Abstract
The food-borne pathogen Listeria monocytogenes is adapted to a diversity of environments, such as soil, food, body fluids, and the cytosol of eukaryotic cells. The transition between saprophytic and pathogenic life is mediated through complex regulatory pathways that modulate the expression of virulence factors. Here we examined the expression of inlJ, a recently identified gene encoding a protein of the LPXTG-internalin family and involved in pathogenesis. We show that inlJ expression is controlled neither by the major listerial regulator of virulence genes, PrfA, nor by AxyR, a putative AraC regulator encoded by a gene adjacent to inlJ and divergently transcribed. The InlJ protein is not produced by bacteria grown in vitro in brain heart infusion medium or replicating in the cytosol of tissue-cultured cells. In contrast, it is efficiently produced and localized at the surface of bacteria present in the liver and blood of infected animals. Strikingly, the expression of inlJ by a heterologous promoter in L. monocytogenes or L. innocua promotes bacterial adherence to human cells in vitro. Taken together, these results strongly suggest that InlJ acts as a novel L. monocytogenes sortase-anchored adhesin specifically expressed during infection in vivo.
The food-borne pathogen Listeria monocytogenes is associated with severe invasive infections in humans and animals. The clinical features of listeriosis include meningitis, encephalitis, septicemia, abortion, and perinatal infection, with a high fatality rate for immunocompromised individuals, fetuses, and neonates (17). L. monocytogenes’ ability to invade and multiply in a wide range of mammalian cells and thereby to cross the intestinal, blood-brain, and maternofetal barriers is essential for the development of systemic listeriosis (34, 67).
L. monocytogenes can survive and grow in extreme conditions, such as that encountered in soil, the food chain, the stomach, the gut, body fluids, and the cytosol of mammalian cells (21, 34, 67). This ability to colonize a broad range of ecosystems correlates with an abundance of genes encoding different transport and regulatory proteins (11.6% and 7.3%, respectively, of all predicted genes of L. monocytogenes [27]). As is true for many other pathogenic bacteria, L. monocytogenes needs to tightly control the expression of virulence genes in order to ensure the production of these effectors at specific sites during bacterial infection. Several studies have focused on the mechanisms by which the listerial effectors involved in the cellular infection process are expressed. These proteins include two invasion proteins, InlA and InlB; the pore-forming toxin listeriolysin O (LLO); two phospholipases, PlcA and PlcB; and ActA, which triggers bacterial intracellular motility (29, 34, 67). These virulence factors are produced under the control of the transcriptional regulator PrfA, which belongs to the Crp/Fnr family of transcriptional activators. PrfA is itself regulated by environmental signals important for the transition between the saprophytic and host cell environments (39). For instance, readily metabolizable sugars that Listeria may encounter outside host cells strongly inhibit the activity of PrfA and subsequently repress virulence gene expression (25, 50, 56, 59). Conversely, PrfA-dependent genes are induced when L. monocytogenes grows intracellularly, suggesting that the composition of the host cell cytosol provides activating signals for PrfA (10, 19, 32, 36, 54, 59a, 64). Some virulence genes, such as inlA and inlB, are also controlled by the stress-responsive alternative sigma factor σB (33, 46). Regulators of two-component systems, such as AgrA (1), VirR (44), and DegU (68), are also associated with L. monocytogenes pathogenesis. Finally, recent investigations have shown that the synthesis of at least some virulence factors, including PrfA itself, ActA, InlA, and LLO, is posttranscriptionally regulated. This control relies on the upstream 5′ untranslated regions (5′-UTR) of their transcripts, which can form secondary structures affecting the translation efficiency (30, 63, 65, 71).
Besides the well-known virulence factors mentioned above, postgenomics approaches have revealed additional novel proteins that play key roles in Listeria's interaction with its host (3, 29). Among them is InlJ, a protein of the internalin family (5, 61). As is true for InlA, InlJ contains an N-terminal leucine-rich repeat (LRR) domain and a C-terminal LPXTG motif characteristic of surface proteins covalently anchored to the peptidoglycan by a sortase (4, 13, 23, 61). However, InlJ LRRs have a cysteine-containing consensus sequence different from that of the InlA LRR prototype. In addition, InlJ has four MucBP (MUCin-binding protein) repeats that share similarities with repeats present in MUB, an extracellular mucus-binding protein of Lactobacillus reuteri (60). The inlJ gene is conserved in the genome of L. monocytogenes strains and is absent from nonpathogenic Listeria species (5). An inlJ deletion mutant is significantly attenuated in virulence after intravenous infections of BALB/c mice or oral inoculations of hEcad mice. However, the function of InlJ is unknown (61).
In this study, we aimed to get insight into the expression of inlJ and the function of its product. Our results strongly suggest a mechanism of posttranscriptional regulation, which may silence the expression of inlJ gene outside the mammalian host and induce it specifically during infection to facilitate bacterial adherence to host tissues.
MATERIALS AND METHODS
Bacterial strains, mammalian cells, and growth conditions.
Listeria strains used in this study are listed in Table 1 and were routinely grown in brain heart infusion (BHI) medium (Difco) at 37°C. Erythromycin at 5 μg/ml was added for the growth of strains carrying plasmids. Experiments involving growth under stress conditions were performed as follows. For iron depletion experiments, bacteria were first cultured at 37°C overnight in BHI, diluted to an optical density at 600 nm (OD600) of 0.1, and then grown for 6 h at 37°C in BHI with 100 or 250 μM of dipyridyl (Sigma). For acid stress, hypoxia, and temperature shift experiments, the same parameters were used, but growth was at pH 4.5 or pH 5.5 upon the addition of HCl (acid stress), in 6% O2 atmosphere for 12 h (hypoxia), or in BHI at 16°C, 30°C, 37°C, 39°C, or 42°C (for temperature shift). For bacterial growth in murine blood, bacteria were grown at 37°C in BHI until the OD600 was 0.8 and rinsed twice in phosphate-buffered saline (PBS) and once in RPMI (Gibco). Bacteria (106) were then added in 500 μl of pure or diluted (1:5 in RPMI) murine blood containing 250 units/ml of heparin and incubated for 6 or 24 h at 37°C in 10% CO2. Eukaryotic cells were JEG-3 placental (ATCC HTB-36) or Lovo and HT29 intestinal (ATCC CCL229 and ATCCHTB38) human cells or J774 murine macrophages (ATCC TIB-67) and were grown according to ATCC instructions.
TABLE 1.
Bacterial strains
| Strain | Property | Reference or source |
|---|---|---|
| BUG 1600 | L. monocytogenes EGDe wild-type strain | 27 |
| BUG 1777 | EGDe ΔsrtA strain | 4 |
| BUG 2159 | EGDe ΔinlJ strain | 61 |
| BUG 2214 | EGDe ΔprfA strain | 45 |
| BUG 2354 | EGDe/ΔHTH-axyR | This study |
| BUG 2350 | EGDe/pTprot-inlJ | This study |
| BUG 2351 | EGDe/ΔinlJ-pPL2::prot-inlJ | This study |
| BUG 2353 | EGDe/pPinlJ-gfp | This study |
| BUG 2364 | EGDe/ΔsrtA/pTprot-inlJ | This study |
| BUG 2366 | E. coli BL21Star/pET101-inlJ | This study |
| BUG 2293 | EGDe/pTprot | This study |
| BUG 499 | L. innocua | 27 |
| BUG 2356 | L. innocua expressing pTprot-inlJ | This study |
| BUG 1496 | L. innocua expressing inlA | 41 |
Generation of the EGDe ΔHTH-axyR in-frame deletion mutant.
