Abstract
The findings from a preliminary assessment of a new instrument designed for the inoculation and spreading of specimens for microbiological analysis onto agar plates are described. The study found that the instrument was able to select full or biplates from a number of input cassettes, each containing different agar types. Samples were then inoculated by the instrument onto the agar surfaces and spread by a novel plastic applicator. Following this, the instrument labeled the plates and sorted them into a number of specified output stations. It was found that the instrument was able to inoculate and spread samples over a greater proportion of the agar plate surface than the manual loop-to-plate method. As a consequence, up to 44% more usable colonies were produced per plate from clinical specimens and standard cultures. Viable counts showed that the instrument was able to detect as few as 102 CFU/ml in fluids and also facilitated the enumeration of organisms, particularly in specimens such as urine.
The processing of specimens in many routine diagnostic laboratories is a manual and time-consuming activity. Attempts to gain more efficiency in this area have evolved slowly since 1881, when Robert Koch first published a technique for the isolation and examination of bacterial colonies using pools of gelatin-based media on glass slides (4). At that time, Koch described the inoculation and spreading of samples using pins or vaccination needles sterilized by heat. Koch's technique was revolutionized in 1887, when his assistant, Richard Petri, described a method using two round glass dishes, one for the culture medium and the other, a little larger, to act as a lid (6).
Since that time, apart from the development of molded plastic petri dishes and plastic disposable loops, the main tools used by microbiologists for the inoculation of samples onto agar surfaces have remained relatively unchanged. The development of inoculating and spreading devices, such as spiral platers and wire loop-based robotic systems, have made contributions to automating the process but have not found their place in many routine diagnostic laboratories.
This report describes the preliminary assessment of an automated specimen and agar plate management instrument that utilizes a novel plastic applicator to spread inocula. The instrument is able to select from a variety of cassettes containing different agar types in standard 90- to 100-mm petri dishes, inoculate liquid samples onto the media, spread the samples, label the inoculated plates, and distribute them to a number of specific output cassettes for incubation.
The objectives of the study were to assess the ability of the instrument to inoculate commonly encountered clinical specimens and to compare the microbiological results with the manual loop-to-plate method.
MATERIALS AND METHODS
MicroStreak.
MicroStreak (LabTech Systems, Kent Town, South Australia) is an automated instrument that is able to select standard 90- to 100-mm-diameter full and biplates from a number of input cassettes. The instrument incorporates a precision pipettor using a separate, plugged, disposable pipette tip for each patient, to inoculate defined volumes of liquid sample across the surfaces of selected plates, and another robotic arm applies a single-use, multitined plastic disposable applicator (international patent application PCT/AU2005/000079) to the inoculum. While the applicator is in contact with the inoculum on the agar surface, the instrument rotates the plate at a defined speed. On completion of this spreading cycle, the plates are sorted and delivered to specified output stations for removal by the operator.
Study design.
A prototype MicroStreak instrument was tested at the Institute of Medical and Veterinary Science (Adelaide, South Australia) over a period of 10 weeks. Different types of prepared plate media from a number of manufacturers from Australia, the United States, and France were used, and standard bacterial strains and human clinical specimens were assessed using both the instrument and the traditional manual loop-to-plate inoculation method (7).
Dilution studies.
Standard cultures of Staphylococcus epidermidis ATCC 14990 and Escherichia coli ATCC 25922 were grown overnight in brain heart infusion broth (Oxoid, Australia), and 10-fold dilutions of each culture were prepared in sterile saline to a dilution of 10−7. Ten-microliter samples of each dilution were then dispensed by the instrument onto five Columbia agar plates with 5% horse blood (HBA) (Oxoid, Australia), and each plate was rotated at speeds ranging from 10 to 100 rpm. After overnight aerobic incubation at 35°C, the growth on each plate was described and discrete colonies were counted. Bacterial counts were determined from these dilutions. A similar methodology was followed for biplates containing HBA and cystine-lactose-electrolyte-deficient agar (CLED) (Oxoid, Australia).
Assessment of clinical samples. (i) Enrichment broths.
