Abstract
We describe and validate a novel PCR assay to detect the pandemic hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA) lineage ST 239. Results based on previously uncharacterized isolates from a hospital in northeast Thailand support the view that at least 90% of HA-MRSA isolates in mainland Asia correspond to ST 239 or close relatives.
Methicillin-resistant Staphylococcus aureus (MRSA) strains constitute a considerable health and resource burden in the hospital environment. Although there is currently a paucity of data (17), the evidence available suggests a high frequency of methicillin resistance throughout mainland Asia (9, 14). Two recent papers have used multilocus sequence typing (MLST) to characterize hospital-acquired MRSA (HA-MRSA) isolates from nine Asian countries from Saudi Arabia to the Philippines (4, 15). These data suggest that at least 90% of the cases of HA-MRSA within a region accounting for >60% of the world's population can be accounted for by a single clonal subgroup, ST 239. This genotype has also been detected in 26 countries outside of Asia, corresponding to the EMRSA-1, -4, -7, -9, and -11, Brazilian (1), Portuguese, Hungarian (8), and Viennese clones, although in many countries, it has recently been replaced by other clones (1, 5, 7, 8, 13, 16, 18-20).
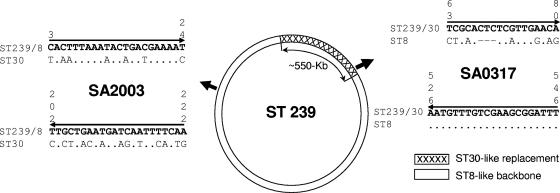
The global dissemination of this clone is consistent with heightened transmissibility, and two recent reports noted an association between ST 239 and increased virulence (2, 10). ST 239 has evolved through a large-scale recombination event involving the import of ∼20% of the genome from CC 30 (ST 30-like), while the remaining 80% (genomic backbone) corresponds to CC 8 (ST 8-like) (21). As these two lineages are unrelated (6), this mosaic structure provides the means to develop a rapid, accurate, and sensitive PCR-based assay. We designed two pairs of discriminatory nonredundant primers based on two variable genes, one of which (SA2003) lies within the ST 8-like backbone, while the other (SA0317) lies within the ST 30-like imported region (21) (Fig. 1). As the primers were designed to amplify products of different sizes (220 bp for SA2003 and 484 bp for SA0317), it is possible to identify cases where both products are amplified from a single PCR. The presence of both amplicons of the predicted sizes indicates the presence of both the ST 30- and ST 8-like sequences in the hybrid genome of ST 239.
FIG. 1.
Primer design. Two pairs of primers were designed based on SA0317 and SA2003 loci (gene codes from the N315 genome and annotated as hypothetical protein and hysA, respectively). These genes were identified as being the most variable of those sequenced previously by Robinson and Enright (21), which facilitates the design of primers specific for ST 30- and ST 8-like sequences. The ∼550-kb ST 30-like replacement spans the origin of replication and is shown as the hatched block. The positions of SA0317 and SA2003 are identified by arrows. SA0317 falls within the ST 30-like replacement, while SA2003 falls outside of it. The primer sequences are shown in boldface type, with the positions given in vertical numbers above the sites. Primer direction is indicated by arrows. SA2003 forward and reverse primers are specific for the ST 239/ST 8-like sequence, while the SA0317 forward primer is specific for the ST 239/ST 30-like sequence (note the 3-base indel in the corresponding ST 8 sequence in this case).
The primer sequences used were ST 239/ST 30-like specific primers 5′-TCGCACTCTCGTTGAACA-3′ (SA0317Forward) and 5′-AAATCCGCTTCGACAAACATT (SA0317Reverse) (Fig. 1) and ST 239/ST 8-like specific primers 5′-CACTTTAAATACTGACGAAAAT-3′ (SA2003Forward) and 5′-TTGAAAATTGATCATTCAGCAA-3′ (SA2003Reverse).
Heteroduplex PCR was carried in a 25-μl mixture consisting of 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 0.7 μM of each of the four primers, 1.25 U of HotStart Taq DNA polymerase, and 2.5 μl of DNA. PCR conditions were 95°C for 15 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and 72°C for 7 min.