Two ∼650-bp DNA fragments flanking the sequence encoding the helix-turn-helix (HTH) domain were amplified by PCR from strain EGDe chromosomal DNA. Primers for the HTH 5′ flanking fragment were axy-1 (5′-CGCGGATCCGGCGCACTCCTTATTCGTTCTCTATAATCA-3′) and axy-2 (5′-ATCATTGACTTCTTTCCTAGCAGAAGATGGAACCCC-3′), and those for the 3′ fragment were axy-3 (5′-GGGGTTCCATCTTCTGCTAGGAAAGAAGTCAATGATCATCATGCGGAAACGAATTTGTTAATAGAC-3′), which is complementary to axy-2 in 5′, and axy-4 (5′-TCCCCCGGGATCAGCATATGTATTTTTCGGCTCAAAAAACAT-3′). The amplified 5′ and 3′ fragments were coligated by PCR using axy-1 and axy-4 and then cloned in BamHI and SmaI sites of the thermosensitive plasmid pMAD, yielding pMAD-ΔHTH-axyR. This plasmid was electroporated into L. monocytogenes strain EGDe and gene replacement was performed as described previously (61).
Construction of pTprot-inlJ, pPL2::prot-inlJ, pPinlJ-gfp, pET101-inlJ, and pB29-inlJ. (i) pTprot-inlJ.
inlJ (from ribosome binding site [RBS] to stop codon) was PCR amplified from EGDe chromosomal DNA by using primers inlJcplm-1 (5′-ACGAGCTCAATAAGGAGTGCGCCAAATTG-3′) and inlJcplm-2 (5′-TCCCCCGGGTCCTGTCCGGCTTTTTTACTATTTTTTC-3′). The DNA fragment was digested with SacI and SmaI and inserted into the SacI and SphI sites in the shuttle plasmid pP1 (14) downstream from the promoter prot, yielding pTprot-inlJ. This plasmid was electroporated into L. monocytogenes strain EGDe.
(ii) pPL2::prot-inlJ.
The prot-inlJ sequence was amplified by PCR by using primers inlJcplm1ppl2 (5′-GGACTAGTGAATTCGATTCGGTCTCCTCT-3′) and inlJcplm-2 (5′-TCCCCCGGGTCCTGTCCGGCTTTTTTACTATTTTTTC-3′), digested with SpeI and SmaI, and inserted in the integrative vector pPL2, yielding pPL2::prot-inlJ, which was integrated in the chromosome as described previously (40).
(iii) pPinlJ-gfp.
A 297-bp DNA fragment located immediately upstream of the RBS of inlJ was amplified by PCR using primers inlJprom-1 (5′-CCGGAATTCGCCCAGAAATAGCAAATACCA-3′) and inlJprom-2 (5′-CGCGGATCCATTCGTTCTCTATAATCATTTATT-3). The gfp gene with an RBS optimized for gram-positive bacteria was PCR amplified from plasmid pNF8 (18) using primers gfp1 (5′-CGGAAAGGAGGTTTATTAAAATGAGTAAAGGAGAAGAACTT) and gfp2 (5′-AAACTGCAGTTATTTGTATAGTTCATCCATGCCA-3′). Amplified fragments were digested with BamH1, coligated, and amplified by PCR with inlJprom-1 and gfp2 primers. The resulting PinlJ-gfp fragment was inserted at EcoRI and PstI sites of the pAT18 vector (66), yielding pPinlJ-gfp.
(iv) pET101-inlJ.
The sequence encoding InlJ (residues 26 to 792) was PCR amplified from EGDe chromosomal DNA by use of primers inlJpet-1 (5′-CACCATGGCAACGAATGATGTTATTGATAATACG-3′) and inlJpet-2 (5′-AGGTTGATCTGTGTTTGTGTTTTTC-3′) and inserted into the pET101 vector (Invitrogen) according to the manufacturer's instructions.
(v) pB29-inlJ.
The sequence encoding InlJ (residues 26 to 769) was PCR amplified and cloned in pB29, a LexA N-terminal fusion vector optimized by Hybrigenics. All constructions were verified by sequencing.
InlJ purification.
pET101-inlJ was used to transform Escherichia coli BL21Star (Invitrogen), and recombinant His-tagged InlJ was produced according to the manufacturer's instructions. Briefly, expression of the recombinant protein was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) during a 2-h period at 37°C. Bacterial pellets were resuspended in buffer A (50 mM Tris-HCl, 150 mM NaCl, pH 7.8) and disrupted with a French press. InlJ was purified from the supernatants by metal affinity chromatography using Ni-nitrilotriacetic acid columns (Novagen). After elution with imidazole, InlJ protein was desalted and concentrated to the desired volume using Centriprep devices (Amicon).
Antibodies, bacterial extracts, and Western blotting techniques.
Polyclonal rabbit anti-InlJ (R133) was generated against recombinant His-tagged InlJ protein and affinity purification of the antibodies was done as described previously (20). Polyclonal rabbit anti-L. monocytogenes EGDe (R97) was generated against 109 heat-killed bacteria as described previously (28). Anti-L. monocytogenes EGDe mouse polyclonal antibodies (MS2) were obtained by immunizing BALB/c mice with 108 heat-killed bacteria in Freund's incomplete adjuvant (Difco). Animals were boosted 3 and 6 weeks after inoculation and bled 10 days after each challenge. We also used rabbit polyclonal antibodies against L. innocua (R6 [16]) and mouse monoclonal antibodies against InlA (L7.7 [48]). InlJ and InlA detection at the surface of bacteria was performed as described previously (4) using primary antibodies and goat anti-rabbit Alexa Fluor 488 or goat anti-mouse Cy3-conjugated (Molecular Probes, Jackson) secondary antibodies and 4′,6′-diamidino-2-phenylindole (DAPI). Bacterial cell wall extracts were obtained as described previously (31). After separation on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, proteins were detected by Western blotting.
RNA purification, primer extension, and rapid amplification of cDNA ends (RACE). (i) RNA purification.
RNA from L. monocytogenes bacteria grown to the mid-log phase was extracted with an RNeasy mini kit (Qiagen) according to the manufacturer's protocol. Residual DNA on the column was removed with Turbo DNase (Ambion). The absence of DNA from RNA samples was verified by PCR amplification of the genes to be assayed with 5 μg of RNA as the template.
Primer extension.
RNAs were used as templates for primer extension reactions using radiolabeled primer inlJ-ext (5′-TTGGCGCACTCCTTATTCGTTCTCTATAATCATTT-3′) as described previously (9).
RACE.
5′ RACE was carried out as described previously (24). Reverse transcription of the 5′-UTR of inlJ mRNA was performed with the inlJ-ext primer, while the inlJ-ext2 primer (5′-CTCTATAATCATTTATTATACGCCGTTTTATTTAC-3′) was used for PCR amplification. PCR products were run on a 3% agarose gel in TAE buffer. Bands present only in the tobacco acid pyrophosphatase (TAP)-treated samples were excised from the gel and were purified by use of a Qiagen gel extraction kit. The eluted DNA was cloned using a TOPO TA cloning kit. pCR2.1-TOPO plasmids containing appropriate PCR fragments were sent to sequencing.
Real-time quantitative PCR (qRT-PCR). (i) cDNA synthesis.
Reverse transcription reaction mixtures (20 μl) containing 200 ng of random hexamers (Roche, Basel, Switzerland), 0.5 mM deoxynucleoside triphosphate, and 5 μg of total RNA were incubated at 65°C for 5 min to remove secondary structures and placed on ice. AMV reverse transcriptase (25 U) and its buffer (Roche, Basel, Switzerland) were then added to the mix. After 10 min at 25°C, the mix was incubated at 42°C for 50 min. Reverse transcriptase was then inactivated by heating at 70°C for 15 min.
(ii) qRT-PCRs.