One hundred sixty selenite broths (Oxoid, Australia) were inoculated with 1 ml of fecal-saline suspensions prepared from samples submitted for routine analysis and incubated aerobically for 18 h at 35°C. Following incubation, 10 μl of each selenite broth was dispensed by the instrument onto xylose-lactose-desoxycholate agar (XLD) (Oxoid, Australia). Fifty of the inoculated plates were rotated at 40 rpm and 110 plates at 50 rpm before being incubated aerobically for 18 h at 35°C. Manually streaked control plates using sterile 10-μl loops were also prepared from each selenite broth (5).
Following incubation, 54 blood samples previously inoculated into Bactec blood culture bottles (Becton Dickinson, County Clare, Ireland) were processed by the instrument and manually. Samples were removed from the Bactec bottles manually using sterile syringes and inoculated onto agar plates for manual streaking. An aliquot was also transferred aseptically to sterile tubes, and 10 μl was inoculated from these tubes by the instrument onto selected plates, which were rotated at 50 rpm. The plates inoculated by both methods were chocolate agar (Oxoid, Australia), HBA, CLED, and HBA/CLED biplates. The CLED plates were incubated in air, the chocolate agar and HBA/CED plates in 5% CO2, and the HBA anaerobically before being examined.
(ii) Swabs.
Thirty-eight swabs submitted for routine microbiological assessment from wounds, ears, genital tracts, and eyes were processed by both the manual method and MicroStreak. All swabs tested had been placed in Amies (gel) transport medium (Eurotube; Deltalab, Rubi, Spain).
Following the preparation of slides for routine stains and microscopy, manual control plates were prepared by rolling each swab onto the surfaces of selected plates and streaked using the manual loop-to-plate method (5). The remaining contents of these swabs were then expressed into 1.0 ml of saline within 1.5-ml microtubes, and 30 μl of these preparations was inoculated by the instrument onto selected plates and rotated at a speed of 40 rpm. A range of plates similar to those used for the blood cultures was inoculated with the addition of Candida ID 2 (bioMérieux, Marcy-l'Etoile, France) for the genital samples.
(iii) Urine.
Five hundred urine specimens submitted for routine assessment were inoculated onto chromogenic urinary tract agar plates from two manufacturers (Oxoid, Australia, and bioMérieux, Marcy-l'Etoile, France).
For the manual control method, the specimens were streaked using standard 1-μl plastic disposable loops to obtain semiquantitative counts (5), and the plates were incubated aerobically at 35°C for 18 h.
With the automated instrument, 10 μl of each specimen was inoculated onto chromogenic agar plates used for the detection and identification of urinary pathogens and spread at a rotation speed of 50 rpm. The plates were incubated at 35°C for 18 h before being examined. Where growth occurred, the numbers of colonies and organism types were recorded, with particular note being made of the colony numbers found in various plate quadrants.
(iv) Feces.
One hundred three fecal samples submitted for the detection of enteric pathogens were processed. To prepare for culture, approximately 1 g of solid feces or 1 ml of liquid specimen was transferred into 5 to 10 ml of sterile saline and mixed. Approximately 10 to 20 μl of these suspensions were inoculated onto desoxycholate citrate agar (DCA) (Oxoid, Australia) and manually streaked using sterile loops. With the automated instrument, 20 μl of each specimen was inoculated onto DCA and spread at a rotation speed of 40 rpm. The plates were incubated aerobically at 35°C for 18 h before being examined.
Assessment for cross-contamination.
A variety of methods were used to determine if any cross-contamination between specimens was evident. Two hundred eighty-nine samples of sterile saline were placed at the start and end and interspersed between samples within each batch of clinical specimens processed by the instrument. Ten microliters of each saline sample was inoculated onto HBA by the instrument, incubated aerobically at 35°C for 18 h, and examined for growth. All plates inoculated with clinical samples and saline were examined for growth that would indicate cross-contamination.
RESULTS
Dilution studies.
Viable counts showed that the concentrations of bacteria in the nutrient broths were 108 CFU/ml for S. epidermidis and 109 CFU/ml for E. coli. The lowest concentration of bacteria detected in any of the diluted broths was 102 CFU/ml.