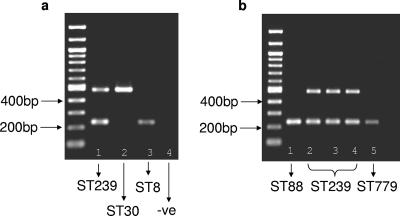
Figure 2a shows the initial validation of this assay against well-characterized strains from the United Kingdom (11). As predicted, only the larger product was amplified in the ST 30 strain (C507), only the smaller product was amplified in the ST 8 strain (C125), and both products were amplified in the ST 239 strain (EMRSA-4). We further validated the assay using 95 HA-MRSA isolates recovered from a tertiary hospital in Guangzhou, People's Republic of China, 90 (94.7%) of which correspond to ST 239 (B. Xu et al., unpublished data). Both bands were amplified for all 90 ST 239 isolates, but only the larger band was amplified for single isolates corresponding to ST 217, ST 188, and ST 864, all of which are unrelated to ST 239 (not shown). Both bands were also amplified for the two ST 865 isolates. This genotype is a single-locus variant of ST 239, differing at only a single nucleotide site at arcC, and is assumed to have inherited the mosaic structure.
FIG. 2.
(a) Agarose gel electrophoresis of amplicons using initial control strains. Lane 1, EMRSA-4 (ST 239); lane 2, C507 (ST 30); lane 3, C125 (ST 8); lane 4, negative control (−ve) (no DNA). Both amplicons are present for ST 239, only the larger one is present for ST 30, and the smaller one is present for ST 8. The sizes of the amplicons are as predicted. (b) Agarose gel electrophoresis of five Thai isolates characterized by MLST. Lane 1, MSSA (ST 88) (only the small band amplifies); lanes 2 to 4, MRSA (ST 239) (both bands amplify); lane 5, MRSA (ST 779) (only the small band amplifies).
We then applied the assay to 27 MRSA and 26 methicillin-sensitive S. aureus (MSSA) isolates assembled as part of an ongoing prospective study of serious S. aureus infections in patients presenting to Sappasithiprasong Hospital, Thailand. Ethical permission was granted by the Ethics Committee of the Ministry of Public Health, Royal Government of Thailand, and the Oxford Tropical Research Ethics Committee.
None of the 26 MSSA isolates produced both bands, indicating the absence of ST 239. This was expected, as there are no examples of MSSA ST 239 in the database (http://www.mlst.net). In contrast, both bands amplified in 25 of 27 (∼93%) MRSA isolates. This suggests that ST 239 accounts for at least 90% of HA-MRSA isolates in northeast Thailand, which is highly consistent with the results for south China and for continental Asia in general. Four of the MRSA isolates and one MSSA isolate were characterized by MLST. Three of the MRSA isolates which had amplified both bands were confirmed to be ST 239 (Fig. 2b). Amplification of only the smaller product was noted in the remaining MRSA isolate and the single MSSA isolate, and these isolates were found to correspond to ST 88 and ST 779, respectively. These genotypes are unrelated to ST 239, hence providing further validation for the assay.
In all, the PCR assay was tested against 103 isolates with known MLST genotypes and achieved 100% specificity and 100% sensitivity. Our results are consistent with the dominance of this clone in HA-MRSA infection (∼93%) throughout Asia (excluding Japan and South Korea) (4, 15). If this epidemiological pattern is confirmed, MLST will prove to be an inefficient tool for epidemiological studies of HA-MRSA throughout Asia, where resources may be relatively poor. Instead, an initial screening based on the PCR assay described here will allow the rapid detection of ST 239 isolates. Highly discriminatory typing procedures, based on hypervirulent loci or single-nucleotide polymorphism discovery, should be specifically tailored for this clone to facilitate high-resolution epidemiological data. Such an approach should provide evidence concerning the introduction and spread of ST 239 into Asia and other parts of the world. Finally, recent evidence raises the possibility that ST 239 will in time be replaced by other clones (3, 12), and the use of this PCR assay should prove to be instrumental in capturing the dynamics of this process.