Primers were selected with the Beacon Designer 4.02 software (Premier Biosoft International, Palo Alto, CA) for inlJ (5′-AGTTTGTCTTGCGTAAATGC-3′ and 5′-CTGGATTGTCTGTGCTGAG-3′), inlA (5′-ACACGGTCTCACAAACAG-3′ and 5′-TCAAGTATTCCACTCCATCG-3′), gyrA (5′-GCGATGAGTGTAATTGTTG-3′ and 5′-ATCAGAAGTCATACCTAAGTC-3′), and rpoB (5′-CACCTGGAGTAAACCAATTAGTACG-3′ and 5′-TAGTGGGTTAAGCATGATATCAACA-3′). qRT-PCRs were performed in a 25-μl reaction volume containing 5 μl of a 1/100 dilution of cDNA, 1 μl of gene-specific primers (10 μM), and 12.5 μl of iQ SYBR green supermix (Bio-Rad, Hercules, CA) with the MyiQ single-color RT iCycler PCR detection system and the MyiQ optical system software (Bio-Rad, Hercules, CA). PCR conditions included an initial denaturation step at 95°C for 3 min followed by a 40-cycle amplification (95°C for 15 s, 55°C for 15 s, 72°C for 15 s). Specificity of the amplified product and primer dimer formation were checked by generating a melting curve with a final step of 80 cycles consisting of a 0.5°C temperature incrementation each 10 s, beginning at 55°C. The absence of contaminating genomic DNA was verified by testing each sample in control reactions without a prior reverse transcription step. The critical threshold cycle (CT) was defined for each sample. Expression levels of the tested genes were normalized using the rpoB or gyrA genes of L. monocytogenes as an internal standard. Each assay was performed in quadruplicate and repeated with at least two independent RNA samples. The n-fold change of the transcript level was calculated using the following equations: ΔCT = CT(test DNA) − CT(reference cDNA); ΔΔCT = ΔCT(target gene) − ΔCT(16S rRNA); ratio = 2−ΔΔCT (42). Statistical significance between the mean expressions of genes was evaluated by Student's t test. A P value of <0.05 was considered significant.
Yeast two-hybrid screening.
pB29-inlJ was transformed into the L40DGAL4 yeast (Saccharomyces cerevisiae) strain and yeast two-hybrid screening was performed by Hybrigenics, S.A., Paris, France. Briefly, a human placenta randomly primed cDNA library (RP4) transformed into the Y187 yeast strain and containing 10 million independent fragments was used for mating. The screen was first performed on a small scale to adapt the selective pressure to the intrinsic property of the bait. Neither toxicity nor autoactivation of the bait was observed. Then, the full-scale screen, allowing 104 million interactions to be tested, was performed.
Adherence and invasion assays.
L. innocua or L. innocua/pTprot-inlJ grown at an OD600 of 0.6 was diluted in modified Eagle's medium and added at an ≈20:1 ratio to JEG-3 confluent cells or HT29 monolayers of polarized cells that had been differentiated for 3 weeks. After 4 h of incubation, nonadherent bacteria were removed by washing 5 times with modified Eagle's medium. Cells were fixed with 4% paraformaldehyde (wt/vol in PBS) for 20 min and washed three times again with PBS. Bacteria were sequentially labeled with anti-Listeria innocua antibodies diluted 1:1,000 in 5% bovine serum albumin-PBS and a 488-labeled goat anti-rabbit immunoglobulin G (Molecular Probes) diluted 1:1,000. Cell nuclei were stained with DAPI. Each assay was carried out in triplicate and repeated two times. Adherent bacteria were counted by fluorescent microscopy, examining ≈1,000 cells in randomly picked microscopic fields.
Quantification of bacterial entry in JEG-3 cell monolayers was performed with a classical gentamicin assay as described previously (4). To study inlJ expression in intracellular bacteria in tissue-cultured cells, EGDe or EGDe/pPinlJ-gfp was incubated with cells for 1 hour then washed twice with cell growth medium and incubated for 1 h, 7 h, or 23 h with cell culture medium containing gentamicin at 20 μg/ml to kill extracellular bacteria. Cells were then fixed with 4% paraformaldehyde (wt/vol in PBS), permeabilized with 0.4% Triton X-100 in PBS for 5 min at room temperature, and processed for immunofluorescence.
Infections in animals and organ treatments.
All animals were treated in accordance with Institut Pasteur guidelines for laboratory animal husbandry. Bacterial growth in mice was studied by injecting 4- to 6-week-old specific-pathogen-free female BALB/c mice (Charles River) intravenously with a sublethal bacterial inoculum, 104 CFU. Seventy-two hours after infection, livers and spleens were sterilely dissected and the numbers of CFU were determined by plating serial dilutions of organ (liver and spleen) homogenates on BHI agar medium. Blood was resuspended in heparin (250 units/ml) and 100-μl portions were plated on BHI agar medium. Four animals were used for each experiment. Statistical analyses were performed using the Student t test. P values of <0.05 were considered statistically different. To study inlJ expression in bacteria present in blood or liver, the L. monocytogenes wild-type and ΔinlJ strains were inoculated intravenously into BALB/c mice with 104 and 105 CFU, respectively. Following 72 h of infection, blood samples and livers were recovered. Blood streams were fixed on coverslips with 4% paraformaldehyde (wt/vol in PBS) for 20 min, and blood cells were permeabilized with 0.3% Triton X-100 in PBS for 5 min and then processed for immunofluorescence. Optimal cutting temperature TissueTek compound (VWR Scientific)-embedded blocks of liver were cryosectioned (7-μm-thick sections), fixed in ethanol (−20°C for 5 min), and processed for immunofluorescence. Blocking steps, antibody dilutions, and washes were performed in 1% bovine serum albumin in PBS.
Histoimmunochemistry.
Paraffin-embedded blocks of liver and spleen tissues from mice infected intravenously by L. monocytogenes were formalin fixed and sliced. Sections of tissues were stained with anti-EGDe or with anti-InlJ primary antibodies followed by horseradish peroxidase-conjugated anti-rabbit immunoglobulin. Antigen-antibody complexes were visualized by using reagents supplied in the Envision kit (Dakocytomation, Carpinteria, CA).
RESULTS
inlJ transcription occurs independent of AxyR- or PrfA-mediated regulation.
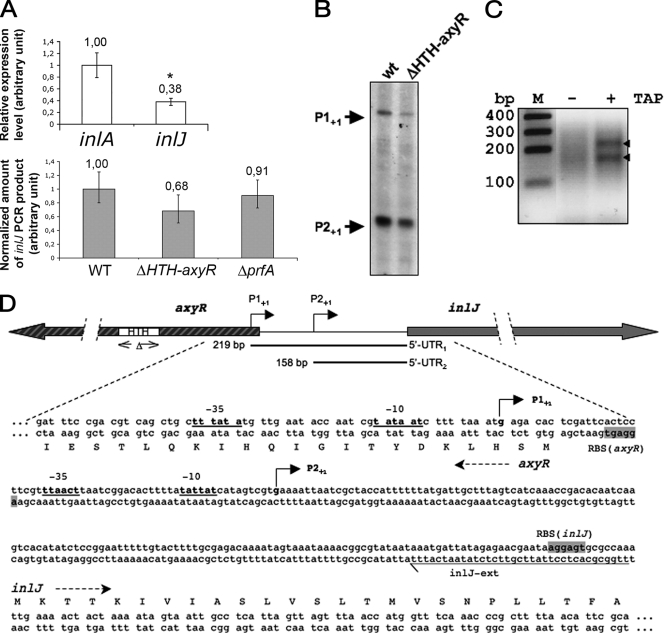
We previously showed by semiquantitative RT-PCR that inlJ is transcribed in bacteria grown in BHI rich medium (61). This result was further supported by qRT-PCR experiments indicating that the amount of inlJ transcript is approximately one-half the amount of the inlA transcript, which allows production of the surface invasion protein InlA (Fig. 1A). Primer extension reactions revealed two inlJ transcription start sites upstream of the start codon of inlJ (Fig. 1B). However, since the smaller transcript may represent a processed RNA species derived from cleavage of the longer transcript, the transcriptional start sites of inlJ were subsequently mapped by 5′ RACE (see Materials and Methods). The nucleotide sequences of the 5′ RACE PCR products also revealed two transcriptional start sites, P1+1 and P2+1, located 219 and 158 bp upstream of the inlJ translational start codon, respectively. Both are preceded by −35 and −10 regions characteristic of listerial RpoD-dependent promoters (Fig. 1C and D). The inlJ P1 promoter is located at the beginning of the coding sequence of the divergently transcribed lmo2820 gene.
FIG. 1.