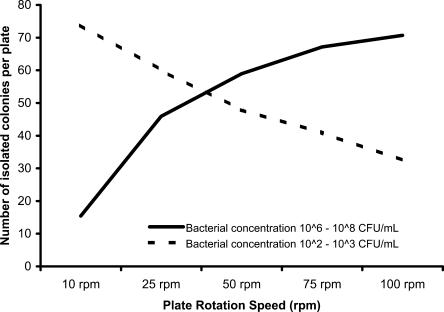
With a total of 600 plates from the S. epidermidis and E. coli broths and dilutions counted, the number of isolated colonies produced, in addition to areas of confluence, was found to be dependent on the rotation speed of the plates (Fig. 1). At bacterial concentrations in the range from 106 to 108 CFU/ml, an average of more than 50 isolated colonies were produced per plate at speeds above 50 rpm. It was also noted that when bacteria were present in smaller numbers, particularly in the samples with bacterial concentrations in the range from 102 to 103 CFU/ml, optimal results were obtained at rotation speeds of <50 rpm. For the biplates, similar observations were made, with the exception that the optimal rotation speed was found to be 75 rpm.
FIG. 1.
Influence of plate rotation speed on the formation of isolated colonies from broths and saline dilutions of S. epidermidis ATCC 14990 and E. coli ATCC 25922.
Enrichment broths.
Of the 160 selenite enrichment broths, 20 showed no growth by both methods and 140 yielded growth of a number of pathogenic and nonpathogenic organisms. Within the samples that demonstrated growth, six cases of Salmonella enterica were detected by both methods. The automated method produced colonies on XLD with classic morphology and the expected fermentation reactions.
For the XLD plates rotated at 40 rpm that demonstrated growth, colony counting showed a 17% advantage for the automated method, with the manually streaked plates growing an average of 23 isolated colonies per plate compared with 27 for the automated method. When the rotation speed was increased to 50 rpm, the manually streaked plates had an average of 26 isolated colonies per plate compared with 36 for the automated method, a 38% increase using the automated method.
Of the 54 blood culture broths subcultured, 28 were found to grow one or more organisms and 26 did not demonstrate any growth. There were no discrepancies between the manual and automated methods. MicroStreak produced colonies with classic morphology, the expected hemolytic reactions, and an average of 32% more discrete, useable colonies per plate than the manual method. The manually streaked plates grew an average of 34 isolated colonies per plate compared with 45 for the automated method. The organisms found included Clostridium perfringens, Staphylococcus aureus (Fig. 2), Micrococcus spp., S. epidermidis, Haemophilus influenzae, Proteus spp., E. coli, Enterobacter spp., viridans streptococci, and several uncharacterized yeasts.
FIG. 2.
S. aureus from a wound swab preparation plated on Columbia agar with 5% horse blood. The plate on the left was inoculated by the manual loop-to-plate method and the plate on the right by MicroStreak.
Swabs.
A large number of organisms were isolated from the 57 clinical samples, and no discrepancies were noted between the manual and automated methods. The organisms isolated included those listed above for blood cultures plus Candida albicans, a variety of mixed anaerobes, and beta hemolytic streptococci. The manually streaked plates produced an average of 25 isolated colonies per plate compared with 36 for the automated method, a 44% increase using MicroStreak.
Urine.
One of the first observations made was that in cases where specimens with >105 CFU/ml were found by the manual method, a large number of organisms were seen to be evenly distributed around the plates prepared by the instrument (Fig. 3). Conversely, specimens with <105 CFU/ml by the manual method showed sparse growth in the first two quadrants from the inoculation line on the instrument-prepared plates.
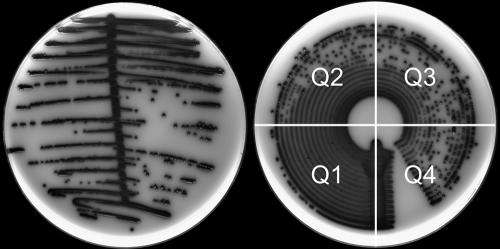
FIG. 3.
Isolate of the Klebsiella-Enterobacter-Serratia group from a urine sample plated onto chromogenic urinary tract agar plates (Oxoid, Australia). The plate on the left was inoculated by the manual loop-to-plate method and the plate on the right by MicroStreak. Shown are the plate quadrants (Q1 to Q4) that are useful in the enumeration of bacteria in urine.
On reviewing this information and comparing colony counts from the quadrants, a relationship between the number of colonies in the third quadrant from the inoculation line and the finding of >105 CFU/ml was noted. In these cases, the counts showed that the presence of 30 or more colonies in quadrant 3 correlated with loop-based counts of >105 CFU/ml (Table 1). This relationship showed a diagnostic sensitivity of 99% and a specificity of 99.7%.
TABLE 1.