Acknowledgments
We are grateful for the support of the medical, nursing, and laboratory staff at Sappasithiprasong Hospital and staff at the Mahidol-Oxford Tropical Medicine Research Unit, including Direk Limmathurotsakul, Gumphol Wongsuvan, Maliwan Hongsuwan, Sukallaya Paengmee, and Nittaya Teerawattanasook. We also gratefully acknowledge Ye Huifen and Zhou Xiaomian for kindly providing MRSA strains collected from Guangzhou First Municipal People's Hospital. The EMRSA-4 isolate was kindly donated by Mark Enright.
W.P. acknowledges support from a Sun Yat-sen University grant (36000-3253285). S.J.P. was supported by a Wellcome Trust career development award in clinical tropical medicine. This study was funded by the Wellcome Trust.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Aires de Sousa, M., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 362590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral, M. M., L. R. Coelho, R. P. Flores, R. R. Souza, M. C. Silva-Carvalho, L. A. Teixeira, B. T. Ferreira-Carvalho, and A. M. Figueiredo. 2005. The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J. Infect. Dis. 192801-810. [DOI] [PubMed] [Google Scholar]
- 3.Amorim, M. L., N. A. Faria, D. C. Oliveira, C. Vasconcelos, J. C. Cabeda, A. C. Mendes, E. Calado, A. P. Castro, M. H. Ramos, J. M. Amorim, and H. de Lencastre. 2007. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J. Clin. Microbiol. 452881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 501001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conceicao, T., M. Aires-de-Sousa, M. Fuzi, A. Toth, J. Paszti, E. Ungvari, W. B. van Leeuwen, A. van Belkum, H. Grundmann, and H. de Lencastre. 2007. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13971-979. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, J. E., and E. J. Feil. 2006. The phylogeny of Staphylococcus aureus—which genes make the best intra-species markers? Microbiology 1521297-1305. [DOI] [PubMed] [Google Scholar]
- 7.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 2987-106. [DOI] [PubMed] [Google Scholar]
- 8.de Lencastre, H., E. P. Severina, H. Milch, M. K. Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3289-296. [DOI] [PubMed] [Google Scholar]
- 9.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl 2)S114-S132. [DOI] [PubMed] [Google Scholar]
- 10.Edgeworth, J. D., G. Yadegarfar, S. Pathak, R. Batra, J. D. Cockfield, D. Wyncoll, R. Beale, and J. A. Lindsay. 2007. An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin. Infect. Dis. 44493-501. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 1853307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, L. Y., N. Loomba-Chlebicka, Y. L. Koh, T. Y. Tan, P. Krishnan, R. T. Lin, N. W. Tee, D. A. Fisher, and T. H. Koh. 2007. Evolving EMRSA-15 epidemic in Singapore hospitals. J. Med. Microbiol. 56376-379. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48143-144. [DOI] [PubMed] [Google Scholar]
- 14.Kim, H. B., H. C. Jang, H. J. Nam, Y. S. Lee, B. S. Kim, W. B. Park, K. D. Lee, Y. J. Choi, S. W. Park, M. D. Oh, E. C. Kim, and K. W. Choe. 2004. In vitro activities of 28 antimicrobial agents against Staphylococcus aureus isolates from tertiary-care hospitals in Korea: a nationwide survey. Antimicrob. Agents Chemother. 481124-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51516-521. [DOI] [PubMed] [Google Scholar]
- 17.Nickerson, E. K., V. Wuthiekanun, N. P. Day, W. Chaowagul, and S. J. Peacock. 2006. Methicillin-resistant Staphylococcus aureus in rural Asia. Lancet Infect. Dis. 670-71. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira, D., I. Santos-Sanches, R. Mato, M. Tamayo, G. Ribeiro, D. Costa, and H. de Lencastre. 1998. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the ‘Iberian’ and the ‘Brazilian’ clones of MRSA. Clin. Microbiol. Infect. 4373-384. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2180-189. [DOI] [PubMed] [Google Scholar]
- 20.Potel, C., M. Alvarez, P. Alvarez, I. Otero, and E. Fluiters. 2007. Evolution, antimicrobial susceptibility and assignment to international clones of methicillin-resistant Staphylococcus aureus isolated over a 9-year period in two Spanish hospitals. Clin. Microbiol. Infect. 13728-730. [DOI] [PubMed] [Google Scholar]
- 21.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 1861060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]