Studies of inJ promoter region. (A) Studies of inlJ transcription by qRT-PCR. (Top) qRT-PCR with inlA or inlJ internal primers on total RNA extracts from cultures of wild-type L. monocytogenes grown in BHI. inlA and inlJ transcription levels are significantly different (*, P < 0.05 by Student's t test). (Bottom) qRT-PCR with inlJ internal primers on total RNA extracts from cultures of L. monocytogenes wild-type, ΔHTH-axyR, and ΔprfA strains grown in BHI. The amounts of inlJ transcripts in ΔHTH-axyR and ΔprfA mutants are not significantly different from that in the wild-type strain (P = 0.07 and P = 0.3, respectively, by Student's t test). (B) Primer extension analysis of the inlJ transcript. Primer extensions were performed on total RNAs extracted from cultures of wild-type and ΔHTH-axyR strains grown in BHI to an OD600 of 0.8 by use of the inlJ-ext primer. The arrows indicate putative transcription starts of the inlJ transcript. (C) RACE mapping of inlJ transcript 5′ end. Total RNA was linked to a 5′ adaptor RNA without or after treatment with TAP (TAP− and TAP+ lanes, respectively). TAP converts the 5′-triphosphate of an RNA to a monophosphate. Thus, a comparison can distinguish between transcription start site (carrying a 5′-triphosphate) and processing site (carrying a 5′-monophosphate). InlJ-specific cDNA was made and the 5′-UTR of InlJ mRNA was amplified using gene- and adaptor-specific primers. PCR products were separated on a 3% agarose gel. Bands indicated by arrows were excised, cloned, and sequenced. Two transcription start site were mapped and the corresponding position are shown in panel A. (D) Physical map of the inlJ and axyR gene regions. The locations of the two mapped transcription start sites (P1+1 and P2+1) of the inlJ promoters are shown by arrows, as are the two 5′-UTRs. Deduced −10 and −35 hexamers of putative RpoD-dependent promoters are underlined. The inlJ and axyR RBS are framed in gray. The HTH domain deleted from the ΔHTH-axyR mutant is underlined by double arrows. Primer inlJ-ext is underlined.
lmo2820 encodes a putative bimodular protein with an N-terminal region sharing similarities with transcriptional AraC/XylS regulators (22) and a C-terminal region sharing similarities with β-d-xylosidases (12, 72) that we renamed AxyR (AraC-xylosidase putative regulator). Like other AraC/XylS family members, AxyR contains an ∼100-amino-acid conserved domain (HTH domain) that comprises two potential HTH DNA-binding motifs. HTH domains, as for other regulatory proteins, are required for transcription regulation by AraC regulators at targeted promoters (22). Some members of the AraC/XylS family regulate virulence genes. It was therefore tempting to examine a role of AxyR in inlJ regulation. To avoid alteration of the inlJ promoter region that would occur in an axyR full deletion mutant, we constructed an axyR mutant in which only the sequence encoding the 100-amino-acid HTH domain is deleted (Fig. 1). inlJ transcription was first examined in primer extension assays on total RNA extracts from the resulting ΔHTH-axyR strain grown in BHI at 37°C. The two potential transcription start sites of inlJ were still detected in the ΔHTH-axyR strain, as in the wild-type strain (Fig. 1B). Moreover, inlJ mRNA levels in the ΔHTH-axyR strain were not significantly different from those in the wild-type strain, as measured by qRT-PCR (P > 0.05) (Fig. 1A).
We then examined whether PrfA, the major regulator of virulence genes in L. monocytogenes, regulated inlJ. Similar amounts of inlJ transcript were detected in RNA extracts from L. monocytogenes wild-type and ΔprfA strains by qRT-PCR (Fig. 1A). This observation is consistent with the absence of a PrfA putative binding site in the inlJ promoter region and with data from transcriptome studies that did not reveal inlJ as a gene regulated by PrfA (51). Of note was the fact that none of the other regulatory sequences that are known to play a role in L. monocytogenes regulation of virulence genes, such as a Fur box (2, 58), a VirR DNA-binding site (44), or a sigma B-specific promoter signature (33), were detected in the inlJ promoter region. Taken together, these results indicate that the inlJ virulence gene is not part of the PrfA regulon and is apparently not regulated by AxyR, the product of its adjacent gene.
InlJ is not produced by bacteria grown in rich medium unless inlJ is expressed from a heterologous promoter.
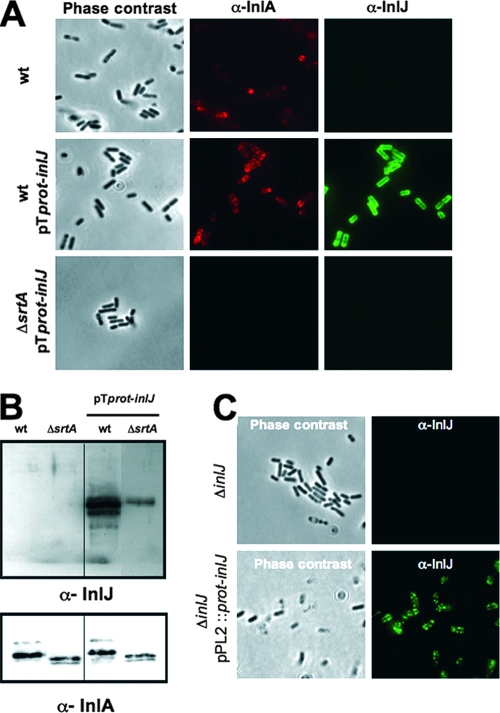
InlJ is an LPXTG surface protein predicted to be anchored to the peptidoglycan by sortase A. To address whether L. monocytogenes bacteria produce InlJ, we generated an antibody against a purified recombinant InlJ protein and used it to detect the protein at the bacterial surface by immunofluorescence. Strikingly, bacteria that had been grown in BHI medium were not stained by the InlJ antibody, while they were stained by an InlA antibody, suggesting that InlJ protein was poorly or not produced, in contrast to InlA (Fig. 2A). Furthermore, InlJ was not detected in bacterial total extracts (data not shown) or cell wall extracts (Fig. 2B) of bacteria grown in BHI. These results are in agreement with data indicating that InlJ is not present in the cell wall proteome of L. monocytogenes EGDe grown in BHI medium (57). The absence of InlJ protein in L. monocytogenes bacteria grown in rich medium, although the inlJ gene is transcribed, suggested a mechanism of posttranscriptional regulation.
FIG. 2.
Bacteria grown in BHI medium do not produce InlJ protein unless inlJ is under the control of the constitutive promoter prot. Immunofluorescence microscopy (A) or immunoblotting of cell wall extracts (B) of L. monocytogenes EGDe wild type (wt), EGDe/pTprot-inlJ (wt pTprot-inlJ), or the ΔsrtA mutant expressing pTprot-inlJ (ΔsrtA pTprot-inlJ) grown in BHI medium with anti-InlA- or anti-InlJ-specific antibodies. InlJ is not detected at the surface of wild-type bacteria, while it is anchored at the surface of bacteria expressing pTprot-inlJ. As for InlA, this anchoring depends on SrtA. (C) InlJ is produced and present at the surface of ΔinlJ/pPL2::prot-inlJ bacteria carrying the fused sequence prot-inlJ in the chromosome. As expected, it is absent at the surface of bacteria of the ΔinlJ strain.
To verify that the inlJ open reading frame encoded a surface protein, inlJ was expressed under the control of the promoter region (prot) of the protease gene from Lactococcus lactis subsp. cremoris (38), which is active in Listeria (4, 6, 14), in a listerial plasmid (pTprot-inlJ). InlJ antibodies efficiently labeled InlJ at the surface of EGDe bacteria expressing inlJ fused to the prot region (wt pTprot-inlJ) and detected the protein in cell wall extracts (Fig. 2B). By contrast, as for InlA, InlJ was not detected by immunofluorescence at the surface of a sortase mutant (ΔsrtA mutant [4]) harboring plasmid pTprot-inlJ (Fig. 2A). Furthermore, the amount of InlJ in cell wall extracts from this strain was significantly smaller than that from the wild-type strain (Fig. 2B). We also examined the expression of inlJ by the heterologous promoter region when the prot-inlJ fusion is present as only one copy in the chromosome. The prot-inlJ sequence was inserted into pPL2, a listerial site-specific integration vector (40), and integrated into the tRNA-Arg site on the chromosome of the ΔinlJ mutant. InlJ was produced and present at the surface of ΔinlJ/pPL2::prot-inlJ bacteria grown in BHI (Fig. 2C). The labeling was specific, as the InlJ antibody did not label inlJ mutant bacteria.