Comparison of MicroStreak with manual urine bacterial counts from 500 clinical samplesa
| Bacterial counts in urine specimens by the manual method (n = 5) (CFU/ml) | No. of patients with >30 colonies in quadrant 3 of plates prepared by MicroStreak/total |
|---|---|
| 0 to 103 | 0/167 |
| 103 to 104 | 0/42 |
| 104 to 105 | 1/90 |
| >105 | 199/201 |
Comparison of incidences of patients with >30 bacterial colonies observed in quadrant 3 of plates prepared by MicroStreak with manual urine bacterial counts from 500 clinical samples.
Of the 500 urine samples processed, 201 were found by the manual method to have >105 CFU/ml and 136 showed no growth. The remaining 163 showed growth of organisms within the range from 102 to 104 CFU/ml. A wide variety of organisms were detected, including E. coli, Enterobacter spp., Proteus spp., Klebsiella spp., Pseudomonas aeruginosa, Enterococcus faecalis, Streptococcus agalactiae, and Staphylococcus spp. Three discrepant results among the 500 specimens were found, and each showed >105 CFU/ml by the manual method and 104 CFU/ml by the automated method.
Feces.
Of the manually prepared DCA plates, 86/103 showed growth of enteric organisms while 87/103 plates prepared by MicroStreak showed growth. The discrepant specimen produced a few colonies of a nonpathogenic organism on the instrument-prepared plate while showing no growth on the manually prepared plate.
Colony counting showed a 23% advantage for the automated method, with the manually streaked plates growing an average of 22 isolated colonies per plate compared with 27 for the automated method.
Cross-contamination.
Throughout the study, 874 clinical specimens and 650 individual cultures of standard bacterial strains were processed by the instrument. All samples shown to be sterile by the manual method were sterile when processed by the MicroStreak instrument, and no evidence of cross-contamination was found on plates prepared from clinical samples or standard bacterial strains. None of the 289 HBA plates inoculated with sterile saline interspersed between the clinical samples and cultures of the ATCC strains showed growth.
DISCUSSION
The MicroStreak instrument and applicators were shown to effectively inoculate and spread samples on a variety of solid media. In doing so, a greater proportion of the plate surface was used, and on average, more colonies were isolated per plate than with the manual method. With the selenite enrichment broths, the automated method produced up to 38% more colonies than the manual loop-to-plate method. For swabs from a number of sites, 44% more isolated colonies were obtained and 32% more from blood cultures.
During the study, it was shown that plate rotation speed was a significant factor in the spreading of samples across the agar surface. The observation that high bacterial numbers were more effectively separated at speeds between 50 and 100 rpm and low bacterial numbers were more effectively separated at speeds of less than 50 rpm is therefore problematic in determining an optimal speed for use with clinical samples. Although further work is still required, it appears that a compromise rotation speed of 50 rpm should be recommended for samples such as urine and enrichment broths that are likely to contain 105 or more CFU/ml. A slower speed of 40 rpm is recommended for diluted swab preparations in which lower numbers of organisms could be present. For samples inoculated onto biplates, the studies showed an optimal rotation speed of 75 rpm. The mechanics of this are not understood, but the need for a higher rotation speed could be due to the ability of the instrument to reach 75 rpm in the reduced space available.
Throughout this assessment, the instrument was programmed to use sample volumes of 10, 20, and 30 μl. The determination of an optimal sample volume will require more extensive work, particularly with swabs, where dilution is required to render the specimen into a liquid, useable form. The 10-μl inoculation volume used by the instrument for urine samples compared with 1 μl for the traditional manual method appears to result in more accurate results, particularly in the range from 102 to 104 CFU/ml. The consistent and precise dispensing obtained with the automated pipette is also likely to be a major factor associated with the accuracy and reproducibility of results and may explain the three discrepant results from the 500 urine samples tested. With one discrepant fecal sample, the instrument also produced growth where the manual method did not, again most probably due to the consistency of the pipettor over the manual method.
