. Infect. Immun. 661460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai, W. H., C. W. Chang, W. J. Chuang, Y. S. Lin, J. J. Wu, C. C. Liu, W. T. Chang, and M. T. Lin. 2004. Streptococcal pyrogenic exotoxin B-induced apoptosis in A549 cells is mediated by a receptor- and mitochondrion-dependent pathway. Infect. Immun. 727055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Kerkhof, P., C. M. Alves dos Santos, M. Sachse, J. Klumperman, G. Bu, and G. J. Strous. 2001. Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell 122556-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viera, N. T., M. J. Romero, M. K. Montero, J. Rincon, and J. A. Mosquera. 2001. Streptococcal erythrogenic toxin B induces apoptosis and proliferation in human leukocytes. Kidney Int. 59950-958. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, T., M. Maruyama, T. Fujita, K. Miyabayashi, C. Shinoda, Y. Kawagishi, T. Fujishita, R. Hayashi, T. Miwa, N. Arai, S. Matsui, E. Sugiyama, and M. Kobayashi. 2006. Ionizing radiation suppresses FAP-1 mRNA level in A549 cells via p53 activation. FEBS Lett. 5804387-4391. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, X., F. R. Murphy, N. Gehdu, J. Zhang, J. P. Iredale, and R. C. Benyon. 2004. Engagement of αvβ3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J. Biol. Chem. 27923996-24006. [DOI] [PubMed] [Google Scholar]