Taken together, these results suggest that the inlJ regulatory region lying upstream of the mRNA coding sequence prevents inlJ transcript translation in bacteria grown in BHI. When inlJ is expressed from the heterologous prot promoter region, the InlJ protein is produced, secreted, and anchored at the bacterial surface by a sortase A-dependent process.
inlJ expression does not increase upon stress or during infection of cultured cells.
To investigate further the mechanism underlying inlJ regulation, we constructed a reporter plasmid, pPinlJ-gfp, in which the 300-bp DNA sequence lying upstream of the inlJ RBS was fused to gfp. The fluorescence of L. monocytogenes live bacteria harboring the pPinlJ-gfp plasmid and grown in BHI was only 2-fold greater than that of control nonfluorescent bacteria carrying the empty vector and was 10-fold lower than that of bacteria harboring pNF8, a plasmid in which gfp is under the control of the strong listerial promoter Pdlt (18), indicating that the green fluorescent protein was not efficiently produced (results not shown). This result was consistent with the absence of InlJ production in this growth condition. We then tested the effect of different stresses or growth conditions on the activation of inlJ expression. Bacteria were grown in BHI at different temperature (16°C, 30°C, 37°C, 39°C, and 42°C) or with iron depletion or under hypoxia or acidic conditions. None of these stresses stimulated the fluorescence of L. monocytogenes/pPinlJ-gfp bacteria or allowed the detection of InlJ at the surface of wild-type EGDe bacteria (data not shown). Variation of glucose concentration had also no effect on inlJ expression.
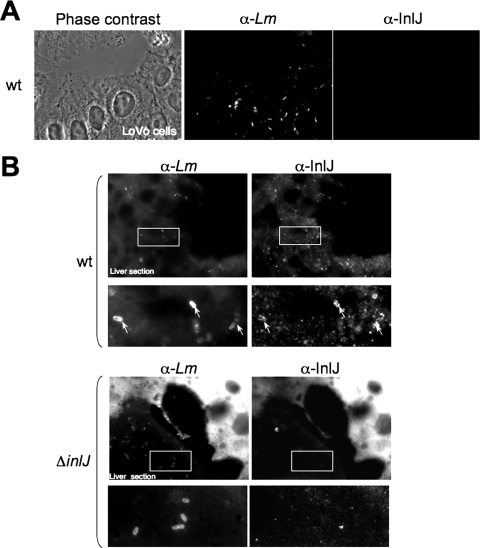
We then studied the ability of the InlJ protein to be specifically produced by bacteria during cell infection of tissue-cultured cells, such as human JEG-3 placental cells, Lovo enterocytes, or J774 murine macrophages, or during growth in blood. Cells were infected with L. monocytogenes EGDe or EGDe/pPinlJ-gfp strains for 2, 8, or 24 h. InlJ protein was still not detected at the surface of intracellular wild-type bacteria (Fig. 3A) and none of the intracellular EGDe/pPinlJ-gfp bacteria appeared fluorescent. Likewise, inlJ was not expressed in bacteria grown in murine blood in vitro (data not shown). These results indicated that the expression of inlJ, in contrast to what is seen for many listerial virulence genes, is not dependent on temperature, on entry into host cell cytosol, or on the presence of blood components (7, 30, 36, 64).
FIG. 3.
inlJ is not expressed by bacteria in tissue-cultured cells but is efficiently expressed by bacteria present in the livers of infected mice. (A) Lovo cells were infected with L. monocytogenes EGDe wild-type bacteria (wt) for 24 h and analyzed by immunofluorescence. InlJ is not detected at the surface of intracellular bacteria. (B) L. monocytogenes EGDe wild-type and ΔinlJ strains were inoculated intravenously into BALB/c mice with an inlJ mutant inoculum 10 times larger than that of the wild-type strain to compensate for the attenuation in the virulence of the ΔinlJ strain. Following 72 h of infection, thin liver frozen sections were analyzed by immunofluorescence microscopy. InlJ is present at the surface of wild-type bacteria (indicated by arrows) but not for of the inlJ mutant (ΔinlJ) within livers of infected mice. The intensity of the fluorescence background varies relative to the thickness and the angle of the section. Bacteria are labeled with the L. monocytogenes MS2.1 antibody (α-Lm) and with the InlJ antibody (α-InlJ). Framed regions indicate the positions of the fields magnified in the lower images.
InlJ is efficiently produced during L. monocytogenes infection of mice.
Since inlJ plays a role in listerial infections in mice (61), it is likely induced in vivo. To confirm this hypothesis, we searched for the presence of the InlJ protein at the surface of L. monocytogenes present in blood or organs of infected mice. Mice were inoculated intravenously either with the wild type or with the ΔinlJ strain. We used an inlJ mutant inoculum 10 times larger than that for the wild-type strain to compensate for the attenuation in the virulence of the ΔinlJ strain (61). InlJ protein was not produced by wild-type bacteria present in the inoculum (data not shown). By contrast, at 72 h postinfection InlJ was present at the surface of wild-type bacteria in liver (Fig. 3B) or in blood (Fig. 4A and B) from infected animals, while it was not detected at the surface of ΔinlJ bacteria.
FIG. 4.
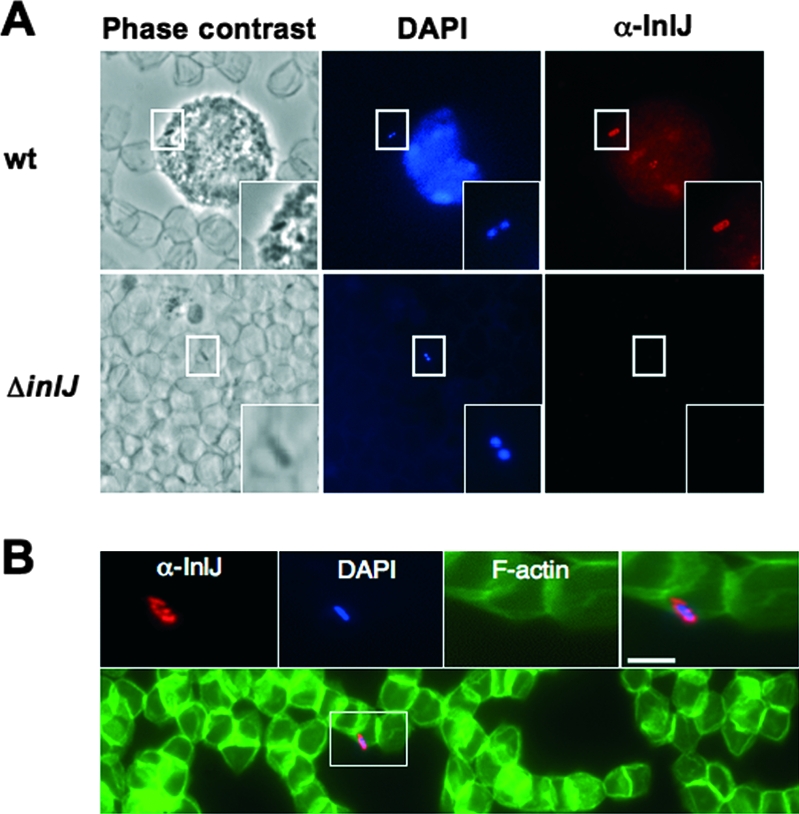
Bacteria present in the blood of infected mice produce InlJ. L. monocytogenes wild-type (wt) or ΔinlJ strain bacteria were inoculated intravenously into BALB/c mice. Following 72 h of infection, blood was recovered and fixed blood streams were labeled with InlJ antibody (in red) and DAPI, which detects white blood cells and bacterial DNA (in blue). (A) InlJ is present at the surface of a wild-type bacterium associated with a polynuclear cell (labeled with DAPI) and is absent at the surface of an inlJ mutant bacterium associated with erythrocytes (visible in phase-contrast image). (B) A wild-type L. monocytogenes bacterium in contact with erythrocytes expresses InlJ at its surface. Fluorescent phalloidin-488 labels actin in erythrocytes (in green). Scale bar, 2 μm. Rectangular regions indicate the positions of the magnified inset fields. α, anti-.