The enumeration of bacteria in urine is important in determining the significance of isolates (3) and constitutes a significant proportion of the workload in clinical microbiological laboratories. In most laboratories, the surface streak method is employed and requires the inoculation of 1 μl of urine onto plates using a calibrated metal or plastic disposable loop (7). An important shortcoming associated with this method is the technique-sensitive variation of up to 50% in dispensed volume (1). MicroStreak showed an ability to differentiate microbial counts in urine samples of >105 CFU/ml from those that were <105 CFU/ml. The instrument was also able to show good discrimination for specimens in which counts were between 102 and 104 CFU/ml, a range that has been shown to be significant in some patients (8). These preliminary findings indicate that the MicroStreak system will be able to offer an alternative to the manual method in the enumeration of urinary isolates in clinical samples.
Results from the swab samples processed showed the feasibility of automating this most time-consuming activity. This confirms the findings from an earlier study (9) in which swabs from a variety of body sites were suspended in fluid before being inoculated onto plates by an automated streaking device. The nature of any diluent used for the preparation of swabs requires further assessment. Previous work has shown a reduction in the number of beta-hemolytic streptococci and Neisseria gonorrhoeae when swab contents were suspended in a variety of diluents, including phosphate-buffered saline and liquid Stuart's medium (9). In the current study, the use of saline for emulsifying swabs did not appear to diminish the recovery of organisms. It is possible that this was due to the period of less than 15 min between the suspension of swab contents and inoculation onto plates by the instrument. Recent studies have also shown that Copan tubes with liquid Amies transport medium will support bacteria, including some fastidious species, for periods of between 6 and 48 h (2). Further studies using MicroStreak with this and other media are planned.
For any automated system to be accepted into a laboratory, microbiologists will need to have complete confidence that samples will not be contaminated within the instrument by any mechanical processes. Throughout this assessment, there was no evidence of contamination between samples. This confirmed that the use of plugged, sterile pipette tips and a sterile applicator for each sample prevented any possibility of contamination from specimen to specimen.
Diagnostic laboratories are being required to manage increasing demands for services while responding to strong pressures for cost containment. Microbiology laboratories are facing the same pressures but have been constrained by the slow development of automated specimen management and analytical systems compared with the variety available within the other diagnostic disciplines.
The application of the technology described has the potential to allow the automation of laborious tasks, such as plate selection, labeling, inoculation, and streaking. This will provide technologists with time to concentrate on higher-level activities more closely associated with adding value to laboratory services. Additionally, with its ability to produce more isolated colonies per plate, the technology appears to offer an additional advantage by enhancing opportunities to better identify pathogens within mixed bacterial populations.
Acknowledgments
We are grateful to David H. Pincus (bioMérieux, Hazelwood, MO) and Andrew Butcher (Institute of Medical and Veterinary Science, Adelaide, South Australia) for their helpful direction regarding the manuscript.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Albers, C. A., and R. D. Fletcher. 1983. Accuracy of the calibrated loop technique. J. Clin. Microbiol. 1840-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake, C., J. Barefanger, J. Lawhorn, and S. Verhulst. 2005. Comparison of Easy-Flow Copan Liquid Stuart's and Starplex Swab Transport Systems for recovery of fastidious aerobic bacteria. J. Clin. Microbiol. 431301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kass, E. H. 1956. Asymptomatic infections of the urinary tract. Trans. Assoc. Am. Physicians 6956-63. [PubMed] [Google Scholar]
- 4.Koch, R. 1881. Zur Untersuchung von pathogen Organismen. Mitt. Kaiserlichen Gesundheitsamte. 125-26. [Google Scholar]
- 5.Koneman, E. W., S. D. Allen, W. M. Janda, P. C. Schreckenberger, and C. W. Washington. 1997. Processing of cultures, p. 93-98. In Color atlas and textbook of diagnostic microbiology, 5th ed. Lippincott, Philadelphia, PA.
- 6.Petri, R. J. 1887. Eine kliene Modification des Koch'shen Platten-verfahrens. Zentralbl. Bakteriol. Parasitenkd. 1279-280. [Google Scholar]
- 7.Reisner, B. S., G. L. Woods, R. B. Thompson, D. H. Lorne, L. S. Garcia, and R. Y. Shimuzu. 1999. Specimen processing, p. 75-76. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 8.Stamm, W. E., G. W. Counts, K. R. Running, S. Finn, M. Turck, and K. K Holmes. 1982. Diagnosis of coliform infection in acutely dysuric women. N. Engl. J. Med. 307463-468. [DOI] [PubMed] [Google Scholar]
- 9.Tilton, R. C., and R. W. Ryan. 1978. Evaluation of an automated agar plate streaker. J. Clin. Microbiol. 7298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]