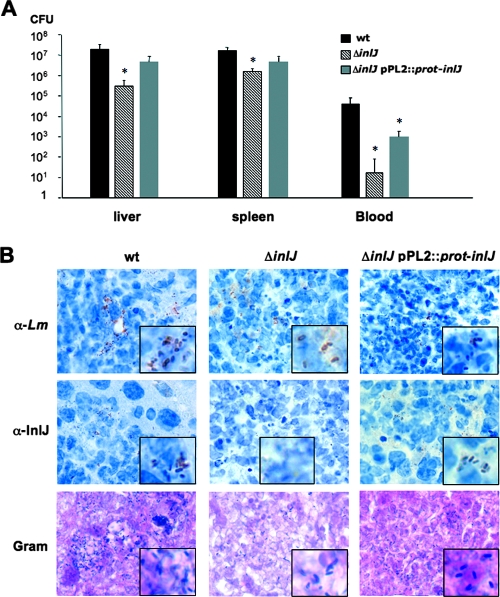
To address the importance of the inlJ regulatory region in InlJ function in vivo, the virulence of the ΔinlJ/pPL2::prot-inlJ strain, in which inlJ is expressed under the control of the heterologous prot regulatory region, was compared to that of the wild-type and ΔinlJ strains. It was assessed by determining bacterial counts in liver and spleen of infected mice as well as by histological studies. As expected from our previous studies (61), the inlJ mutant strain showed 2 log10 and 1 log10 unit decreases in colonizations of the liver and spleen, respectively, and a 3 log10 unit decrease in bacteremia in blood, compared to the wild-type strain (Fig. 5A). Interestingly, the ΔinlJ/pPL2::prot-inlJ strain showed an phenotype intermediate between those of the wild-type and ΔinlJ strains in colonizing organs (Fig. 5A) and in terms of abscess induction in liver when examined by histochemistry. The difference between the wild-type and ΔinlJ/pPL2::prot-inlJ strain phenotypes in vivo was not due to a defect in InlJ production, since InlJ protein was detected at the surface of ΔinlJ/pPL2::prot-inlJ bacteria as for the surface of wild-type bacteria (Fig. 5B). These results suggest that inlJ's own regulatory region is important for a full-virulence phenotype, enabling the production of InlJ virulence protein at the right time and at the right place during the infection process.
FIG. 5.
Importance of inlJ regulatory region for InlJ function in vivo. (A) The L. monocytogenes wild-type (wt), ΔinlJ, and ΔinlJ/pPL2::prot-inlJ strains were intravenously inoculated into BALB/c mice at 104 CFU. Animals were euthanized 72 h after infection and organs and blood were recovered, homogenized, and plated. The number of bacteria able to colonize liver and spleen or survive in blood is expressed as log10 CFU. ΔinlJ/pPL2::prot-inlJ bacteria are not as efficient as the wild-type strain in organ colonization. (B) Analysis of InlJ expression at the surface of bacteria present in livers of infected mice by immunohistochemistry. Bacteria were labeled with the L. monocytogenes R97 antibody (α-Lm) or with the InlJ antibody (α-InlJ) or were Gram stained. The InlJ antibody stained wild-type and ΔinlJ/pPL2::prot-inlJ bacteria equally efficiently, whereas ΔinlJ bacteria were not labeled, as expected. Rectangular inset regions are magnified.
InlJ behaves as an adhesin.
We previously reported that an inlJ mutant and its parental strain behaved similarly in terms of bacterial adherence, entry, replication, and spreading in tissue-cultured cells (61). However, we show here that bacteria do not express inlJ in mammalian cell lines in vitro, thus precluding any function identification. To address InlJ function, we first searched for a mammalian interactor by a two-hybrid screen against a human cDNA library, since InlJ as other LRR-containing proteins is likely to interact with a protein partner (37). However, among 104 million interactions tested, no partner was found.
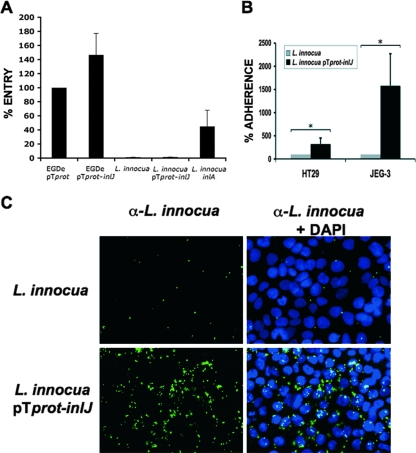
To further investigate a role of InlJ in the cellular infection process, we quantified entry into placental JEG-3 cells of L. monocytogenes expressing inlJ from the prot promoter, in which InlJ is efficiently produced and anchored at the bacterial surface. Interestingly, entry of EGDe/pTprot-inlJ increased by 46% ± 30% from that of control bacteria carrying the empty vector (EGDe/pTprot) (Fig. 6A). To address whether this stimulation of invasion resulted from increased adhesion to host cells or increased internalization per se, we then expressed inlJ in the noninvasive species L. innocua. Like in L. monocytogenes, InlJ was efficiently produced and anchored at the surface of L. innocua carrying plasmid pTprot-inlJ (data not shown). inlJ expression in L. innocua did not promote bacterial entry into JEG-3 cells (Fig. 6A) or HT29 cells (data not shown), in contrast to expression of the invasion gene inlA (Fig. 6A) (40). However, inlJ expression in L. innocua stimulated adherence 16-fold in JEG-3 cells and 3-fold in HT29 cells (Fig. 6B and C). Taken together, these results indicate that InlJ stimulates bacterial adherence to host cells, which then favors bacterial internalization.
FIG. 6.
InlJ stimulates bacterial adherence to eukaryotic cells. (A) Gentamicin invasion assays in JEG-3 cells of L. monocytogenes EGDe/pTprot and EGDe/pTprot-inlJ, L. innocua, L. innocua expressing pTprot-inlJ or inlA. The level of entry of L. monocytogenes EGDe/pTprot was reported as 100. The levels of entry of the other bacteria are given as relative values. For L. innocua and L. innocua expressing pTprot-inlJ, the percentages of entry are 0.29% ± 0.09% and 0.77% ± 0.2%, respectively. Results are means and standard deviations from four different experiments. (B and C) Expression of inlJ in L. innocua stimulates bacterial adherence to JEG-3 cells. Cells were incubated for 4 h at 37°C with L. innocua or L. innocua expressing inlJ at a multiplicity of infection of ≈20 bacteria per cell, extensively washed, and processed for immunofluorescence and quantification of bacterial adherence. (B) Results are expressed as the mean (± standard deviation) number of adherent bacteria per cell. The level of adherence of the L. innocua strain has been artificially reported as 100, and the level of adherence of L. innocua expressing pTprot-inlJ is reported as a relative value. Asterisks indicate that differences are statistically significant (P < 0.05). (C) An immunofluorescence experiment is shown, with bacteria labeled with an anti-L. innocua antibody (in green) and cell nuclei detected with DAPI (in blue). α-, anti-.
DISCUSSION
We report here the characterization of InlJ, an L. monocytogenes surface protein of the internalin family involved in virulence. We show that InlJ is exposed on the listerial surface by a sortase A-mediated process and contributes to bacterial adherence to host cells. Furthermore, we show that InlJ is specifically produced and anchored at the bacterial surface during the infection process in vivo, whereas it is not produced by bacteria grown in BHI medium, within the cytosol of tissue-cultured cells, or in blood in vitro.
This observation contrasts with the situation for other major listerial virulence factors, such as InlA, InlB, LLO, ActA, PlcA, and PlcB. First, all these proteins are detectable in extracts of L. monocytogenes strain EGDe grown in BHI medium. Second, they are all upregulated during bacterial infection of the host cell cytosol in vitro (7, 10, 36, 64), whereas InlJ is not.
inlJ expression may be regulated at the transcriptional level by as-yet-unknown regulators. Signatures for major listerial regulatory proteins such as PrfA, VirR, Fur, and sigma B are not present in the inlJ regulatory region, and the inactivation of PrfA has no effect on inlJ transcription. Furthermore, no role in inlJ regulation could be attributed to AxyR, encoded by a putative regulator gene adjacent to inlJ. The only signal that appears to regulate inlJ transcription is temperature, as revealed by a recent study examining temperature-dependent expression of some listerial internalins which shows that the level of inlJ transcript increases upon a shift from 16°C to 30°C (47). However, as we show here, increasing temperature does not lead to a stimulation of InlJ protein synthesis. The inlJ mRNA is detected in bacteria grown in vitro, while the InlJ protein is not produced. Taken together, all results converge to establish that inlJ is submitted to a posttranscriptional control.
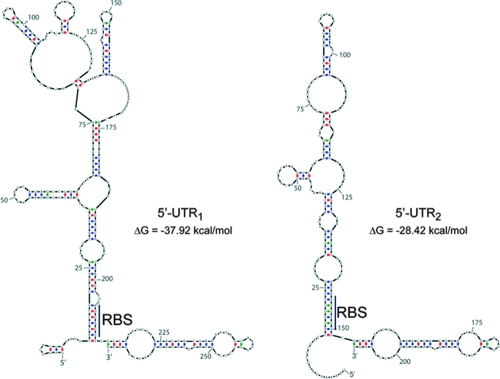
Promoter mapping indicates the existence of two inlJ transcriptional starts, generating 219-bp and 158-bp 5′-UTRs that each could fold into a structure sequestering the inlJ RBS (Fig. 7). Such structures could mediate inlJ posttranscriptional regulation, as described previously in the cases of prfA, actA, inlA, and hly (30, 63, 65, 71). Changes in RNA secondary structures can occur upon the binding of a specific component, as in the case of riboswitches (11, 53, 55, 69, 70). It is possible that during host infection by Listeria a metabolite or another molecule could be specifically released from the inlJ 5′-UTR, providing a quick switch-on of InlJ synthesis. Another possibility is that the regulation of inlJ expression could be coupled to axyR transcription because of the overlap between inlJ and axyR promoter regions. Future work will address this hypothesis.
FIG. 7.
Predictive folding of inlJ 5′-UTR RNAs. inlJ 5′-UTR1 and 5′-UTR2 RNAs predicted from the two transcriptional starts (Fig. 1) may fold into structures in which the RBS could be sequestered by a sequence present at the 5′ extremity of the UTR. The energetically most stable structure is shown. The RNA sequence includes the 5′-UTR and 50 bases inside the mRNA coding sequence. Watson-Crick base pairing is indicated in red for cytosine and guanine and in blue for adenine and uracil. Wobble base pairing (guanine with uracil) is indicated in green. (Pairing was done according to Mfold [http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi].).
We show here that when inlJ is expressed from a heterologous promoter, the InlJ protein is clearly exposed at the surface of bacteria and acts as an adhesin. This result suggests that InlJ interacts with a receptor present at the surface of the host cell. A two-hybrid screening approach did not succeed in finding an InlJ human receptor. This may be explained by several hypothesis: (i) InlJ may not interact with a protein; (ii) the cDNA encoding the InlJ receptor may not be present in the cDNA library; (iii) the interaction may require posttranslational modifications of the receptor, such as glycosylations; (iv) the receptor may be a membrane protein, as the two-hybrid screen is known to fail in detecting membrane-interacting proteins; and (v) the function of InlJ in adherence may be indirect, for example in stabilizing or activating another adhesin on the cell surface. Techniques other than two-hybrid screening will be used to search for an InlJ mammalian or bacterial partner.
Adhesion and invasion are crucial in initiating L. monocytogenes infection in intestinal, uteroplacental, hepatic, and neurological phases of infection. There are only a few adhesion-associated genes known for L. monocytogenes, although genome annotation suggests the existence of several other candidates (3). These genes encode the autolysin Ami (52), the fibronectin binding protein (FbpA) (15, 26), and the Listeria adhesion protein LAP (35). Furthermore, by interacting with the adhesion molecule E-cadherin, the well-characterized internalin InlA acts as an adhesin before playing a role in invasion (49). An LRR protein from group B streptococci, LrrG, was also shown to adhere to epithelial cells, suggesting that it may function as an adhesion factor (62). InlJ is thus a novel example of an LRR bacterial protein with adhesive properties produced by a pathogen. L. innocua cell lines expressing inlJ were found to bind eukaryotic cells with efficiencies that differed between cell lines and were high in placental cells, suggesting that InlJ binds to receptor molecules that are variably expressed by eukaryotic cells. This issue warrants further studies with a larger panel of cell lines, as well as investigation of the role of InlJ as a colonizing factor for placental infection in vivo. Of note is the fact that an InlJ-like protein recently identified in the oral pathogen Porphyromonas gingivalis is involved in biofilm formation (8). Whether listerial InlJ plays a role in biofilm formation on tissues or on abiotic surfaces is still unknown.
In conclusion, our report is the first description of a listerial virulence protein detected at the surface of bacteria present in infected organs and undetectable on bacteria infecting tissue-cultured cells. A strict regulation is likely to enable bacteria to rapidly adjust inlJ gene expression to environmental changes, especially during the infection process in vivo to stimulate adhesion to tissues. This regulation may involve the inlJ 5′-UTR. Given the large number of listerial genes with long 5′-UTRs (43), inlJ may be the first member of a list of genes specifically expressed by L. monocytogenes in vivo.
ADDENDUM IN PROOF
The X-ray structure of the InlJ internalin domain has recently been determined (M. Bublitz, C. Holland, C. Sabet, J. Reichelt, P. Cossart, D. W. Heinz, H. Bierne, and W.-D. Schubert, J. Mol. Biol., in press).
Acknowledgments
We are grateful to Michel Huerre and Huot Khun for histoimmunochemistry work and to Roger Zenon and Laurence Maranghi for technical help. We thank Olivier Dussurget for help in animal facilities. We are grateful to Tarek Msadek (Institut Pasteur), by whose group part of this work was carried out.
Work in P. Cossart's laboratory received financial support from the Pasteur Institute (GPH no. 9), INRA (AIP291), INSERM, ERA-NET (Spatelis), and the Ministère de l'Education Nationale, de la Recherche et de la Technologie. O. Poupel and S. Dubrac were supported by research funds from the European Commission (BaSysBio; LSHG-CT-2006-037469). C. Sabet received financial support from FRM and Université Pierre et Marie Curie. A. Toledo-Arana is an EMBO long-term fellow. P. Cossart is an international research scholar of the Howard Hughes Medical Institute. H. Bierne is on the INRA staff.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Autret, N., C. Raynaud, I. Dubail, P. Berche, and A. Charbit. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun. 714463-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 1845826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierne, H., and P. Cossart. 2007. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 4423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43869-881. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 91156-1166. [DOI] [PubMed] [Google Scholar]
- 6.Braun, L., S. Dramsi, P. Dehoux, H. Bierne, G. Lindahl, and P. Cossart. 1997. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25285-294. [DOI] [PubMed] [Google Scholar]
- 7.Bubert, A., Z. Sokolovic, S. K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261323-336. [DOI] [PubMed] [Google Scholar]
- 8.Capestany, C. A., M. Kuboniwa, I. Y. Jung, Y. Park, G. D. Tribble, and R. J. Lamont. 2006. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect. Immun. 743002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 1837295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 741323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppins, R. L., K. B. Hall, and E. A. Groisman. 2007. The intricate world of riboswitches. Curr. Opin. Microbiol. 10176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czjzek, M., A. Ben David, T. Bravman, G. Shoham, B. Henrissat, and Y. Shoham. 2005. Enzyme-substrate complex structures of a GH39 beta-xylosidase from Geobacillus stearothermophilus. J. Mol. Biol. 353838-846. [DOI] [PubMed] [Google Scholar]
- 13.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 393725-3733. [DOI] [PubMed] [Google Scholar]
- 14.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16251-261. [DOI] [PubMed] [Google Scholar]
- 15.Dramsi, S., F. Bourdichon, D. Cabanes, M. Lecuit, H. Fsihi, and P. Cossart. 2004. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53639-649. [DOI] [PubMed] [Google Scholar]
- 16.Dramsi, S., S. Levi, A. Triller, and P. Cossart. 1998. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect. Immun. 664461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortineau, N., P. Trieu-Cuot, O. Gaillot, E. Pellegrini, P. Berche, and J. L. Gaillard. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 151353-360. [DOI] [PubMed] [Google Scholar]
- 19.Freitag, N. E., and K. E. Jacobs. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect. Immun. 671844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friederich, E., E. Gouin, R. Hellio, C. Kocks, P. Cossart, and D. Louvard. 1995. Targeting of Listeria monocytogenes ActA protein to the plasma membrane as a tool to dissect both actin-based cell morphogenesis and ActA function. EMBO J. 142731-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gahan, C. G., and C. Hill. 2005. Gastrointestinal phase of Listeria monocytogenes infection. J. Appl. Microbiol. 981345-1353. [DOI] [PubMed] [Google Scholar]
- 22.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garandeau, C., H. Reglier-Poupet, I. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 701382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhart, E., H. Wagner, and J. Vogel. 2005. Approaches to identify novel non-messenger RNAs in bacteria and to investigate their biological functions: functional analysis of identified non-mRNAs, p. 614-642. In R. K. Hartmann, A. Bindereif, A. Schön, and E. Westhof (ed.), Handbook of RNA biochemistry, vol. 2. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 25.Gilbreth, S. E., A. K. Benson, and R. W. Hutkins. 2004. Catabolite repression and virulence gene expression in Listeria monocytogenes. Curr. Microbiol. 4995-98. [DOI] [PubMed] [Google Scholar]
- 26.Gilot, P., Y. Jossin, and J. Content. 2000. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 49887-896. [DOI] [PubMed] [Google Scholar]
- 27.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294849-852. [DOI] [PubMed] [Google Scholar]
- 28.Gouin, E., P. Dehoux, J. Mengaud, C. Kocks, and P. Cossart. 1995. iactA of Listeria ivanovii, although distantly related to Listeria monocytogenes actA, restores actin tail formation in an L. monocytogenes actA mutant. Infect. Immun. 632729-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4423-434. [DOI] [PubMed] [Google Scholar]
- 30.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110551-561. [DOI] [PubMed] [Google Scholar]
- 31.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34902-914. [DOI] [PubMed] [Google Scholar]
- 32.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 1855722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khelef, N., M. Lecuit, C. Buchrieser, D. Cabanes, O. Dussurget, and P. Cossart. 2005. Listeria monocytogenes and the genus Listeria, 3rd ed. Springer, New York, NY.
- 35.Kim, K. P., B. Jagadeesan, K. M. Burkholder, Z. W. Jaradat, J. L. Wampler, A. A. Lathrop, M. T. Morgan, and A. K. Bhunia. 2006. Adhesion characteristics of Listeria adhesion protein (LAP)-expressing Escherichia coli to Caco-2 cells and of recombinant LAP to eukaryotic receptor Hsp60 as examined in a surface plasmon resonance sensor. FEMS Microbiol. Lett. 256324-332. [DOI] [PubMed] [Google Scholar]
- 36.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13585-597. [DOI] [PubMed] [Google Scholar]
- 37.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11725-732. [DOI] [PubMed] [Google Scholar]
- 38.Kok, J., K. J. Leenhouts, A. J. Haandrikman, A. M. Ledeboer, and G. Venema. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291145-157. [DOI] [PubMed] [Google Scholar]
- 40.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 1844177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 655309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 43.Loh, E., J. Gripenland, and J. Johansson. 2006. Control of Listeria monocytogenes virulence by 5′-untranslated RNA. Trends Microbiol. 14294-298. [DOI] [PubMed] [Google Scholar]
- 44.Mandin, P., H. Fsihi, O. Dussurget, M. Vergassola, E. Milohanic, A. Toledo-Arana, I. Lasa, J. Johansson, and P. Cossart. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 571367-1380. [DOI] [PubMed] [Google Scholar]
- 45.Mandin, P., F. Repoila, M. Vergassola, T. Geissmann, and P. Cossart. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35962-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGann, P., M. Wiedmann, and K. J. Boor. 2007. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 732919-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGann, P., R. Ivanek, M. Wiedmann, and K. J. Boor. 2007. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl. Environ. Microbiol. 732806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mengaud, J., M. Lecuit, M. Lebrun, F. Nato, J. C. Mazie, and P. Cossart. 1996. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect. Immun. 645430-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84923-932. [DOI] [PubMed] [Google Scholar]
- 50.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 231075-1085. [DOI] [PubMed] [Google Scholar]
- 51.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 471613-1625. [DOI] [PubMed] [Google Scholar]
- 52.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 391212-1224. [DOI] [PubMed] [Google Scholar]
- 53.Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111747-756. [DOI] [PubMed] [Google Scholar]
- 54.Moors, M. A., B. Levitt, P. Youngman, and D. A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nudler, E. 2006. Flipping riboswitches. Cell 12619-22. [DOI] [PubMed] [Google Scholar]
- 56.Park, S. F., G. S. Stewart, and R. G. Kroll. 1992. The use of bacterial luciferase for monitoring the environmental regulation of expression of genes encoding virulence factors in Listeria monocytogenes. J. Gen. Microbiol. 1382619-2627. [DOI] [PubMed] [Google Scholar]
- 57.Pucciarelli, M. G., E. Calvo, C. Sabet, H. Bierne, P. Cossart, and F. Garcia-del Portillo. 2005. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics 54808-4817. [DOI] [PubMed] [Google Scholar]
- 58.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renzoni, A., A. Klarsfeld, S. Dramsi, and P. Cossart. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 651515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Renzoni, A., P. Cossart, and S. Dramsi. 1999. PrfA, the transcriptional activator of virulence genes, is upregulated during interaction of Listeria monocytogenes with mammalian cells and in eukaryotic cell extracts. Mol. Microbiol. 34552-561. [DOI] [PubMed] [Google Scholar]
- 60.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148433-442. [DOI] [PubMed] [Google Scholar]
- 61.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 736912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seepersaud, R., S. B. Hanniffy, P. Mayne, P. Sizer, R. Le Page, and J. M. Wells. 2005. Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect. Immun. 731671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen, A., and D. E. Higgins. 2005. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol. Microbiol. 571460-1473. [DOI] [PubMed] [Google Scholar]
- 64.Shetron-Rama, L. M., H. Marquis, H. G. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 701087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stritzker, J., C. Schoen, and W. Goebel. 2005. Enhanced synthesis of internalin A in aro mutants of Listeria monocytogenes indicates posttranscriptional control of the inlAB mRNA. J. Bacteriol. 1872836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 10299-104. [DOI] [PubMed] [Google Scholar]
- 67.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams, T., B. Joseph, D. Beier, W. Goebel, and M. Kuhn. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252287-298. [DOI] [PubMed] [Google Scholar]
- 69.Winkler, W., A. Nahvi, and R. R. Breaker. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419952-956. [DOI] [PubMed] [Google Scholar]
- 70.Winkler, W. C., and R. R. Breaker. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59487-517. [DOI] [PubMed] [Google Scholar]
- 71.Wong, K. K., H. G. Bouwer, and N. E. Freitag. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell. Microbiol. 6155-166. [DOI] [PubMed] [Google Scholar]
- 72.Yang, J. K., H. J. Yoon, H. J. Ahn, B. I. Lee, J. D. Pedelacq, E. C. Liong, J. Berendzen, M. Laivenieks, C. Vieille, G. J. Zeikus, D. J. Vocadlo, S. G. Withers, and S. W. Suh. 2004. Crystal structure of beta-d-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. J. Mol. Biol. 335155-165. [DOI] [PubMed] [Google Scholar]